Microtubules in Influenza Virus Entry and Egress
Abstract
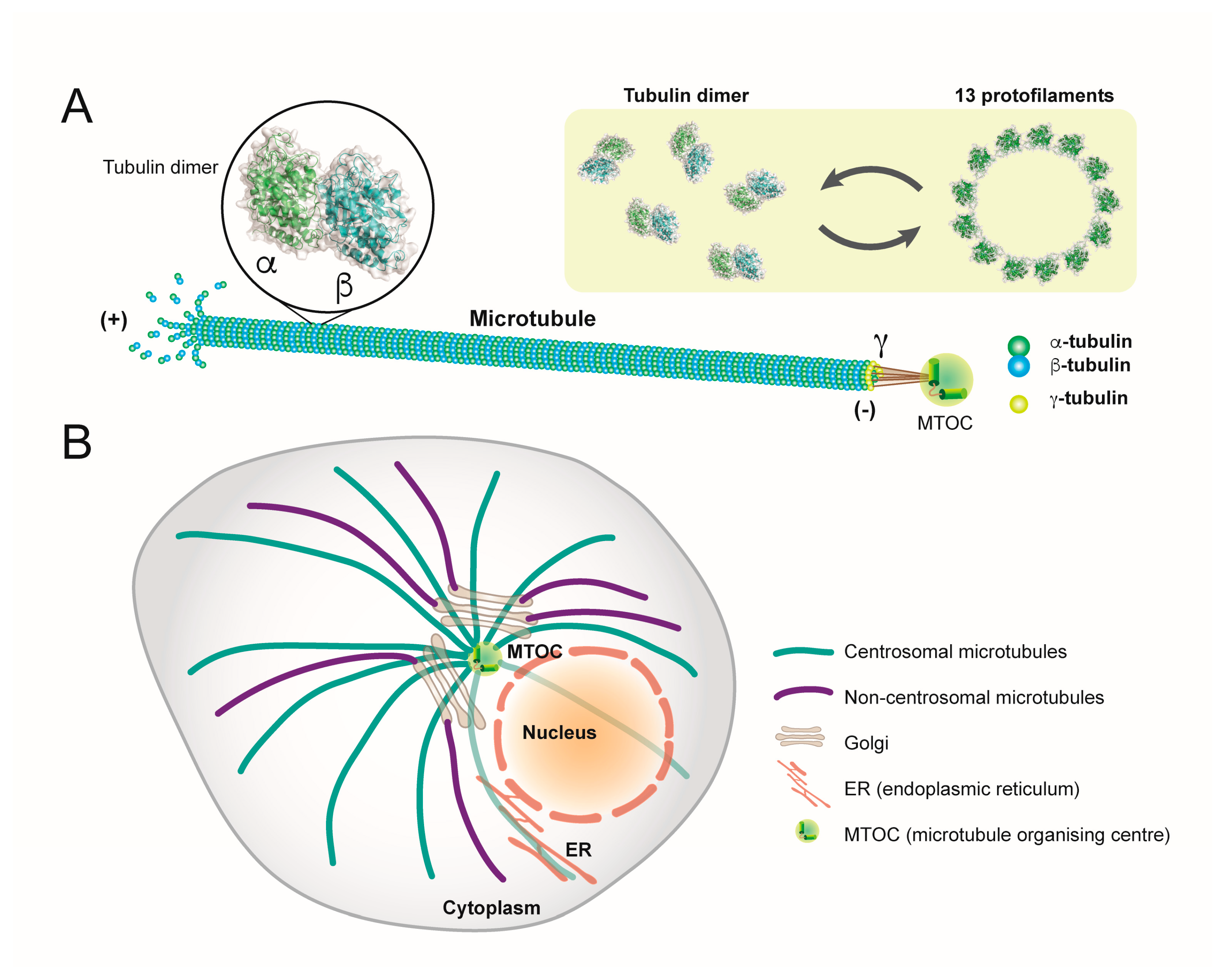
1. Microtubules: Structure, Function and Organisation
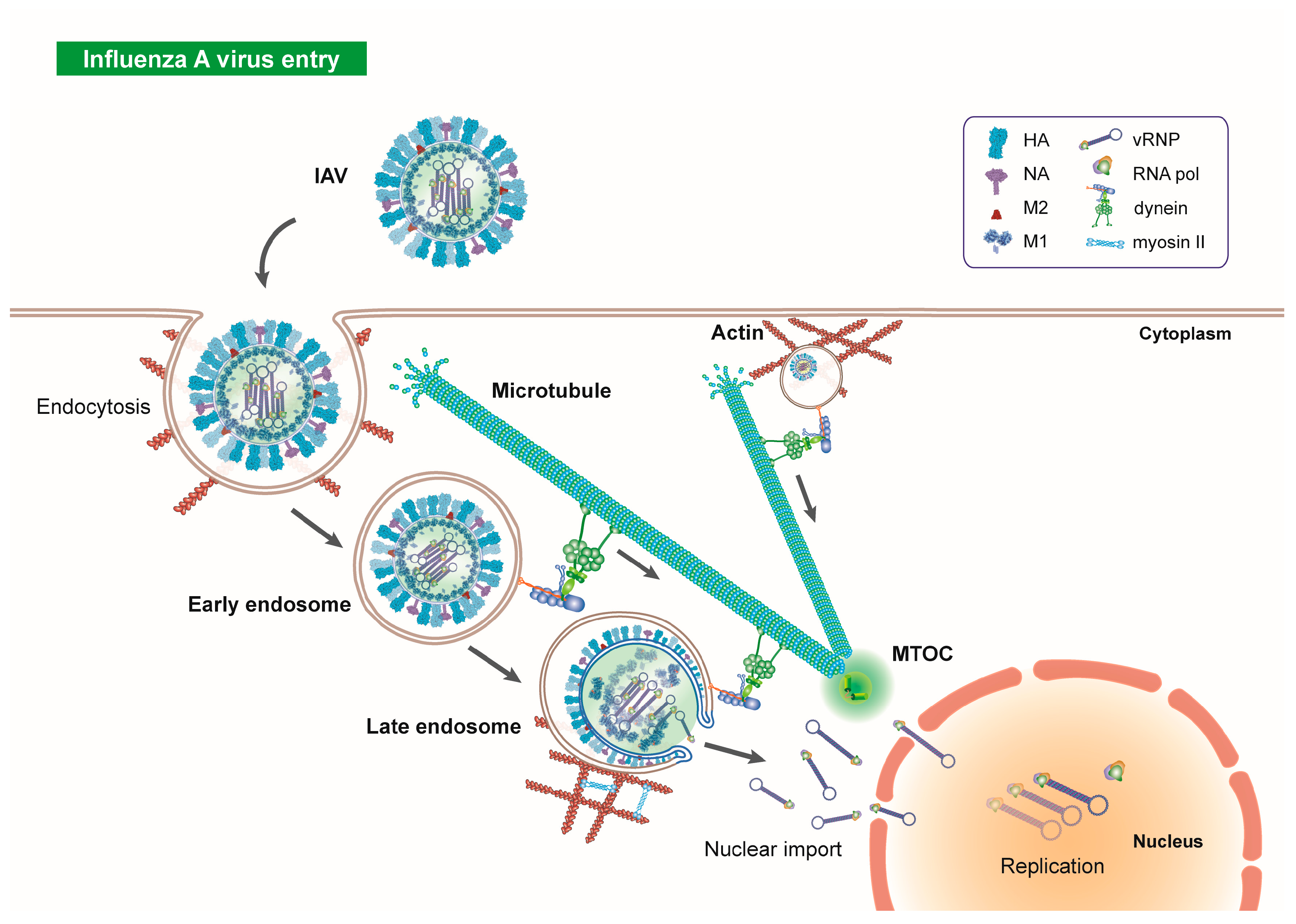
2. Microtubules in Influenza Virus Entry
3. HDACs, Microtubules and Endocytosis
4. Influenza Virus Priming in Endosomes
5. Influenza Virus Uncoating and Aggresome Processing
6. vRNP Nuclear Import
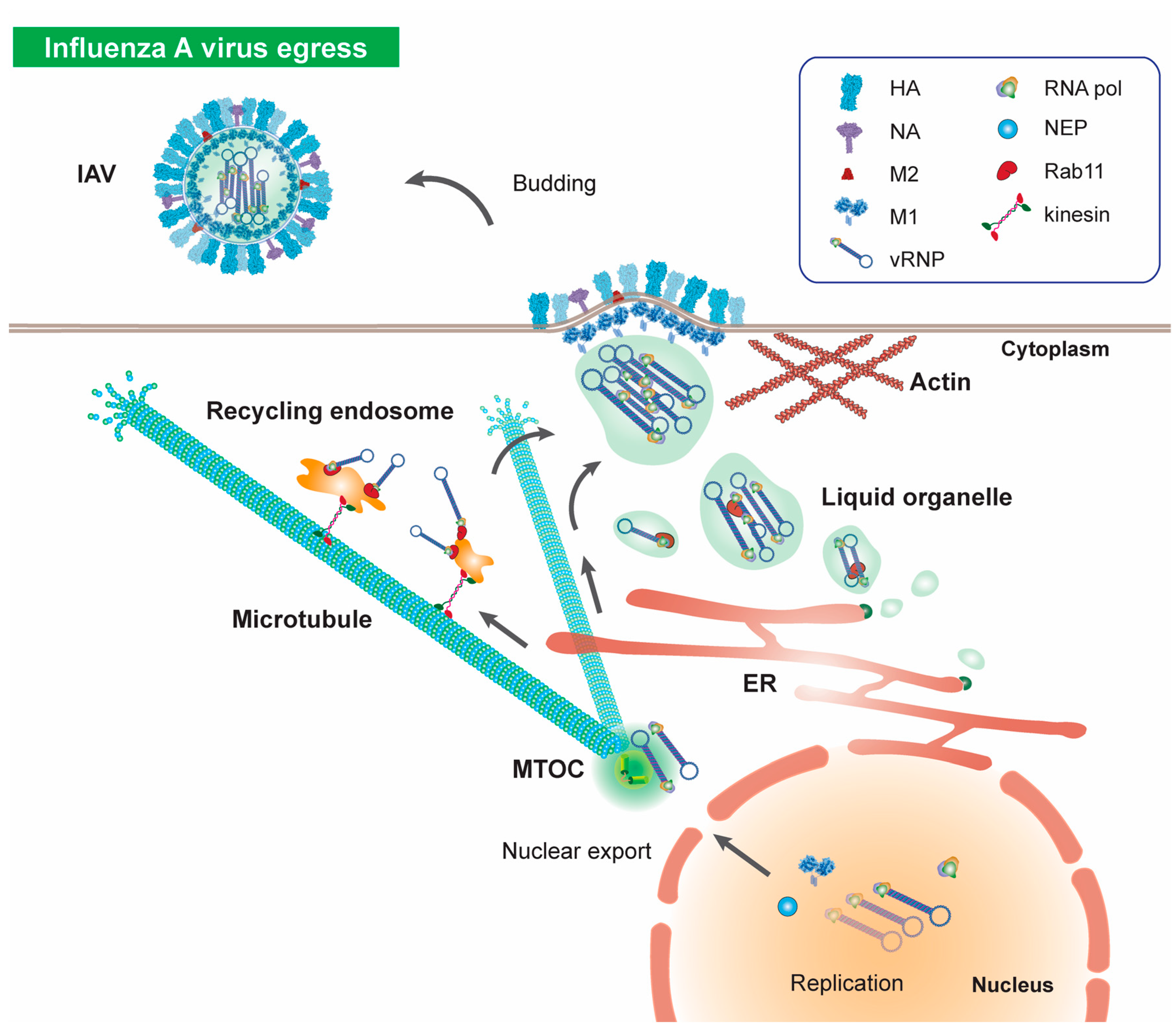
7. Microtubules in Influenza Virus Egress
8. Conclusions
Acknowledgments
Conflicts of Interest
References
- Bedi, S.; Ono, A. Friend or Foe: The Role of the Cytoskeleton in Influenza A Virus Assembly. Viruses 2019, 11, 46. [Google Scholar] [CrossRef] [PubMed]
- Muroyama, A.; Lechler, T. Microtubule organization, dynamics and functions in differentiated cells. Development 2017, 144, 3012–3021. [Google Scholar] [CrossRef] [PubMed]
- Wickstead, B.; Gull, K. The evolution of the cytoskeleton. J. Cell Biol. 2011, 194, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Akhmanova, A.; Steinmetz, M.O. Control of microtubule organization and dynamics: Two ends in the limelight. Nat. Rev. Mol. Cell Biol. 2015, 16, 711–726. [Google Scholar] [CrossRef]
- Cai, D.; McEwen, D.P.; Martens, J.R.; Meyhofer, E.; Verhey, K.J. Single molecule imaging reveals differences in microtubule track selection between Kinesin motors. PLoS Biol. 2009, 7, e1000216. [Google Scholar] [CrossRef]
- Mohan, N.; Sorokina, E.M.; Verdeny, I.V.; Alvarez, A.S.; Lakadamyali, M. Detyrosinated microtubules spatially constrain lysosomes facilitating lysosome-autophagosome fusion. J. Cell Biol. 2019, 218, 632–643. [Google Scholar] [CrossRef]
- Perdiz, D.; Mackeh, R.; Pous, C.; Baillet, A. The ins and outs of tubulin acetylation: More than just a post-translational modification? Cell. Signal. 2011, 23, 763–771. [Google Scholar] [CrossRef]
- Creppe, C.; Malinouskaya, L.; Volvert, M.L.; Gillard, M.; Close, P.; Malaise, O.; Laguesse, S.; Cornez, I.; Rahmouni, S.; Ormenese, S.; et al. Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell 2009, 136, 551–564. [Google Scholar] [CrossRef]
- Ohkawa, N.; Sugisaki, S.; Tokunaga, E.; Fujitani, K.; Hayasaka, T.; Setou, M.; Inokuchi, K. N-acetyltransferase ARD1-NAT1 regulates neuronal dendritic development. Genes Cells 2008, 13, 1171–1183. [Google Scholar] [CrossRef]
- Chu, C.W.; Hou, F.; Zhang, J.; Phu, L.; Loktev, A.V.; Kirkpatrick, D.S.; Jackson, P.K.; Zhao, Y.; Zou, H. A novel acetylation of beta-tubulin by San modulates microtubule polymerization via down-regulating tubulin incorporation. Mol. Biol. Cell 2011, 22, 448–456. [Google Scholar] [CrossRef]
- Akella, J.S.; Wloga, D.; Kim, J.; Starostina, N.G.; Lyons-Abbott, S.; Morrissette, N.S.; Dougan, S.T.; Kipreos, E.T.; Gaertig, J. MEC-17 is an alpha-tubulin acetyltransferase. Nature 2010, 467, 218–222. [Google Scholar] [CrossRef] [PubMed]
- Shida, T.; Cueva, J.G.; Xu, Z.; Goodman, M.B.; Nachury, M.V. The major alpha-tubulin K40 acetyltransferase alphaTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc. Natl. Acad. Sci. USA 2010, 107, 21517–21522. [Google Scholar] [CrossRef] [PubMed]
- Topalidou, I.; Keller, C.; Kalebic, N.; Nguyen, K.C.; Somhegyi, H.; Politi, K.A.; Heppenstall, P.; Hall, D.H.; Chalfie, M. Genetically separable functions of the MEC-17 tubulin acetyltransferase affect microtubule organization. Curr. Biol. 2012, 22, 1057–1065. [Google Scholar] [CrossRef] [PubMed]
- Hubbert, C.; Guardiola, A.; Shao, R.; Kawaguchi, Y.; Ito, A.; Nixon, A.; Yoshida, M.; Wang, X.F.; Yao, T.P. HDAC6 is a microtubule-associated deacetylase. Nature 2002, 417, 455–458. [Google Scholar] [CrossRef] [PubMed]
- North, B.J.; Marshall, B.L.; Borra, M.T.; Denu, J.M.; Verdin, E. The Human Sir2 Ortholog, SIRT2, is an NAD+-Dependent Tubulin Deacetylas. Mol. Cell 2003, 11, 437–444. [Google Scholar] [CrossRef]
- Eshun-Wilson, L.; Zhang, R.; Portran, D.; Nachury, M.V.; Toso, D.B.; Lohr, T.; Vendruscolo, M.; Bonomi, M.; Fraser, J.S.; Nogales, E. Effects of alpha-tubulin acetylation on microtubule structure and stability. Proc. Natl. Acad. Sci. USA 2019, 116, 10366–10371. [Google Scholar] [CrossRef]
- Hammond, J.; Dawen, C.; Verhey, K.J. Tubulin modifications and their cellular functions. Curr. Opin. Cell Biol. 2008, 20, 71–76. [Google Scholar] [CrossRef]
- Jin, H.; White, S.R.; Shida, T.; Schulz, S.; Aguiar, M.; Gygi, S.P.; Bazan, J.F.; Nachury, M.V. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell 2010, 141, 1208–1219. [Google Scholar] [CrossRef]
- Reed, N.A.; Cai, D.; Blasius, T.L.; Jih, G.T.; Meyhofer, E.; Gaertig, J.; Verhey, K.J. Microtubule acetylation promotes kinesin-1 binding and transport. Curr. Biol. 2006, 16, 2166–2172. [Google Scholar] [CrossRef]
- L’Hernault, S.W.; Rosenbaum, J.L. Chlamydomonas alpha-tubulin is posttranslationally modified by acetylation on the epsilon-amino group of a lysine. Biochemistry 1985, 24, 258–263. [Google Scholar] [CrossRef]
