Genetic Diversity of Sapoviruses among Inpatients in Germany, 2008−2018
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Analysis of Local Temperature
2.3. Nucleic Acid Extraction and Sapovirus RNA Detection
2.4. Nucleic Acid Sequencing
2.5. Phylogenetic Analysis and VP1 Genotyping
2.6. Nosocomial Sapovirus Infections
2.7. Ethical Considerations
3. Results
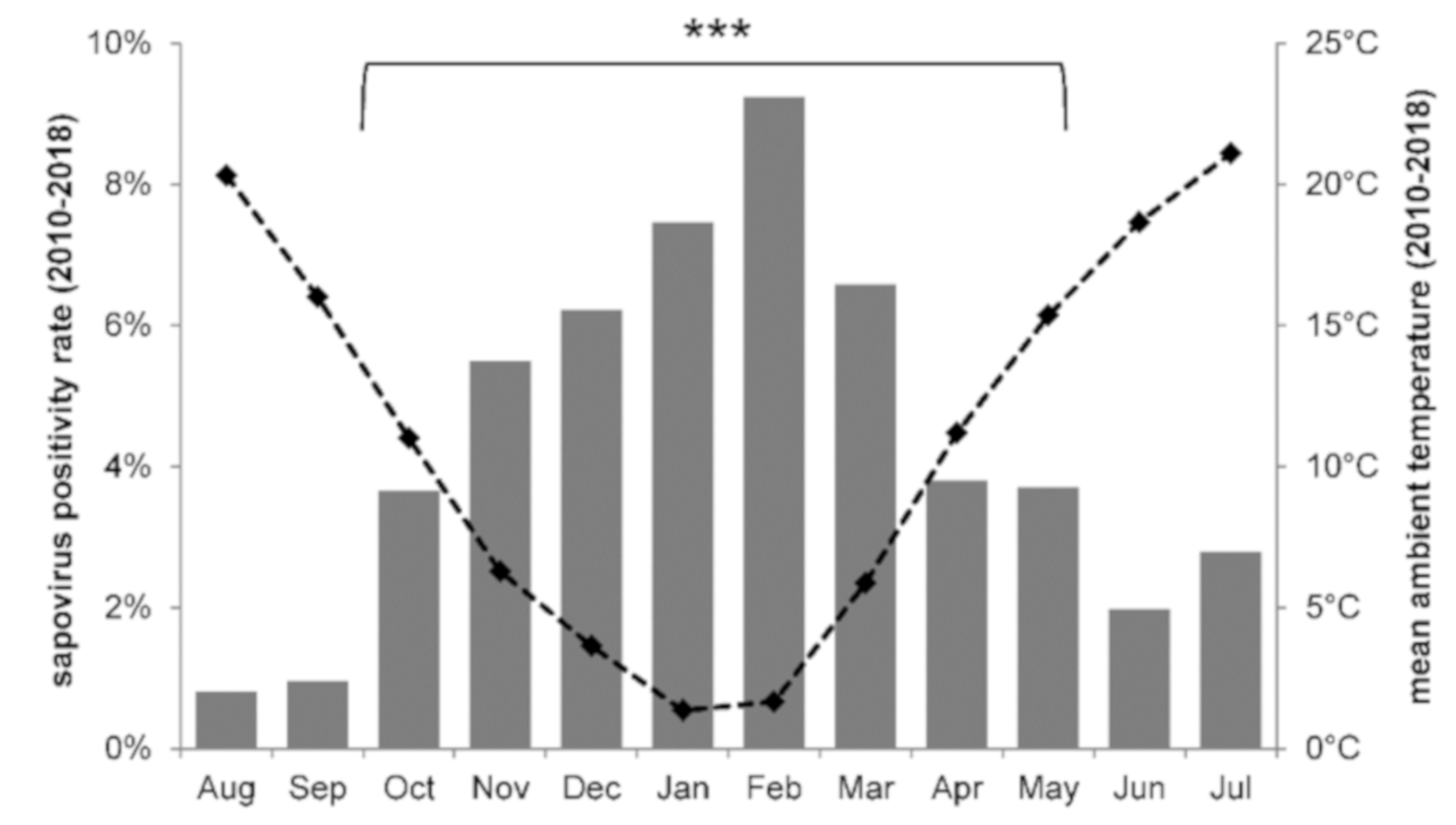
3.1. Sapovirus Stool Positivity
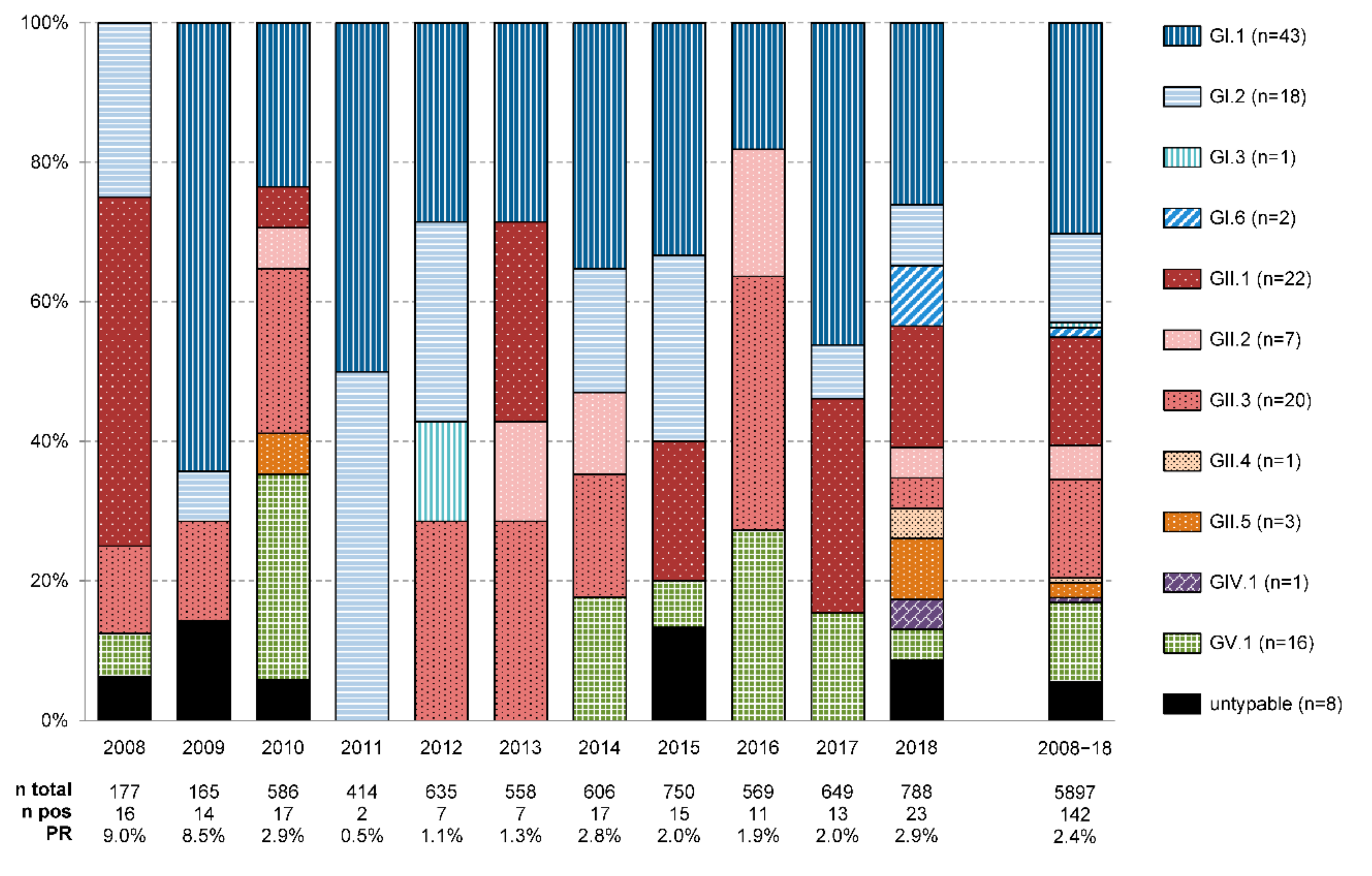
3.2. Sapovirus Genotypes
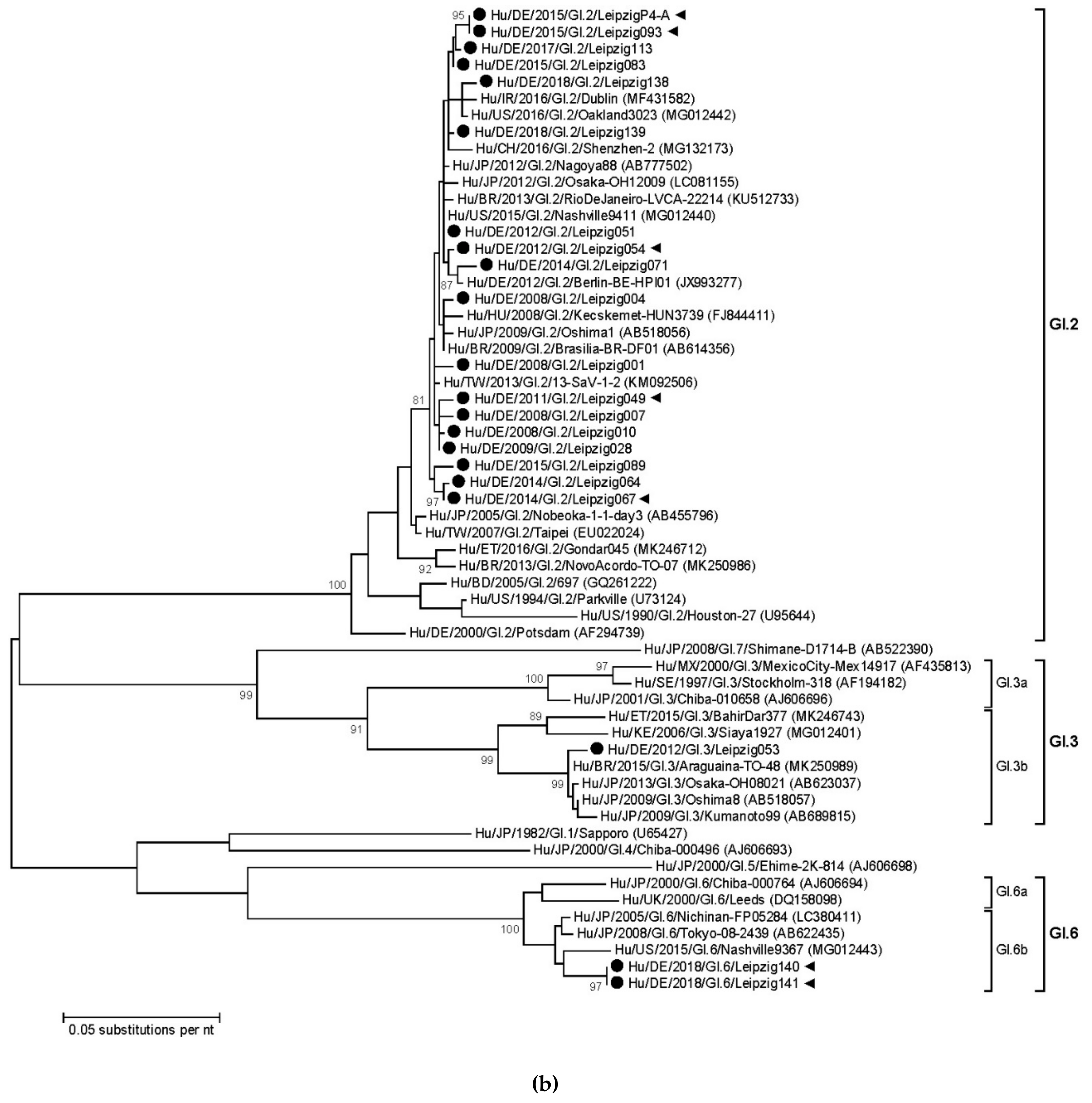
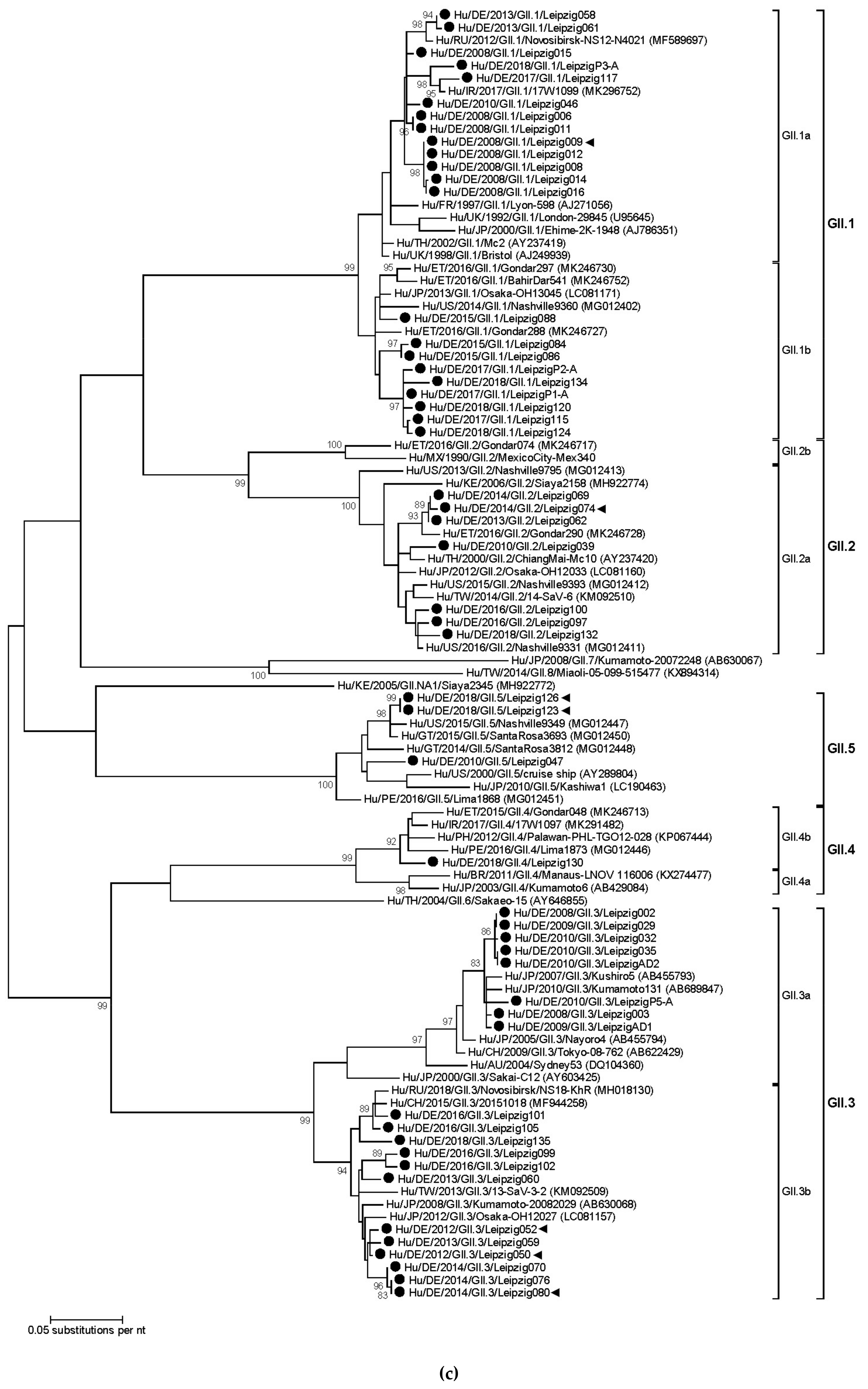
3.3. Sapovirus Phylogenetic Analysis
3.4. Nosocomial Sapovirus Infections
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Thongprachum, A.; Khamrin, P.; Maneekarn, N.; Hayakawa, S.; Ushijima, H. Epidemiology of gastroenteritis viruses in Japan: Prevalence, seasonality, and outbreak. J. Med. Virol. 2016, 88, 551–570. [Google Scholar] [CrossRef] [PubMed]
- Oka, T.; Wang, Q.; Katayama, K.; Saif, L.J. Comprehensive review of human sapoviruses. Clin. Microbiol. Rev. 2015, 28, 32–53. [Google Scholar] [CrossRef] [PubMed]
- Pitkanen, O.; Vesikari, T.; Hemming-Harlo, M. The role of the sapovirus infection increased in gastroenteritis after national immunisation was introduced. Acta Paediatr. 2019, 108, 1338–1344. [Google Scholar] [CrossRef] [PubMed]
- Sakai, Y.; Nakata, S.; Honma, S.; Tatsumi, M.; Numata-Kinoshita, K.; Chiba, S. Clinical severity of Norwalk virus and Sapporo virus gastroenteritis in children in Hokkaido, Japan. Pediatr. Infect. Dis. J. 2001, 20, 849–853. [Google Scholar] [CrossRef] [PubMed]
- Madeley, C.R.; Cosgrove, B.P. Letter: Caliciviruses in man. Lancet 1976, 1, 199–200. [Google Scholar] [CrossRef]
