Cucurbit Chlorotic Yellows Virus p22 Protein Interacts with Cucumber SKP1LB1 and Its F-Box-Like Motif Is Crucial for Silencing Suppressor Activity
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid Construction
2.2. Plant Materials and Virus Inoculation
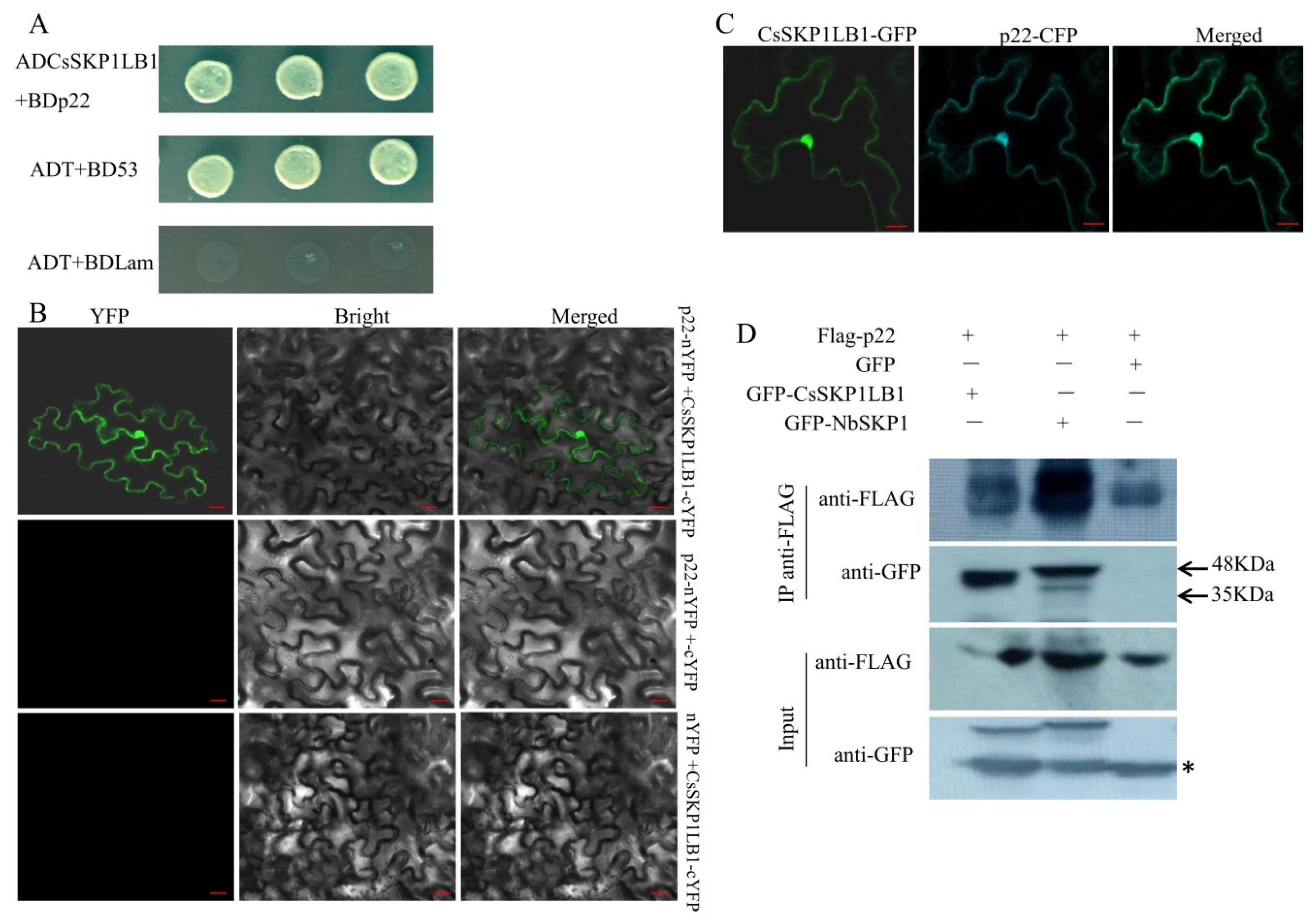
2.3. Yeast Two-Hybrid Screen and Interaction Assay
2.4. Confocal Laser Scanning Microscopy
2.5. Co-Immunoprecipitation In Vivo
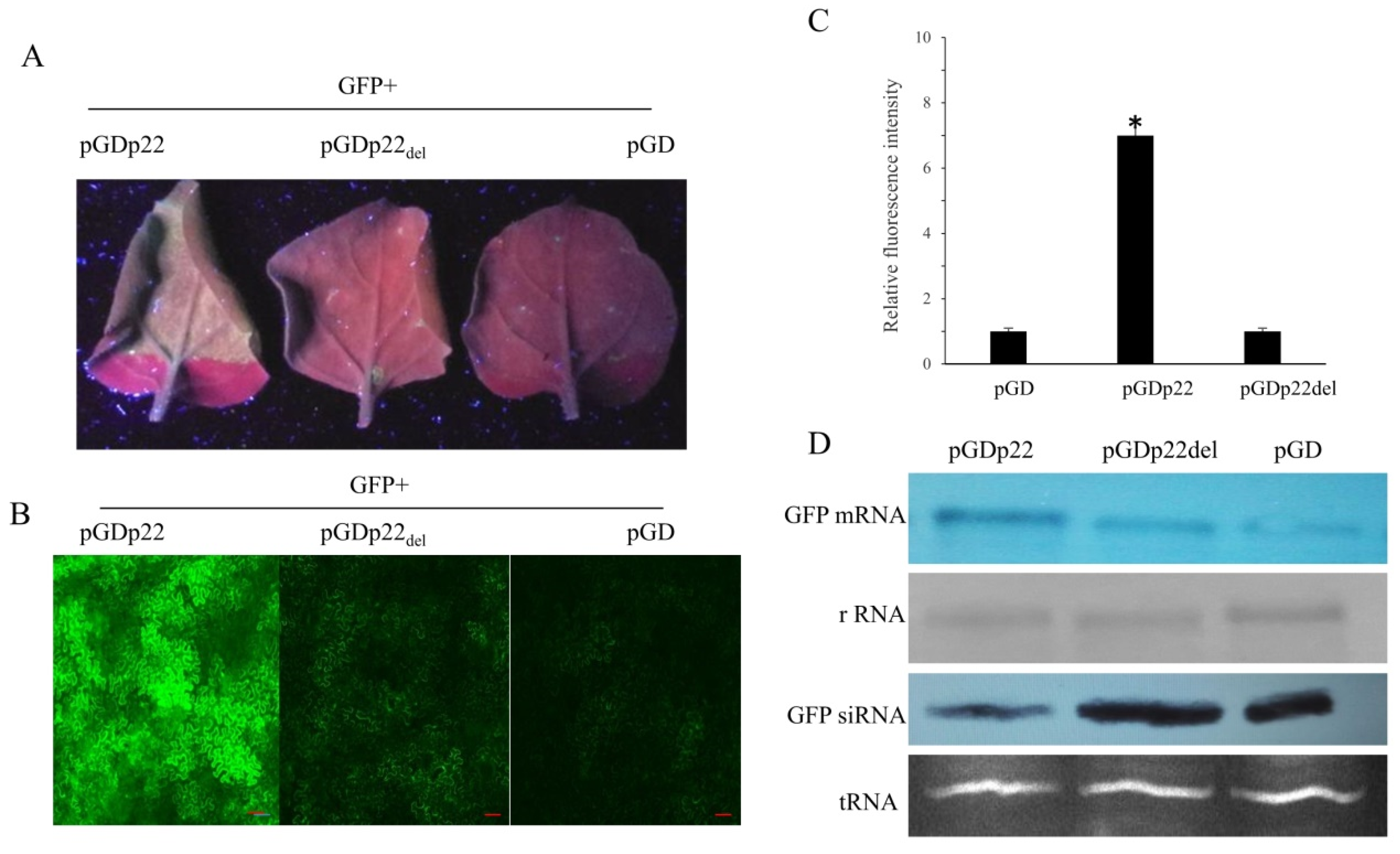
2.6. Quantification of GFP Fluorescence Intensity
2.7. Northern Blot Analysis
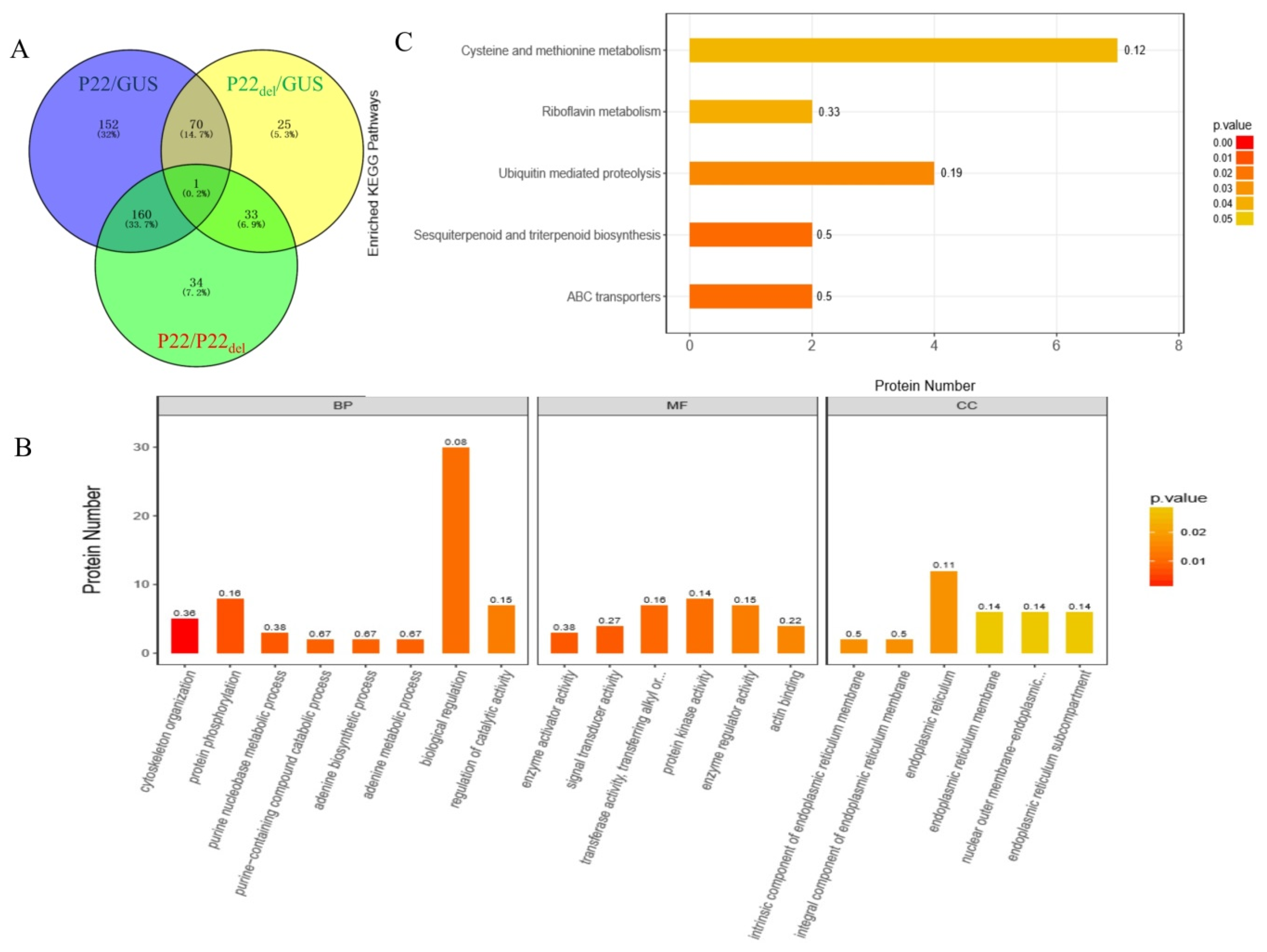
2.8. Proteomic Analysis
2.9. Quantitative RT-PCR
3. Results
3.1. Identification of CCYV p22 as a Silencing Suppressor
3.2. Identification of a p22-interacting SKP1 Protein from Cucumber
3.3. Examining the Interaction between p22 and Other SKP1 Proteins and Mapping the SKP1 Interacting Domain
3.4. The F-Box-Like Motif Is Essential for p22-Mediated Viral Pathogenicity
3.5. Deletion of the p22 F-Box Motif Leads to Inhibition of Silencing Suppressor Activity
3.6. Effects of the Deletion of F-Box-Like Motif on the Expression of Different Proteins
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wisler, G.C.; Duffus, J.E.; Liu, H.Y.; Li, R.H. Ecology and epidemiology of whitefly-transmitted closteroviruses. Plant Dis. 1998, 82, 270–280. [Google Scholar] [CrossRef] [PubMed]
- Navas-Castillo, J.; Fiallo-Olive, E.; Sanchez-Campos, S. Emerging virus diseases transmitted by whiteflies. Annu. Rev. Phytopathol. 2011, 49, 219–248. [Google Scholar] [CrossRef] [PubMed]
- Tzanetakis, I.E.; Martin, R.R.; Wintermantel, W.M. Epidemiology of criniviruses: An emerging problem in world agriculture. Front. Microbio. 2013, 4, 119. [Google Scholar] [CrossRef] [PubMed]
- Gu, Q.S.; Liu, Y.H.; Wang, Y.H.; Huangfu, W.G.; Gu, H.; Xu, L.; Song, F.M.; Brown, L.K. First report of Cucurbit chlorotic yellows virus in cucumber, melon, and watermelon in China. Plant Dis. 2011, 95, 73. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.H.; Tseng, H.H.; Li, J.T.; Chen, T.C. First report of cucurbit chlorotic yellows virus infecting cucurbits in Taiwan. Plant Dis. 2010, 94, 1168. [Google Scholar] [CrossRef] [PubMed]