- Shigematsu, H.; Imasaki, T.; Doki, C.; Sumi, T.; Aoki, M.; Uchikubo-Kamo, T.; Sakamoto, A.; Tokuraku, K.; Shirouzu, M.; Nitta, R. Structural insight into microtubule stabilization and kinesin inhibition by Tau family MAPs. J. Cell Biol. 2018, 217, 4155–4163. [Google Scholar] [CrossRef] [PubMed]
- Kadavath, H.; Hofele, R.V.; Biernat, J.; Kumar, S.; Tepper, K.; Urlaub, H.; Mandelkow, E.; Zweckstetter, M. Tau stabilizes microtubules by binding at the interface between tubulin heterodimers. Proc. Natl. Acad. Sci. USA 2015, 112, 7501–7506. [Google Scholar] [CrossRef] [PubMed]
- Panda, D.; Samuel, J.C.; Massie, M.; Feinstein, S.C.; Wilson, L. Differential regulation of microtubule dynamics by three- and four-repeat tau: Implications for the onset of neurodegenerative disease. Proc. Natl. Acad. Sci. USA 2003, 100, 9548–9553. [Google Scholar] [CrossRef] [PubMed]
- Monroy, B.Y.; Sawyer, D.L.; Ackermann, B.E.; Borden, M.M.; Tan, T.C.; Ori-McKenney, K.M. Competition between microtubule-associated proteins directs motor transport. Nat. Commun. 2018, 9, 1487. [Google Scholar] [CrossRef] [PubMed]
- Tokuraku, K.; Noguchi, T.Q.; Nishie, M.; Matsushima, K.; Kotani, S. An isoform of microtubule-associated protein 4 inhibits kinesin-driven microtubule gliding. J. Biochem. 2007, 141, 585–591. [Google Scholar] [CrossRef] [PubMed]
- Hagiwara, H.; Yorifuji, H.; Sato-Yoshitake, R.; Hirokawa, N. Competition between motor molecules (kinesin and cytoplasmic dynein) and fibrous microtubule-associated proteins in biding to microtubules. J. Biol. Chem. 1994, 269, 3581–3589. [Google Scholar] [PubMed]
- Jourdain, L.; Curmi, P.; Sobel, A.; Pantaloni, D.; Carlier, M.F. Stathmin: A tubulin-sequestering protein which forms a ternary T2S complex with two tubulin molecules. Biochemistry 1997, 36, 10817–10821. [Google Scholar] [CrossRef]
- Howell, B.; Deacon, H.; Cassimeris, L. Decreasing oncoprotein 18/stathmin levels reduces microtubule catastrophes and increases microtubule polymer in vivo. J. Cell Sci. 1999, 112, 3713–3722. [Google Scholar]
- Rubin, C.I.; Atweh, G.F. The role of stathmin in the regulation of the cell cycle. J. Cell Biochem. 2004, 93, 242–250. [Google Scholar] [CrossRef]
- Ross, J.L.; Ali, M.Y.; Warshaw, D.M. Cargo transport: Molecular motors navigate a complex cytoskeleton. Curr. Opin. Cell Biol. 2008, 20, 41–47. [Google Scholar] [CrossRef]
- Brinkley, W. Microtubules: A brief historical perspective. J. Struct. Biol. 1997, 118, 84–86. [Google Scholar] [CrossRef] [PubMed]
- Greber, U.F.; Way, M. A superhighway to virus infection. Cell 2006, 124, 741–754. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.J.; Kon, T.; Knight, P.J.; Sutoh, K.; Burgess, S.A. Functions and mechanics of dynein motor proteins. Nat. Rev. Mol. Cell Biol. 2013, 14, 713–726. [Google Scholar] [CrossRef] [PubMed]
- Miki, H.; Okada, Y.; Hirokawa, N. Analysis of the kinesin superfamily: Insights into structure and function. Trends Cell Biol. 2005, 15, 467–476. [Google Scholar] [CrossRef]
- Hirokawa, N.; Noda, Y.; Tanaka, Y.; Niwa, S. Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 2009, 10, 682–696. [Google Scholar] [CrossRef]
- She, Z.Y.; Pan, M.Y.; Tan, F.Q.; Yang, W.X. Minus end-directed kinesin-14 KIFC1 regulates the positioning and architecture of the Golgi apparatus. Oncotarget 2017, 8, 36469–36483. [Google Scholar] [CrossRef]
- Wiese, C.; Zheng, Y. Microtubule nucleation: Gamma-tubulin and beyond. J. Cell Sci. 2006, 119, 4143–4153. [Google Scholar] [CrossRef]
- Keating, T.J.; Borisy, G.G. Centrosomal and non-centrosomal microtubules. Biol. Cell 2000, 91, 321–329. [Google Scholar] [CrossRef]
- Bornens, M. The Centrosome in Cells and Organisms. Science 2012, 335, 422–426. [Google Scholar] [CrossRef]
- Efimov, A.; Kharitonov, A.; Efimova, N.; Loncarek, J.; Miller, P.M.; Andreyeva, N.; Gleeson, P.; Galjart, N.; Maia, A.R.; McLeod, I.X.; et al. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev. Cell 2007, 12, 917–930. [Google Scholar] [CrossRef]
- Chabin-Brion, K.; Marceiller, J.; Perez, F.; Settegrana, C.; Drechou, A.; Durand, G.; Pous, C. The Golgi complex is a microtubule-organizing organelle. Mol. Biol. Cell 2001, 12, 2047–2060. [Google Scholar] [CrossRef] [PubMed]
- Fuller, S.D.; Gowen, B.E.; Reinsch, S.; Sawyer, A.; Buendia, B.; Wepf, R.; Karsenti, E. The core of the mammalian centriole contains γ-tubulin. Curr. Biol. 1995, 5, 1384–1393. [Google Scholar] [CrossRef]
- Shu, H.B.; Li, Z.; Palacios, M.J.; Li, Q.; Joshi, H.C. A transient association of gamma-tubulin at the midbody is required for the completion of cytokinesis during the mammalian cell division. J. Cell Sci. 1995, 108, 2047–2060. [Google Scholar]