- Chiba, S.; Sakuma, Y.; Kogasaka, R.; Akihara, M.; Horino, K.; Nakao, T.; Fukui, S. An outbreak of gastroenteritis associated with calicivirus in an infant home. J. Med. Virol. 1979, 4, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Desselberger, U. Caliciviridae Other Than Noroviruses. Viruses 2019, 11, 286. [Google Scholar] [CrossRef]
- Liu, B.L.; Clarke, I.N.; Caul, E.O.; Lambden, P.R. Human enteric caliciviruses have a unique genome structure and are distinct from the Norwalk-like viruses. Arch. Virol. 1995, 140, 1345–1356. [Google Scholar] [CrossRef]
- Oka, T.; Lu, Z.; Phan, T.; Delwart, E.L.; Saif, L.J.; Wang, Q. Genetic Characterization and Classification of Human and Animal Sapoviruses. PLoS ONE 2016, 11, e0156373. [Google Scholar] [CrossRef]
- Xue, L.; Cai, W.; Gao, J.; Jiang, Y.; Wu, H.; Zhang, L.; Zuo, Y.; Dong, R.; Pang, R.; Zeng, H.; et al. Genome characteristics and molecular evolution of the human sapovirus variant GII.8. Infect. Genet. Evol. 2019, 73, 362–367. [Google Scholar] [CrossRef]
- Yoneda, M.; Nakano, M.; Sugimoto, D.; Inada, M.; Fujitani, M.; Kitahori, Y. Epidemiological Characteristics of Sapovirus and Human Astrovirus Detected among Children in Nara Prefecture, Japan, during the 2009/2010–2014/2015 Seasons. Jpn. J. Infect. Dis. 2017, 70, 87–91. [Google Scholar] [CrossRef]
- Harada, S.; Tokuoka, E.; Kiyota, N.; Katayama, K.; Oka, T. Phylogenetic analysis of the nonstructural and structural protein encoding region sequences, indicating successive appearance of genomically diverse sapovirus strains from gastroenteritis patients. Jpn. J. Infect. Dis. 2013, 66, 454–457. [Google Scholar] [CrossRef]