- Abrahamian, P.E.; Sobh, H.; AbouJawdah, Y. First report of Cucurbit chlorotic yellows virus on cucumber in Lebanon. Plant Dis. 2012, 96, 1704. [Google Scholar] [CrossRef]
- Okuda, M.; Okazaki, S.; Yamasaki, S.; Okuda, S.; Sugiyama, M. Host range and complete genome sequence of Cucurbit chlorotic yellows virus, a new member of the genus Crinivirus. Phytopathology 2010, 100, 560–566. [Google Scholar] [CrossRef]
- Kreuze, J.F.; Savenkov, E.I.; Cuellar, W.; Li, X.; Valkonen, J.P.T. Viral Class 1 RNase III Involved in Suppression of RNA Silencing. J. Virol. 2005, 79, 7227–7238. [Google Scholar] [CrossRef]
- Cañizares, M.C.; Navas-Castillo, J.; Moriones, E. Multiple suppressors of RNA silencing encoded by both genomic RNAs of the crinivirus, Tomato chlorosis virus. Virology 2008, 379, 168–174. [Google Scholar] [CrossRef]
- Zheng, N.; Schulman, B.A.; Song, L.; Miller, J.J.; Jeffrey, P.D.; Wang, P.; Chu, C.; Koepp, D.M.; Elledge, S.J.; Pagano, M.; et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 2002, 416, 703–709. [Google Scholar] [CrossRef]
- Cardozo, T.; Pagano, M. The SCF ubiquitin ligase: Insights into a molecular machine. Nat. Rev. Mol. Cell Bio. 2004, 5, 739–751. [Google Scholar] [CrossRef] [PubMed]
- Petroski, M.D.; Deshaies, R.J. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell 2005, 123, 1107–1120. [Google Scholar] [CrossRef] [PubMed]
- Lechner, E.; Achard, P.; Vansiri, A.; Potuschak, T.; Genschik, P. F-box proteins everywhere. Curr. Opin Plant Biol. 2006, 9, 631–638. [Google Scholar] [CrossRef] [PubMed]
- Alcaide-Loridan, C.; Jupin, I. Ubiquitin and plant viruses, let’s play together! Plant Physiol. 2012, 160, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Verchot, J. Plant Virus Infection and the Ubiquitin Proteasome Machinery: Arms Race along the Endoplasmic Reticulum. Viruses 2016, 8, 314. [Google Scholar] [CrossRef] [PubMed]