- Wiese, C.; Zheng, Y. A New Function for the Gamma-tubulin Ring Complex as a Microtubule Minus-End Cap. Nat. Cell Biol. 2000, 2, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Lüders, J.; Stearns, T. Microtubule-organizing centres: A re-evaluation. Nat. Rev. Mol. Cell Biol. 2007, 8, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Dohner, K.; Nagel, C.H.; Sodeik, B. Viral stop-and-go along microtubules: Taking a ride with dynein and kinesins. Trends Microbiol. 2005, 13, 320–327. [Google Scholar] [CrossRef]
- Radtke, K.; Dohner, K.; Sodeik, B. Viral interactions with the cytoskeleton: A hitchhiker’s guide to the cell. Cell. Microbiol. 2006, 8, 387–400. [Google Scholar] [CrossRef]
- Smith, A.E.; Helenius, A. How viruses enter animal cells. Science 2004, 304, 237–242. [Google Scholar] [CrossRef]
- Cossart, P.; Helenius, A. Endocytosis of viruses and bacteria. Cold Spring Harb. Perspect. Biol. 2014, 6, a016972. [Google Scholar] [CrossRef]
- Naghavi, M.H.; Walsh, D. Microtubule Regulation and Function during Virus Infection. J. Virol. 2017, 91, e00538-17. [Google Scholar] [CrossRef]
- Huotari, J.; Helenius, A. Endosome maturation. EMBO J. 2011, 30, 3481–3500. [Google Scholar] [CrossRef] [PubMed]
- Elkin, S.R.; Lakoduk, A.M.; Schmid, S.L. Endocytic pathways and endosomal trafficking: A primer. Wien. Med. Wochenschr. 2016, 166, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Burd, C.; Cullen, P.J. Retromer: A master conductor of endosome sorting. Cold Spring Harb. Perspect. Biol. 2014, 6, a016774. [Google Scholar] [CrossRef] [PubMed]
- Sodeik, B.; Ebersold, M.W.; Helenius, A. Microtubule-mediated Transport of Incoming Herpes Simplex Virus 1 Capsids to the Nucleus. J. Cell Biol. 1997, 136, 1007–1021. [Google Scholar] [CrossRef]
- Seisenberger, G.; Ried, M.U.; Endre, T.; Bu¨ning, H.; Hallek, M.; Bra¨uchle, C. Real-Time Single-Molecule Imaging of the Infection Pathway of an Adeno-Associated Virus. Science 2001, 294, 1929–1932. [Google Scholar] [CrossRef]
- Sanjuan, N.; Porrás, A.; Otero, J. Microtubule-dependent intracellular transport of murine polyomavirus. Virology 2003, 313, 105–116. [Google Scholar] [CrossRef][Green Version]
- Suomalainen, M.; Nakano, M.Y.; Keller, S.; Boucke, K.; Stidwill, R.P.; Greber, U.F. Microtubule-dependent plus- and minus end-directed motilities are competing processes for nuclear targeting of adenovirus. J. Cell Biol. 1999, 144, 657–672. [Google Scholar] [CrossRef]
- Lakadamyali, M.; Rust, M.J.; Babcock, H.P.; Zhuang, X. Visualizing infection of individual influenza viruses. Proc. Natl. Acad. Sci. USA 2003, 100, 9280–9285. [Google Scholar] [CrossRef]
- Granger, E.; McNee, G.; Allan, V.; Woodman, P. The role of the cytoskeleton and molecular motors in endosomal dynamics. Semin. Cell Dev. Biol. 2014, 31, 20–29. [Google Scholar] [CrossRef]
- Zhang, L.J.; Xia, L.; Liu, S.L.; Sun, E.Z.; Wu, Q.M.; Wen, L.; Zhang, Z.L.; Pang, D.W. A “Driver Switchover” Mechanism of Influenza Virus Transport from Microfilaments to Microtubules. ACS Nano 2018, 12, 474–484. [Google Scholar] [CrossRef]
- Murray, J.W.; Bananis, E.; Wolkof, A.W. Reconstitution of ATP-dependent Movement of Endocytic Vesicles Along Microtubules In Vitro: An Oscillatory Bidirectional Process. Mol. Biol. Cell 2000, 11, 419–433. [Google Scholar] [CrossRef] [PubMed]
- Bomsel, M.; Parton, R.; Kuznetsov, S.A.; Schroer, T.A.; Gruenberg, J. Microtubule- and Motor-Dependent Fusion In Vitro between Apical and Basolateral Endocytic Vesicles from MDCK Cells. Cell 1990, 62, 719–731. [Google Scholar] [CrossRef]
- Aniento, F.; Emans, N.; Grittiths, G.; Gruenberg, J. Cytoplasmic Dynein-dependent Vesicular Transport from Early to Late Endosomes. J. Cell Biol. 1993, 123, 1373–1387. [Google Scholar] [CrossRef] [PubMed]
- Hunt, S.D.; Townley, A.K.; Danson, C.M.; Cullen, P.J.; Stephens, D.J. Microtubule motors mediate endosomal sorting by maintaining functional domain organization. J. Cell Sci. 2013, 126, 2493–2501. [Google Scholar] [CrossRef] [PubMed]
- Goodsell, D.S.; Autin, L.; Olson, A.J. Illustrate: Software for Biomolecular Illustration. Structure 2019, 27, 1716–1720. [Google Scholar] [CrossRef] [PubMed]
- Bayer, N.; Schober, D.; Prchla, E.; Murphy, R.F.; Blaas, D.; Fuchs, R. Effect of bafilomycin A1 and nocodazole on endocytic transport in HeLa cells: Implications for viral uncoating and infection. J. Virol. 1998, 72, 9645–9655. [Google Scholar] [CrossRef] [PubMed]
- Marsh, M.; Helenius, A. Virus Entry into Animal Cells. Adv. Virus Res. 1989, 36, 107–151. [Google Scholar]
- Mercer, J.; Schelhaas, M.; Helenius, A. Virus entry by endocytosis. Annu. Rev. Biochem. 2010, 79, 803–833. [Google Scholar] [CrossRef]
- Rust, M.J.; Lakadamyali, M.; Zhang, F.; Zhuang, X. Assembly of endocytic machinery around individual influenza viruses during viral entry. Nat. Struct. Mol. Biol. 2004, 11, 567–573. [Google Scholar] [CrossRef]