- Murray, T.Y.; Nadan, S.; Page, N.A.; Taylor, M.B. Diverse sapovirus genotypes identified in children hospitalised with gastroenteritis in selected regions of South Africa. J. Clin. Virol. 2016, 76, 24–29. [Google Scholar] [CrossRef]
- Sanchez, G.J.; Mayta, H.; Pajuelo, M.J.; Neira, K.; Xiaofang, L.; Cabrera, L.; Ballard, S.B.; Crabtree, J.E.; Kelleher, D.; Cama, V.; et al. Epidemiology of Sapovirus Infections in a Birth Cohort in Peru. Clin. Infect. Dis. 2018, 66, 1858–1863. [Google Scholar] [CrossRef]
- Lasure, N.; Gopalkrishna, V. Epidemiological profile and genetic diversity of sapoviruses (SaVs) identified in children suffering from acute gastroenteritis in Pune, Maharashtra, Western India, 2007–2011. Epidemiol. Infect. 2017, 145, 106–114. [Google Scholar] [CrossRef]
- Ike, A.C.; Hartelt, K.; Oehme, R.M.; Brockmann, S.O. Detection and characterization of sapoviruses in outbreaks of gastroenteritis in southwest Germany. J. Clin. Virol. 2008, 43, 37–41. [Google Scholar] [CrossRef]
- Moser, O.; Luck, S.; Dilloo, D.; Eis-Hubinger, A.M.; Simon, A. Sapovirus as a gastrointestinal pathogen in febrile pediatric patients with cancer. J. Med. Virol. 2011, 83, 2233–2236. [Google Scholar] [CrossRef]
- Oka, T.; Katayama, K.; Hansman, G.S.; Kageyama, T.; Ogawa, S.; Wu, F.T.; White, P.A.; Takeda, N. Detection of human sapovirus by real-time reverse transcription-polymerase chain reaction. J. Med. Virol. 2006, 78, 1347–1353. [Google Scholar] [CrossRef]
- Kitajima, M.; Oka, T.; Haramoto, E.; Katayama, H.; Takeda, N.; Katayama, K.; Ohgaki, S. Detection and genetic analysis of human sapoviruses in river water in Japan. Appl. Environ. Microbiol. 2010, 76, 2461–2467. [Google Scholar] [CrossRef]