- Aronson, M.N.; Meyer, A.D.; Gyorgyey, J.; Katul, L.; Vetten, H.J.; Gronenborn, B.; Timchenko, T. Clink, a nanovirus-encoded protein, binds both pRB and SKP1. J. Virol. 2000, 74, 2967–2972. [Google Scholar] [CrossRef] [PubMed]
- Pazhouhandeh, M.; Dieterle, M.; Marrocco, K.; Lechner, E.; Berry, B.; Brault, V.; Hemmer, O.; Kretsch, T.; Richards, K.E.; Genschik, P.; et al. F-box-like domain in the polerovirus protein P0 is required for silencing suppressor function. Proc. Natl. Acad. Sci. USA 2006, 103, 1994–1999. [Google Scholar] [CrossRef]
- Jia, Q.; Liu, N.; Xie, K.; Dai, Y.; Han, S.; Zhao, X.; Qian, L.; Wang, Y.; Zhao, J.; Gorovits, J.; et al. CLCuMuB βC1 Subverts Ubiquitination by Interacting with NbSKP1s to Enhance Geminivirus Infection in Nicotiana benthamiana. PLoS Pathog. 2016, 12, e1005668. [Google Scholar] [CrossRef]
- Tao, T.; Zhou, C.J.; Wang, Q.; Chen, X.R.; Sun, Q.; Zhao, T.Y.; Ye, J.C.; Wang, Y.; Zhang, Z.Y.; Zhang, Y.L.; et al. Rice black streaked dwarf virus P7-2 forms a SCF complex through binding to Oryza sativa SKP1-like proteins, and interacts with GID2 involved in the gibberellin pathway. PLoS ONE 2017, 12, e0177518. [Google Scholar] [CrossRef]
- Li, Y.; Sun, Q.; Zhao, T.; Xiang, H.; Zhang, X.; Wu, Z.; Zhou, C.; Wang, Y.; Zhang, Y.; Wang, X.; et al. Interaction between Brassica yellows virus silencing suppressor P0 and plant SKP1 facilitates stability of P0 in vivo against degradation by proteasome and autophagy pathways. New Phytol 2019, 222, 1458–1473. [Google Scholar] [CrossRef]
- Baumberger, N.; Tsai, C.H.; Lie, M.; Havecker, E.; Baulcombe, D.C. The Polerovirus Silencing Suppressor P0 Targets ARGONAUTE Proteins for Degradation. Curr. Biol. 2007, 17, 1609–1614. [Google Scholar] [CrossRef] [PubMed]