- Yamauchi, Y.; Boukari, H.; Banerjee, I.; Sbalzarini, I.F.; Horvath, P.; Helenius, A. Histone deacetylase 8 is required for centrosome cohesion and influenza A virus entry. PLoS Pathog. 2011, 7, e1002316. [Google Scholar] [CrossRef]
- Yamauchi, Y.; Helenius, A. Virus entry at a glance. J. Cell Sci. 2013, 126, 1289–1295. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.L.; Zhang, Z.L.; Tian, Z.Q.; Zhao, H.S.; Liu, H.; Sun, E.Z.; Xiao, G.F.; Zhang, W.; Wang, H.Z.; Pang, D.W. Effectively and Efficiently Dissecting the Infection of Influenza Virus by Quantum-Dot-Based Single-Particle Tracking. ACS Nano 2012, 6, 141–150. [Google Scholar] [CrossRef] [PubMed]
- De Vries, E.; Tscherne, D.M.; Wienholts, M.J.; Cobos-Jimenez, V.; Scholte, F.; Garcia-Sastre, A.; Rottier, P.J.; de Haan, C.A. Dissection of the influenza A virus endocytic routes reveals macropinocytosis as an alternative entry pathway. PLoS Pathog. 2011, 7, e1001329. [Google Scholar] [CrossRef] [PubMed]
- Maxson, M.E.; Grinstein, S. The vacuolar-type H+-ATPase at a glance—More than a proton pump. J. Cell Sci. 2014, 127, 4987–4993. [Google Scholar] [CrossRef]
- Nishi, T.; Forgac, M. The vacuolar (H+)-ATPases—Nature’s most versatile proton pumps. Nat. Rev. Mol. Cell Biol. 2002, 3, 94–103. [Google Scholar] [CrossRef]
- Marshansky, V.; Rubinstein, J.L.; Gruber, G. Eukaryotic V-ATPase: Novel structural findings and functional insights. Biochim. Biophys. Acta 2014, 1837, 857–879. [Google Scholar] [CrossRef]
- Maxfield, F.R.; McGraw, T.E. Endocytic recycling. Nat. Rev. Mol. Cell Biol. 2004, 5, 121–132. [Google Scholar] [CrossRef]
- Pungercar, J.R.; Caglic, D.; Sajid, M.; Dolinar, M.; Vasiljeva, O.; Pozgan, U.; Turk, D.; Bogyo, M.; Turk, V.; Turk, B. Autocatalytic processing of procathepsin B is triggered by proenzyme activity. FEBS J. 2009, 276, 660–668. [Google Scholar] [CrossRef]
- Li, S.; Sieben, C.; Ludwig, K.; Hofer, C.T.; Chiantia, S.; Herrmann, A.; Eghiaian, F.; Schaap, I.A. pH-Controlled two-step uncoating of influenza virus. Biophys. J. 2014, 106, 1447–1456. [Google Scholar] [CrossRef]
- Bui, M.; Whittaker, G.; Helenius, A. Effect of M1 Protein and Low pH on Nuclear Transport of Influenza Virus Ribonucleoproteins. J. Virol. 1996, 70, 8391–8401. [Google Scholar] [CrossRef]
- Stauffer, S.; Feng, Y.; Nebioglu, F.; Heilig, R.; Picotti, P.; Helenius, A. Stepwise priming by acidic pH and a high K+ concentration is required for efficient uncoating of influenza A virus cores after penetration. J. Virol. 2014, 88, 13029–13046. [Google Scholar] [CrossRef] [PubMed]
- Martin, K.; Helenius, A. Transport of Incoming Influenza Virus Nucleocapsids into the Nucleu. J. Virol. 1991, 65, 232–244. [Google Scholar] [CrossRef]
- Vonderheit, A.; Helenius, A. Rab7 associates with early endosomes to mediate sorting and transport of Semliki forest virus to late endosomes. PLoS Biol. 2005, 3, e233. [Google Scholar] [CrossRef]
- Mesaki, K.; Tanabe, K.; Obayashi, M.; Oe, N.; Takei, K. Fission of tubular endosomes triggers endosomal acidification and movement. PLoS ONE 2011, 6, e19764. [Google Scholar] [CrossRef]
- Momose, F.; Kikuchi, Y.; Komase, K.; Morikawa, Y. Visualization of microtubule-mediated transport of influenza viral progeny ribonucleoprotein. Microbes Infect. 2007, 9, 1422–1433. [Google Scholar] [CrossRef] [PubMed]
- Arts, J.; de Schepper, S.; Emelen, K.V. Histone Deacetylase Inhibitors: From Chromatin Remodeling to Experimental Cancere Therapeutics. Curr. Med. Chem. 2003, 10, 2343–2350. [Google Scholar] [CrossRef] [PubMed]
- Drazic, A.; Myklebust, L.M.; Ree, R.; Arnesen, T. The world of protein acetylation. Biochim. Biophys. Acta 2016, 1864, 1372–1401. [Google Scholar] [CrossRef] [PubMed]
- de Ruijter, A.J.M.; van Gennip, A.H.; Caron, H.N.; Kemp, S.; van Kuilenburg, A.B.P. Histone deacetylases (HDACs): Characterization of the classical HDAC family. Biochem. J. 2003, 370, 737–749. [Google Scholar] [CrossRef]
- Lamb, R.A.; Zebedee, S.L.; Flichardsont, C.D. Influenza Virus M2 Protein Is an Integral Membrane Protein Expressed on the Infeited-Cell Surface. Cell 1985, 40, 627–633. [Google Scholar] [CrossRef]
- Hamilton, B.S.; Whittaker, G.R.; Daniel, S. Influenza virus-mediated membrane fusion: Determinants of hemagglutinin fusogenic activity and experimental approaches for assessing virus fusion. Viruses 2012, 4, 1144–1168. [Google Scholar] [CrossRef]
- Daniels, R.S.; Jeffries, S.; Yates, P.; Schild, G.C.; Rogers, G.N.; Paulson, J.C.; Wharton, S.A.; Douglas, A.R.; Skehel, J.J.; Wiley, D.C. The receptor-binding and membrane-fusion properties of influenza virus variants selected using anti-haemagglutinin monoclonal antibodies. EMBO J. 1987, 6, 1459–1465. [Google Scholar] [CrossRef] [PubMed]