- Pietsch, C.; Liebert, U.G. Intrahost viral evolution during chronic sapovirus infections. J. Clin. Virol. 2019, 113, 1–7. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Fullerton, S.W.; Blaschke, M.; Coutard, B.; Gebhardt, J.; Gorbalenya, A.; Canard, B.; Tucker, P.A.; Rohayem, J. Structural and functional characterization of sapovirus RNA-dependent RNA polymerase. J. Virol. 2007, 81, 1858–1871. [Google Scholar] [CrossRef] [PubMed]
- Varela, M.F.; Rivadulla, E.; Lema, A.; Romalde, J.L. Human Sapovirus among Outpatients with Acute Gastroenteritis in Spain: A One-Year Study. Viruses 2019, 11, 144. [Google Scholar] [CrossRef]
- Medici, M.C.; Tummolo, F.; Albonetti, V.; Abelli, L.A.; Chezzi, C.; Calderaro, A. Molecular detection and epidemiology of astrovirus, bocavirus, and sapovirus in Italian children admitted to hospital with acute gastroenteritis, 2008–2009. J. Med. Virol. 2012, 84, 643–650. [Google Scholar] [CrossRef]
- Johnsen, C.K.; Midgley, S.; Bottiger, B. Genetic diversity of sapovirus infections in Danish children 2005-2007. J. Clin. Virol. 2009, 46, 265–269. [Google Scholar] [CrossRef]
- Gallimore, C.I.; Iturriza-Gomara, M.; Lewis, D.; Cubitt, D.; Cotterill, H.; Gray, J.J. Characterization of sapoviruses collected in the United Kingdom from 1989 to 2004. J. Med. Virol. 2006, 78, 673–682. [Google Scholar] [CrossRef]
- Iritani, N.; Yamamoto, S.P.; Abe, N.; Kubo, H.; Oka, T.; Kaida, A. Epidemics of GI.2 sapovirus in gastroenteritis outbreaks during 2012–2013 in Osaka City, Japan. J. Med. Virol. 2016, 88, 1187–1193. [Google Scholar] [CrossRef]
- Svraka, S.; Vennema, H.; van der Veer, B.; Hedlund, K.O.; Thorhagen, M.; Siebenga, J.; Duizer, E.; Koopmans, M. Epidemiology and genotype analysis of emerging sapovirus-associated infections across Europe. J. Clin. Microbiol. 2010, 48, 2191–2198. [Google Scholar] [CrossRef]