- Csorba, T.; Lozsa, R.; Hutvagner, G.; Burgyan, J. Polerovirus protein P0 prevents the assembly of small RNA-containing RISC complexes and leads to degradation of ARGONAUTE1. Plant J. 2010, 62, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Derrien, B.; Baumberger, N.; Schepetilnikov, M.; Viotti, C.; De Cillia, J.; Ziegler-Graff, V.; Isono, E.; Schumacher, K.; Genschik, P. Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc. Natl. Acad. Sci. USA 2012, 109, 15942–15946. [Google Scholar] [CrossRef] [PubMed]
- Mangwende, T.; Wang, M.L.; Borth, W.; Hu, J.; Moore, P.H.; Mirkov, T.E.; Albert, H.H. The P0 gene of Sugarcane yellow leaf virus encodes an RNA silencing suppressor with unique activities. Virology 2009, 384, 38–50. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.O.; Zhang, X.Y.; Wang, Y.; Li, D.W.; Yu, J.L.; Han, C.G. The Three Essential Motifs in P0 for Suppression of RNA Silencing Activity of Potato leafroll virus Are Required for Virus Systemic Infection. Viruses 2019, 11, 170. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.Q.; Liu, Z.M.; Xu, J.; Zhou, T.; Wang, M.; Chen, Y.T.; Li, H.F.; Fan, Z.F. HC-Pro protein of sugar cane mosaic virus interacts specifically with maize ferredoxin-5 in vitro and in planta. J. Gen. Virol. 2008, 89, 2046–2054. [Google Scholar] [CrossRef]
- Walter, M.; Chaban, C.; Schutze, K.; Batistic, O.; Weckermann, K.; Nake, C.; Blazevic, D.; Grefen, C.; Schumacher, K.; Oecking, C.; et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004, 40, 428–438. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Luber, C.A.; Cox, J.; Lauterbach, H.; Fancke, B.; Selbach, M.; Tschopp, J.; Akira, S.; Wiegand, M.; Hochrein, H.; O’Keeffe, M.; et al. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity 2010, 32, 279–289. [Google Scholar] [CrossRef]
- Schulman, B.A.; Carrano, A.C.; Jeffrey, P.D.; Bowen, Z.; Kinnucan, E.R.; Finnin, M.S.; Elledge, S.J.; Harper, J.W.; Pagano, M.; Pavletich, N.P. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 2000, 408, 381–386. [Google Scholar] [CrossRef]