- Lagache, T.; Sieben, C.; Meyer, T.; Herrmann, A.; Holcman, D. Stochastic Model of Acidification, Activation of Hemagglutinin and Escape of Influenza Viruses from an Endosome. Front. Phys. 2017, 5, 25. [Google Scholar] [CrossRef]
- Schelker, M.; Mair, C.M.; Jolmes, F.; Welke, R.W.; Klipp, E.; Herrmann, A.; Flottmann, M.; Sieben, C. Viral RNA Degradation and Diffusion Act as a Bottleneck for the Influenza A Virus Infection Efficiency. PLoS Comput. Biol. 2016, 12, e1005075. [Google Scholar] [CrossRef] [PubMed]
- Staring, J.; Raaben, M.; Brummelkamp, T.R. Viral escape from endosomes and host detection at a glance. J. Cell Sci. 2018, 131, jcs216259. [Google Scholar] [CrossRef]
- Kawaguchi, Y.; Kovacs, J.J.; McLaurin, A.; Vance, J.M.; Ito, A.; Yao, T.P. The Deacetylase HDAC6 Regulates Aggresome Formation and Cell Viability in Response to Misfolded Protein Stress. Cell 2003, 115, 727–738. [Google Scholar] [CrossRef]
- Johnston, J.A.; Illing, M.E.; Kopito, R.R. Cytoplasmic dynein/dynactin mediates the assembly of aggresomes. Cell Motil. Cytoskelet. 2002, 53, 26–38. [Google Scholar] [CrossRef]
- Banerjee, I.; Miyake, Y.; Nobs, S.P.; Schneider, C.; Horvath, P.; Kopf, M.; Matthias, P.; Helenius, A.; Yamauchi, Y. Influenza Avirus uses the aggresome processing machinery forhostcell entry. Science 2014, 346, 473–477. [Google Scholar] [CrossRef]
- Rudnicka, A.; Yamauchi, Y. Ubiquitin in Influenza Virus Entry and Innate Immunity. Viruses 2016, 8, 293. [Google Scholar] [CrossRef]
- Hao, R.; Nanduri, P.; Rao, Y.; Panichelli, R.S.; Ito, A.; Yoshida, M.; Yao, T.P. Proteasomes activate aggresome disassembly and clearance by producing unanchored ubiquitin chains. Mol. Cell 2013, 51, 819–828. [Google Scholar] [CrossRef]
- Johnston, J.A.; Ward, C.L.; Kopito, R.R. Aggresomes: A cellular response to misfolded proteins. J. Cell Biol. 1998, 143, 1883–1898. [Google Scholar] [CrossRef]
- Garcia-Mata, R.; Bebok, Z.; Sorscher, E.J.; Sztul, E.S. Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J. Cell Biol. 1999, 146, 1239–1254. [Google Scholar] [CrossRef] [PubMed]
- Wigley, W.C.; Fabunmi, R.P.; Lee, M.G.; Marino, C.R.; Muallem, S.; DeMartino, G.N.; Thomas, P.J. Dynamic association of proteasomal machinery with the centrosome. J. Cell Biol. 1999, 145, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Miyake, Y.; Keusch, J.J.; Decamps, L.; Ho-Xuan, H.; Iketani, S.; Gut, H.; Kutay, U.; Helenius, A.; Yamauchi, Y. Influenza virus uses transportin 1 for vRNP debundling during cell entry. Nat. Microbiol. 2019, 4, 578–586. [Google Scholar] [CrossRef] [PubMed]
- Miyake, Y.; Keusch, J.J.; Wang, L.; Saito, M.; Hess, D.; Wang, X.; Melancon, B.J.; Helquist, P.; Gut, H.; Matthias, P. Structural insights into HDAC6 tubulin deacetylation and its selective inhibition. Nat. Chem. Biol. 2016, 12, 748–754. [Google Scholar] [CrossRef]
- Strunze, S.; Engelke, M.F.; Wang, I.H.; Puntener, D.; Boucke, K.; Schleich, S.; Way, M.; Schoenenberger, P.; Burckhardt, C.J.; Greber, U.F. Kinesin-1-mediated capsid disassembly and disruption of the nuclear pore complex promote virus infection. Cell Host Microbe 2011, 10, 210–223. [Google Scholar] [CrossRef]
- Myers, K.A.; Tint, I.; Nadar, C.V.; He, Y.; Black, M.M.; Baas, P.W. Antagonistic forces generated by cytoplasmic dynein and myosin-II during growth cone turning and axonal retraction. Traffic 2006, 7, 1333–1351. [Google Scholar] [CrossRef]
- Freitas, N.; Cunha, C. Mechanisms and Signals for the Nuclear Import of Proteins. Curr. Genomics 2009, 10, 550–557. [Google Scholar] [CrossRef]
- Weis, K. Regulating Access to the Genome: Nucleocytoplasmic Transport throughout the Cell Cycle. Cell 2003, 112, 441–451. [Google Scholar] [CrossRef]
- Alber, F.; Dokudovskaya, S.; Veenhoff, L.M.; Zhang, W.; Kipper, J.; Devos, D.; Suprapto, A.; Karni-Schmidt, O.; Williams, R.; Chait, B.T.; et al. The molecular architecture of the nuclear pore complex. Nature 2007, 450, 695–701. [Google Scholar] [CrossRef]
- Kabachinski, G.; Schwartz, T.U. The nuclear pore complex—Structure and function at a glance. J. Cell Sci. 2015, 128, 423–429. [Google Scholar] [CrossRef]
- Li, J.; Yu, M.; Zheng, W.; Liu, W. Nucleocytoplasmic shuttling of influenza A virus proteins. Viruses 2015, 7, 2668–2682. [Google Scholar] [CrossRef] [PubMed]
- Mosammaparast, N.; Pemberton, L.F. Karyopherins: From nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 2004, 14, 547–556. [Google Scholar] [CrossRef] [PubMed]
- Kalderon, D.; Roberts, B.L.; Richardson, W.D.; Smith, A.E. A Short Amino Acid Sequence Able to Specify Nuclear Location. Cell 1984, 39, 499–509. [Google Scholar] [CrossRef]
- Hicks, G.R.; Raikhel, N.V. Specific binding of nuclear localization sequences to plant nuclei. Plant Cell 1995, 5, 983–994. [Google Scholar]