- Chhabra, P.; Samoilovich, E.; Yermalovich, M.; Chernyshova, L.; Gheorghita, S.; Cojocaru, R.; Shugayev, N.; Sahakyan, G.; Lashkarashvili, M.; Chubinidze, M.; et al. Viral gastroenteritis in rotavirus negative hospitalized children <5 years of age from the independent states of the former Soviet Union. Infect. Genet. Evol. 2014, 28, 283–288. [Google Scholar] [CrossRef]
- Gelaw, A.; Pietsch, C.; Mann, P.; Liebert, U.G. Molecular detection and characterisation of sapoviruses and noroviruses in outpatient children with diarrhoea in Northwest Ethiopia. Epidemiol. Infect. 2019, 147, e218. [Google Scholar] [CrossRef]
- Magwalivha, M.; Kabue, J.-P.; Traore, A.N.; Potgieter, N. Prevalence of Human Sapovirus in Low and Middle Income Countries. Adv. Virol. 2017, 2018, 5986549. [Google Scholar] [CrossRef]
- Pouletty, M.; de Pontual, L.; Lopez, M.; Morin, L.; Poilane, I.; Pham, L.L.; Carbonnelle, E.; Titomanlio, L.; Faye, A.; Bonacorsi, S. Multiplex PCR reveals a high prevalence of multiple pathogens in traveller’s diarrhoea in children. Arch. Dis. Child. 2018. [Google Scholar] [CrossRef]
- Nakamura, N.; Kobayashi, S.; Minagawa, H.; Matsushita, T.; Sugiura, W.; Iwatani, Y. Molecular epidemiology of enteric viruses in patients with acute gastroenteritis in Aichi prefecture, Japan, 2008/09–2013/14. J. Med. Virol. 2016, 88, 1180–1186. [Google Scholar] [CrossRef]
- Brown, J.R.; Shah, D.; Breuer, J. Viral gastrointestinal infections and norovirus genotypes in a paediatric UK hospital, 2014-2015. J. Clin. Virol. 2016, 84, 1–6. [Google Scholar] [CrossRef]
- Bergallo, M.; Galliano, I.; Montanari, P.; Brusin, M.R.; Finotti, S.; Paderi, G.; Gabiano, C. Development of a quantitative real-time PCR assay for sapovirus in children under 5-years-old in Regina Margherita Hospital of Turin, Italy. Can. J. Microbiol. 2017, 63, 296–302. [Google Scholar] [CrossRef]