- Pruss, G.; Ge, X.; Shi, X.M.; Carrington, J.C.; Bowman Vance, V. Plant viral synergism: The potyviral genome encodes a broad-range pathogenicity enhancer that transactivates replication of heterologous viruses. Plant Cell 1997, 9, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Yelina, N.E.; Savenkov, E.I.; Solovyev, A.G.; Morozov, S.Y.; Valkonen, J.P. Long-distance movement, virulence, and RNA silencing suppression controlled by a single protein in hordei- and potyviruses: Complementary functions between virus families. J. Virol. 2002, 76, 12981–12991. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.R.; Vega-Sanchez, M.E.; Zhu, T.; Wang, G.L. Ubiquitination-mediated protein degradation and modification: An emerging theme in plant-microbe interactions. Cell Res. 2006, 16, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Dreher, K.; Callis, J. Ubiquitin, hormones and biotic stress in plants. Ann. Bot. 2007, 99, 787–822. [Google Scholar] [CrossRef] [PubMed]
- Citovsky, V.; Zaltsman, A.; Kozlovsky, S.V.; Gafni, Y.; Krichevsky, A. Proteasomal degradation in plant-pathogen interactions. Semin. Cell Dev. Biol. 2009, 20, 1048–1054. [Google Scholar] [CrossRef] [PubMed]
- Dielen, A.S.; Badaoui, S.; Candresse, T.; German-Retana, S. The ubiquitin/26S proteasome system in plant-pathogen interactions: A never-ending hide-and-seek game. Mol. Plant Pathol. 2010, 11, 293–308. [Google Scholar] [CrossRef] [PubMed]
- Marino, D.; Peeters, N.; Rivas, S. Ubiquitination during plant immune signaling. Plant Physiol. 2012, 160, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Shackelford, J.; Pagano, J.S. Targeting of host-cell ubiquitin pathways by viruses. Essays Biochem. 2005, 41, 139–156. [Google Scholar] [CrossRef] [PubMed]
- Isaacson, M.K.; Ploegh, H.L. Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell Host Microbe 2009, 5, 559–570. [Google Scholar] [CrossRef]
- Randow, F.; Lehner, P.J. Viral avoidance and exploitation of the ubiquitin system. Nat. Cell Biol. 2009, 11, 527–534. [Google Scholar] [CrossRef]