- Görlich, D.; Dabrowski, M.; Bischoff, F.R.; Kutay, U.; Bork, P.; Hartmann, E.; Prehn, S.; Izaurrald, E. A Novel Class of RanGTP Binding Proteins. J. Cell Biol. 1997, 138, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Chook, Y.M.; Blobel, G. Karyopherins and nuclear import. Curr. Opin. Struct. Biol. 1991, 11, 703–715. [Google Scholar] [CrossRef]
- Boulo, S.; Akarsu, H.; Ruigrok, R.W.; Baudin, F. Nuclear traffic of influenza virus proteins and ribonucleoprotein complexes. Virus Res. 2007, 124, 12–21. [Google Scholar] [CrossRef]
- Kalab, P.; Heald, R. The RanGTP gradient—A GPS for the mitotic spindle. J. Cell Sci. 2008, 121, 1577–1586. [Google Scholar] [CrossRef]
- Paradise, A.; Levin, M.K.; Korza, G.; Carson, J.H. Significant Proportions of Nuclear Transport Proteins with Reduced Intracellular Mobilities Resolved by Fluorescence Correlation Spectroscopy. J. Mol. Biol. 2007, 365, 50–65. [Google Scholar] [CrossRef]
- Pockwinse, S.M.; Rajgopal, A.; Young, D.W.; Mujeeb, K.A.; Nickerson, J.; Javed, A.; Redick, S.; Lian, J.B.; van Wijnen, A.J.; Stein, J.L.; et al. Microtubule-dependent nuclear-cytoplasmic shuttling of Runx2. J. Cell Physiol. 2006, 206, 354–362. [Google Scholar] [CrossRef]
- Gleason, E.L.; Hogan, J.C.; Stephens, J.M. Stabilization, not polymerization, of microtubules inhibits the nuclear translocation of STATs in adipocytes. Biochem. Biophys. Res. Commun. 2004, 325, 716–718. [Google Scholar] [CrossRef] [PubMed]
- Brumwell, C.; Antolik, C.; Carson, J.H.; Barbarese, E. Intracellular trafficking of hnRNP A2 in oligodendrocytes. Exp. Cell Res. 2002, 279, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Davey, J.; Dimmock, N.J.; Colman, A. Identification of the Sequence Responsible for the Nuclear Accumulation of the Influenza Virus Nucleoprotein in Xenopus Oocytes. Cell 1985, 40, 667–675. [Google Scholar] [CrossRef]
- Greenspan, D.; Palese, P.; Krystal, M. Two nuclear location signals in the influenza virus NS1 nonstructural protein. J. Virol. 1988, 62, 3020–3026. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, M.; Fujii, K.; Muramoto, Y.; Yamada, S.; Yamayoshi, S.; Takada, A.; Goto, H.; Horimoto, T.; Kawaoka, Y. Contributions of two nuclear localization signals of influenza A virus nucleoprotein to viral replication. J. Virol. 2007, 81, 30–41. [Google Scholar] [CrossRef]
- Wu, W.W.; Sun, Y.H.; Pante, N. Nuclear import of influenza A viral ribonucleoprotein complexes is mediated by two nuclear localization sequences on viral nucleoprotein. Virol. J. 2007, 4, 49. [Google Scholar] [CrossRef]
- Ye, Z.; Robinson, D.; Wagner, R.R. Nucleus-Targeting Domain of the Matrix Protein (M1) of Influenza Virus. J. Virol. 1995, 69, 1964–1970. [Google Scholar] [CrossRef]
- Cao, S.; Liu, X.; Yu, M.; Li, J.; Jia, X.; Bi, Y.; Sun, L.; Gao, G.F.; Liu, W. A nuclear export signal in the matrix protein of Influenza A virus is required for efficient virus replication. J. Virol. 2012, 86, 4883–4891. [Google Scholar] [CrossRef]
- Roth, D.M.; Moseley, G.W.; Glover, D.; Pouton, C.W.; Jans, D.A. A microtubule-facilitated nuclear import pathway for cancer regulatory proteins. Traffic 2007, 8, 673–686. [Google Scholar] [CrossRef]
- Roth, D.M.; Moseley, G.W.; Pouton, C.W.; Jans, D.A. Mechanism of microtubule-facilitated “fast track” nuclear import. J. Biol. Chem. 2011, 286, 14335–14351. [Google Scholar] [CrossRef]
- Lam, M.H.; Thomas, R.J.; Loveland, K.L.; Schilders, S.; Gu, M.; Martin, T.J.; Gillespie, M.T.; Jans, D.A. Nuclear transport of parathyroid hormone (PTH)-related protein is dependent on microtubules. Mol. Endocrinol. 2002, 16, 390–401. [Google Scholar] [CrossRef] [PubMed]
- Giannakakou, P.; Sackett, D.L.; Ward, Y.; Webster, K.R.; Blagosklonny, M.V.; Fojo, T. p53 is associated with cellular microtubules and is transported to the nucleus by dynei. Nat. Cell Biol. 2000, 2, 709–717. [Google Scholar] [CrossRef] [PubMed]
- Radtke, K.; Kieneke, D.; Wolfstein, A.; Michael, K.; Steffen, W.; Scholz, T.; Karger, A.; Sodeik, B. Plus- and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathog. 2010, 6, e1000991. [Google Scholar] [CrossRef] [PubMed]
- Moseley, G.W.; Roth, D.M.; DeJesus, M.A.; Leyton, D.L.; Filmer, R.P.; Pouton, C.W.; Jans, D.A. Dynein light chain association sequences can facilitate nuclear protein import. Mol. Biol. Cell 2007, 18, 3204–3213. [Google Scholar] [CrossRef]
- McDonald, D.; Vodicka, M.A.; Lucero, G.; Svitkina, T.M.; Borisy, G.G.; Emerman, M.; Hope, T.J. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 2002, 159, 441–452. [Google Scholar] [CrossRef]
- Ogawa-Goto, K.; Tanaka, K.; Gibson, W.; Moriishi, E.; Miura, Y.; Kurata, T.; Irie, S.; Sata, T. Microtubule network facilitates nuclear targeting of human cytomegalovirus capsid. J. Virol. 2003, 77, 8541–8547. [Google Scholar] [CrossRef]
- Douglas, M.W.; Diefenbach, R.J.; Homa, F.L.; Miranda-Saksena, M.; Rixon, F.J.; Vittone, V.; Byth, K.; Cunningham, A.L. Herpes simplex virus type 1 capsid protein VP26 interacts with dynein light chains RP3 and Tctex1 and plays a role in retrograde cellular transport. J. Biol. Chem. 2004, 279, 28522–28530. [Google Scholar] [CrossRef]