- Hebbelstrup Jensen, B.; Jokelainen, P.; Nielsen, A.C.Y.; Franck, K.T.; Rejkjaer Holm, D.; Schonning, K.; Petersen, A.M.; Krogfelt, K.A. Children Attending Day Care Centers are a Year-round Reservoir of Gastrointestinal Viruses. Sci. Rep. 2019, 9, 3286. [Google Scholar] [CrossRef]
- Ouedraogo, N.; Kaplon, J.; Bonkoungou, I.J.; Traore, A.S.; Pothier, P.; Barro, N.; Ambert-Balay, K. Prevalence and Genetic Diversity of Enteric Viruses in Children with Diarrhea in Ouagadougou, Burkina Faso. PLoS ONE 2016, 11, e0153652. [Google Scholar] [CrossRef]
- Bucardo, F.; Carlsson, B.; Nordgren, J.; Larson, G.; Blandon, P.; Vilchez, S.; Svensson, L. Susceptibility of children to sapovirus infections, Nicaragua, 2005–2006. Emerg. Infect. Dis. 2012, 18, 1875–1878. [Google Scholar] [CrossRef]
- Yang, B.; Yang, B.; Shan, X.; Li, B.; Ma, X.; Yin, X.; Zhang, Y.; Liu, Y.; Lan, X. Short communication: Immune responses in sows induced by porcine sapovirus virus-like particles reduce viral shedding in suckled piglets. Res. Vet. Sci. 2017, 117, 196–199. [Google Scholar] [CrossRef]
- Farkas, T.; Deng, X.; Ruiz-Palacios, G.; Morrow, A.; Jiang, X. Development of an enzyme immunoassay for detection of sapovirus-specific antibodies and its application in a study of seroprevalence in children. J. Clin. Microbiol. 2006, 44, 3674–3679. [Google Scholar] [CrossRef]
- Thongprachum, A.; Takanashi, S.; Kalesaran, A.F.; Okitsu, S.; Mizuguchi, M.; Hayakawa, S.; Ushijima, H. Four-year study of viruses that cause diarrhea in Japanese pediatric outpatients. J. Med. Virol. 2015, 87, 1141–1148. [Google Scholar] [CrossRef]