- Risseeuw, E.P.; Daskalchuk, T.E.; Banks, T.W.; Liu, E.; Cotelesage, J.; Hellmann, H.; Estelle, M.; Somers, D.E.; Crosby, W.L. Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis. Plant J. 2003, 34, 753–767. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, K.I.; Eskelin, K.; Bašić, M.; De, S.; Lõhmus, A.; Varjosalo, M.; Mäkinen, K. Molecular insights into the function of the viral RNA silencing suppressor HCPro. Plant J. 2016, 85, 30–45. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Wang, Z.; Gu, Q.; Li, H.; Han, W.; Shi, Y. Transcriptome analysis of Cucumis sativus infected by Cucurbit chlorotic yellows virus. Virol. J. 2017, 14, 18. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kubota, K.; Ng, J.C.K. Lettuce chlorosis virus P23 suppresses RNA silencing and induces local necrosis with increased severity at raised temperatures. Virology 2016, 106, 653–662. [Google Scholar]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Sun, X.; Shi, Y.; Wei, Y.; Han, X.; Li, H.; Chen, L.; Sun, B.; Sun, H.; Shi, Y. Cucurbit Chlorotic Yellows Virus p22 Protein Interacts with Cucumber SKP1LB1 and Its F-Box-Like Motif Is Crucial for Silencing Suppressor Activity. Viruses 2019, 11, 818. https://doi.org/10.3390/v11090818
Chen S, Sun X, Shi Y, Wei Y, Han X, Li H, Chen L, Sun B, Sun H, Shi Y. Cucurbit Chlorotic Yellows Virus p22 Protein Interacts with Cucumber SKP1LB1 and Its F-Box-Like Motif Is Crucial for Silencing Suppressor Activity. Viruses. 2019; 11(9):818. https://doi.org/10.3390/v11090818
Chicago/Turabian StyleChen, Siyu, Xinyan Sun, Yajuan Shi, Ying Wei, Xiaoyu Han, Honglian Li, Linlin Chen, Bingjian Sun, Hangjun Sun, and Yan Shi. 2019. "Cucurbit Chlorotic Yellows Virus p22 Protein Interacts with Cucumber SKP1LB1 and Its F-Box-Like Motif Is Crucial for Silencing Suppressor Activity" Viruses 11, no. 9: 818. https://doi.org/10.3390/v11090818
APA StyleChen, S., Sun, X., Shi, Y., Wei, Y., Han, X., Li, H., Chen, L., Sun, B., Sun, H., & Shi, Y. (2019). Cucurbit Chlorotic Yellows Virus p22 Protein Interacts with Cucumber SKP1LB1 and Its F-Box-Like Motif Is Crucial for Silencing Suppressor Activity. Viruses, 11(9), 818. https://doi.org/10.3390/v11090818