- Carnes, S.K.; Zhou, J.; Aiken, C. HIV-1 Engages a Dynein-Dynactin-BICD2 Complex for Infection and Transport to the Nucleus. J. Virol. 2018, 92, e00358-18. [Google Scholar] [CrossRef]
- Malikov, V.; da Silva, E.S.; Jovasevic, V.; Bennett, G.; de Souza Aranha Vieira, D.A.; Schulte, B.; Diaz-Griffero, F.; Walsh, D.; Naghavi, M.H. HIV-1 capsids bind and exploit the kinesin-1 adaptor FEZ1 for inward movement to the nucleus. Nat. Commun. 2015, 6, 6660. [Google Scholar] [CrossRef]
- Babcock, H.P.; Chen, C.; Zhuang, X. Using single-particle tracking to study nuclear trafficking of viral genes. Biophys. J. 2004, 87, 2749–2758. [Google Scholar] [CrossRef]
- Schatz, C.A.; Santarella, R.; Hoenger, A.; Karsenti, E.; Mattaj, I.W.; Gruss, O.J.; Carazo-Salas, R.E. Importin a-regulated nucleation of microtubules by TPX2. EMBO J. 2003, 22, 2060–2070. [Google Scholar] [CrossRef] [PubMed]
- Akarsu, H.; Burmeister, W.P.; Petosa, C.; Petit, I.; Muller, C.W.; Ruigrok, R.W.; Baudin, F. Crystal structure of the M1 protein-binding domain of the influenza A virus nuclear export protein (NEP/NS2). EMBO J. 2003, 22, 4646–4655. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Chen, J.; Chen, Q.; Wang, H.; Yao, Y.; Chen, J.; Chen, Z. A second CRM1-dependent nuclear export signal in the influenza A virus NS2 protein contributes to the nuclear export of viral ribonucleoproteins. J. Virol. 2013, 87, 767–778. [Google Scholar] [CrossRef] [PubMed]
- Neumann, G.; Hughes, M.T.; Kawaoka, Y. Influenza A virus NS2 protein mediates vRNP nuclear export through NES-independent interaction with hCRM1. EMBO J. 2000, 19, 6751–6758. [Google Scholar] [CrossRef]
- O’Neill, R.E.; Talon, J.; Palese, P. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 1998, 17, 288–296. [Google Scholar] [CrossRef]
- Paterson, D.; Fodor, E. Emerging roles for the influenza A virus nuclear export protein (NEP). PLoS Pathog. 2012, 8, e1003019. [Google Scholar] [CrossRef]
- Muhlbauer, D.; Dzieciolowski, J.; Hardt, M.; Hocke, A.; Schierhorn, K.L.; Mostafa, A.; Muller, C.; Wisskirchen, C.; Herold, S.; Wolff, T.; et al. Influenza virus-induced caspase-dependent enlargement of nuclear pores promotes nuclear export of viral ribonucleoprotein complexes. J. Virol. 2015, 89, 6009–6021. [Google Scholar] [CrossRef]
- Amorim, M.J.; Bruce, E.A.; Read, E.K.; Foeglein, A.; Mahen, R.; Stuart, A.D.; Digard, P. A Rab11- and microtubule-dependent mechanism for cytoplasmic transport of influenza A virus viral RNA. J. Virol. 2011, 85, 4143–4156. [Google Scholar] [CrossRef]
- Avilov, S.V.; Moisy, D.; Munier, S.; Schraidt, O.; Naffakh, N.; Cusack, S. Replication-competent influenza A virus that encodes a split-green fluorescent protein-tagged PB2 polymerase subunit allows live-cell imaging of the virus life cycle. J. Virol. 2012, 86, 1433–1448. [Google Scholar] [CrossRef]
- Jo, S.; Kawaguchi, A.; Takizawa, N.; Morikawa, Y.; Momose, F.; Nagata, K. Involvement of vesicular trafficking system in membrane targeting of the progeny influenza virus genome. Microbes Infect. 2010, 12, 1079–1084. [Google Scholar] [CrossRef]
- Manzoor, R.; Igarashi, M.; Takada, A. Influenza A Virus M2 Protein: Roles from Ingress to Egress. Int. J. Mol. Sci. 2017, 18, 2649. [Google Scholar] [CrossRef]
- Alenquer, M.; Vale-Costa, S.; Etibor, T.A.; Ferreira, F.; Sousa, A.L.; Amorim, M.J. Influenza A virus ribonucleoproteins form liquid organelles at endoplasmic reticulum exit sites. Nat. Commun. 2019, 10, 1629. [Google Scholar] [CrossRef]
- Eisfeld, A.J.; Kawakami, E.; Watanabe, T.; Neumann, G.; Kawaoka, Y. RAB11A is essential for transport of the influenza virus genome to the plasma membrane. J. Virol. 2011, 85, 6117–6126. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, A.; Hirohama, M.; Harada, Y.; Osari, S.; Nagata, K. Influenza Virus Induces Cholesterol-Enriched Endocytic Recycling Compartments for Budozone Formation via Cell Cycle-Independent Centrosome Maturation. PLoS Pathog. 2015, 11, e1005284. [Google Scholar] [CrossRef] [PubMed]
- Vale-Costa, S.; Alenquer, M.; Sousa, A.L.; Kellen, B.; Ramalho, J.; Tranfield, E.M.; Amorim, M.J. Influenza A virus ribonucleoproteins modulate host recycling by competing with Rab11 effectors. J. Cell Sci. 2016, 129, 1697–1710. [Google Scholar] [CrossRef] [PubMed]
- Grant, B.D.; Donaldson, J.G. Pathways and mechanisms of endocytic recycling. Nat. Rev. Mol. Cell Biol. 2009, 10, 597–608. [Google Scholar] [CrossRef] [PubMed]
- Sönnichsen, B.; Renzis, S.D.; Nielsen, E.; Rietdorf, J.; Zeria, M. Distinct Membrane Domains on Endosomes in the Recycling Pathway Visualized by Multicolor Imaging of Rab4, Rab5, and Rab11. J. Cell Biol. 2000, 149, 901–914. [Google Scholar] [CrossRef] [PubMed]