- Nakata, S.; Chiba, S.; Terashima, H.; Yokoyama, T.; Nakao, T. Humoral immunity in infants with gastroenteritis caused by human calicivirus. J. Infect. Dis. 1985, 152, 274–279. [Google Scholar] [CrossRef]
- Lauritsen, K.T.; Hansen, M.S.; Johnsen, C.K.; Jungersen, G.; Bottiger, B. Repeated examination of natural sapovirus infections in pig litters raised under experimental conditions. Acta Vet. Scand. 2015, 57, 60. [Google Scholar] [CrossRef]
- Harada, S.; Oka, T.; Tokuoka, E.; Kiyota, N.; Nishimura, K.; Shimada, Y.; Ueno, T.; Ikezawa, S.; Wakita, T.; Wang, Q.; et al. A confirmation of sapovirus re-infection gastroenteritis cases with different genogroups and genetic shifts in the evolving sapovirus genotypes, 2002–2011. Arch. Virol. 2012, 157, 1999–2003. [Google Scholar] [CrossRef]
- Diez-Valcarce, M.; Montmayeur, A.; Tatusov, R.; Vinje, J. Near-Complete Human Sapovirus Genome Sequences from Kenya. Microbiol. Resour. Announc. 2019, 8. [Google Scholar] [CrossRef]
- Oka, T.; Iritani, N.; Yamamoto, S.P.; Mori, K.; Ogawa, T.; Tatsumi, C.; Shibata, S.; Harada, S.; Wu, F.T. Broadly reactive real-time reverse transcription-polymerase chain reaction assay for detection of human sapovirus genotypes. J. Med. Virol. 2018. [Google Scholar] [CrossRef]
- Chanit, W.; Thongprachum, A.; Khamrin, P.; Okitsu, S.; Mizuguchi, M.; Ushijima, H. Intergenogroup recombinant sapovirus in Japan, 2007–2008. Emerg. Infect. Dis. 2009, 15, 1084–1087. [Google Scholar] [CrossRef]
- Dey, S.K.; Mizuguchi, M.; Okitsu, S.; Ushijima, H. Novel recombinant sapovirus in Bangladesh. Clin. Lab. 2011, 57, 91–94. [Google Scholar]


| Genotype | Reverse Primer | Sequence (5’→3’) |
|---|---|---|
| GI.1 | SaV-VP1-GI.1-R | TCTGGTGARACYCCRTTYTCCAT |
| GI.2 | SaV-VP1-GI.2-R | TCAGGTGACACACCATTBTCCAT |