- Vale-Costa, S.; Amorim, M.J. Clustering of Rab11 vesicles in influenza A virus infected cells creates hotspots containing the 8 viral ribonucleoproteins. Small GTPases 2017, 8, 71–77. [Google Scholar] [CrossRef]
- Eisfeld, A.J.; Neumann, G.; Kawaoka, Y. At the centre: Influenza A virus ribonucleoproteins. Nat. Rev. Microbiol. 2015, 13, 28–41. [Google Scholar] [CrossRef]
- Momose, F.; Sekimoto, T.; Ohkura, T.; Jo, S.; Kawaguchi, A.; Nagata, K.; Morikawa, Y. Apical transport of influenza A virus ribonucleoprotein requires Rab11-positive recycling endosome. PLoS ONE 2011, 6, e21123. [Google Scholar] [CrossRef]
- Bruce, E.A.; Digard, P.; Stuart, A.D. The Rab11 pathway is required for influenza A virus budding and filament formation. J. Virol. 2010, 84, 5848–5859. [Google Scholar] [CrossRef] [PubMed]
- De Castro Martin, I.F.; Fournier, G.; Sachse, M.; Pizarro-Cerda, J.; Risco, C.; Naffakh, N. Influenza virus genome reaches the plasma membrane via a modified endoplasmic reticulum and Rab11-dependent vesicles. Nat. Commun. 2017, 8, 1396. [Google Scholar] [CrossRef] [PubMed]
- Saito, M.; Hess, D.; Eglinger, J.; Fritsch, A.W.; Kreysing, M.; Weinert, B.T.; Choudhary, C.; Matthias, P. Acetylation of intrinsically disordered regions regulates phase separation. Nat. Chem. Biol. 2019, 15, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Mellman, I.; Warren, G. The Road Taken: Past and Future Foundations of Membrane Traffic. Cell 2000, 100, 99–112. [Google Scholar] [CrossRef]
- Farhan, H.; Rabouille, C. Signalling to and from the secretory pathway. J. Cell Sci. 2011, 124, 171–180. [Google Scholar] [CrossRef]
- Spang, A. On vesicle formation and tethering in the ER-Golgi shuttle. Curr. Opin. Cell Biol. 2009, 21, 531–536. [Google Scholar] [CrossRef]
- Rios, R.M. The centrosome-Golgi apparatus nexus. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130462. [Google Scholar] [CrossRef]
- Rossman, J.S.; Jing, X.; Leser, G.P.; Lamb, R.A. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell 2010, 142, 902–913. [Google Scholar] [CrossRef]
- Sakaguchi, T.; Leser, G.P.; Lam, R.A. The Ion Channel Activity of the Influenza Virus M2 Protein Affects Transport through the Golgi Apparatus. J. Cell Biol. 1998, 133, 733–747. [Google Scholar] [CrossRef]
- Ichinohe, T.; Pang, I.K.; Iwasaki, A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat. Immunol. 2010, 11, 404–410. [Google Scholar] [CrossRef]
- Nturibi, E.; Bhagwat, A.R.; Coburn, S.; Myerburg, M.M.; Lakdawala, S.S. Intracellular Colocalization of Influenza Viral RNA and Rab11A Is Dependent upon Microtubule Filaments. J. Virol. 2017, 91, e01179-17. [Google Scholar] [CrossRef] [PubMed]
- Kumakura, M.; Kawaguchi, A.; Nagata, K. Actin-myosin network is required for proper assembly of influenza virus particles. Virology 2015, 476, 141–150. [Google Scholar] [CrossRef]
- Husain, M.; Cheung, C.Y. Histone deacetylase 6 inhibits influenza A virus release by downregulating the trafficking of viral components to the plasma membrane via its substrate, acetylated microtubules. J. Virol. 2014, 88, 11229–11239. [Google Scholar] [CrossRef] [PubMed]
- Husain, M.; Harrod, K.S. Influenza A virus-induced caspase-3 cleaves the histone deacetylase 6 in infected epithelial cells. FEBS Lett. 2009, 583, 2517–2520. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, N.; Caron, C.; Matthias, G.; Hess, D.; Khochbin, S.; Matthias, P. HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO J. 2003, 22, 1168–1179. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Qian, Y.; Chen, X.; Ruan, Z.; Ye, Y.; Chen, H.; Babiuk, L.A.; Jung, Y.S.; Dai, J. HDAC6 Restricts Influenza A Virus by Deacetylation of the RNA Polymerase PA Subunit. J. Virol. 2019, 93, e01896-18. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.J.; Lee, H.C.; Kim, J.H.; Park, S.Y.; Kim, T.H.; Lee, W.K.; Jang, D.J.; Yoon, J.E.; Choi, Y.I.; Kim, S.; et al. HDAC6 regulates cellular viral RNA sensing by deacetylation of RIG-I. EMBO J. 2016, 35, 429–442. [Google Scholar] [CrossRef]
- Yamauchi, Y.; Greber, U.F. Principles of Virus Uncoating: Cues and the Snooker Ball. Traffic 2016, 17, 569–592. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Simpson, C.; Yamauchi, Y. Microtubules in Influenza Virus Entry and Egress. Viruses 2020, 12, 117. https://doi.org/10.3390/v12010117
Simpson C, Yamauchi Y. Microtubules in Influenza Virus Entry and Egress. Viruses. 2020; 12(1):117. https://doi.org/10.3390/v12010117
Chicago/Turabian StyleSimpson, Caitlin, and Yohei Yamauchi. 2020. "Microtubules in Influenza Virus Entry and Egress" Viruses 12, no. 1: 117. https://doi.org/10.3390/v12010117
APA StyleSimpson, C., & Yamauchi, Y. (2020). Microtubules in Influenza Virus Entry and Egress. Viruses, 12(1), 117. https://doi.org/10.3390/v12010117