| GI.3 | SaV-VP1-GI.3-R | TCAGGTGACAMYCCRTTYTCCAT |
| GI.6 | SaV-VP1-GI.6-R | TCAGGGGACACACCRTTYTCCAT |
| GI.x | SaV-VP1-GI.x-R | TCRGGKGAVAHNCCRTTBTSCAT |
| GII.1 | SaV-VP1-GII.1-R | GCRGGTGATATCCCATTGTCCAT |
| GII.2 | SaV-VP1-GII.2-R | GCGGGYGAAATTCCATTGTCCAT |
| GII.3 | SaV-VP1-GII.3-R | GCAGGTGATATGCCRTTRTCCAT |
| GII.4 | SaV-VP1-GII.4-R | GCRGGDGAKAYRCCRTTRTCCAT |
| GII.5 | SaV-VP1-GII.5-R | GCRGGTGATATGCCRTTGTCCAT |
| GII.x | SaV-VP1-GII.x-R | GCDGGNGANAYNCCRTTRTCCAT |
| GIV.1 | SaV-VP1-GIV.1-R | GCTGGGYGARARCCCRTTCTCCAT |
| GV.1 | SaV-VP1-GV.1-R | GANGGTGARCCTCCRTTCTCCAT |
| Cohort | 0 to ≤5 Months | 6 Months to ≤2 Years | 3 to ≤4 Years | 5 to 97 Years | |
|---|---|---|---|---|---|
| Retrospective 2008−2009 | n, PR 1 95% CI | 4/123, 3.3% (1.3−8.1%) | 24/187, 12.8% (8.8−18.4%) | 2/32, 6.3% (1.7−20.2%) | n.a. |
| Prospective 2010−2018 | n, PR 1 95% CI | 3/584, 0.5% (0.2−1.5%) | 77/1049, 7.3% (5.9−9.1%) | 9/249, 3.6% (1.9−6.7%) | 23/3673, 0.6% (0.4−0.9%) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mann, P.; Pietsch, C.; Liebert, U.G. Genetic Diversity of Sapoviruses among Inpatients in Germany, 2008−2018. Viruses 2019, 11, 726. https://doi.org/10.3390/v11080726
Mann P, Pietsch C, Liebert UG. Genetic Diversity of Sapoviruses among Inpatients in Germany, 2008−2018. Viruses. 2019; 11(8):726. https://doi.org/10.3390/v11080726
Chicago/Turabian StyleMann, Pia, Corinna Pietsch, and Uwe G. Liebert. 2019. "Genetic Diversity of Sapoviruses among Inpatients in Germany, 2008−2018" Viruses 11, no. 8: 726. https://doi.org/10.3390/v11080726
APA StyleMann, P., Pietsch, C., & Liebert, U. G. (2019). Genetic Diversity of Sapoviruses among Inpatients in Germany, 2008−2018. Viruses, 11(8), 726. https://doi.org/10.3390/v11080726





