Isolation and Characterization T4- and T7-Like Phages that Infect the Bacterial Plant Pathogen Agrobacterium tumefaciens
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Phage Isolation and Purification
2.3. Plaque Assays
2.4. Preparation of Virion DNA, Genome Sequencing, and Genome Assembly
2.5. DNA Restriction Analysis
2.6. Growth Curves
2.7. Transmission Electron Microscopy
2.8. Genome Annotation
2.9. Phylogenetic Analysis
2.10. GenBank Accession Number
3. Results and Discussion
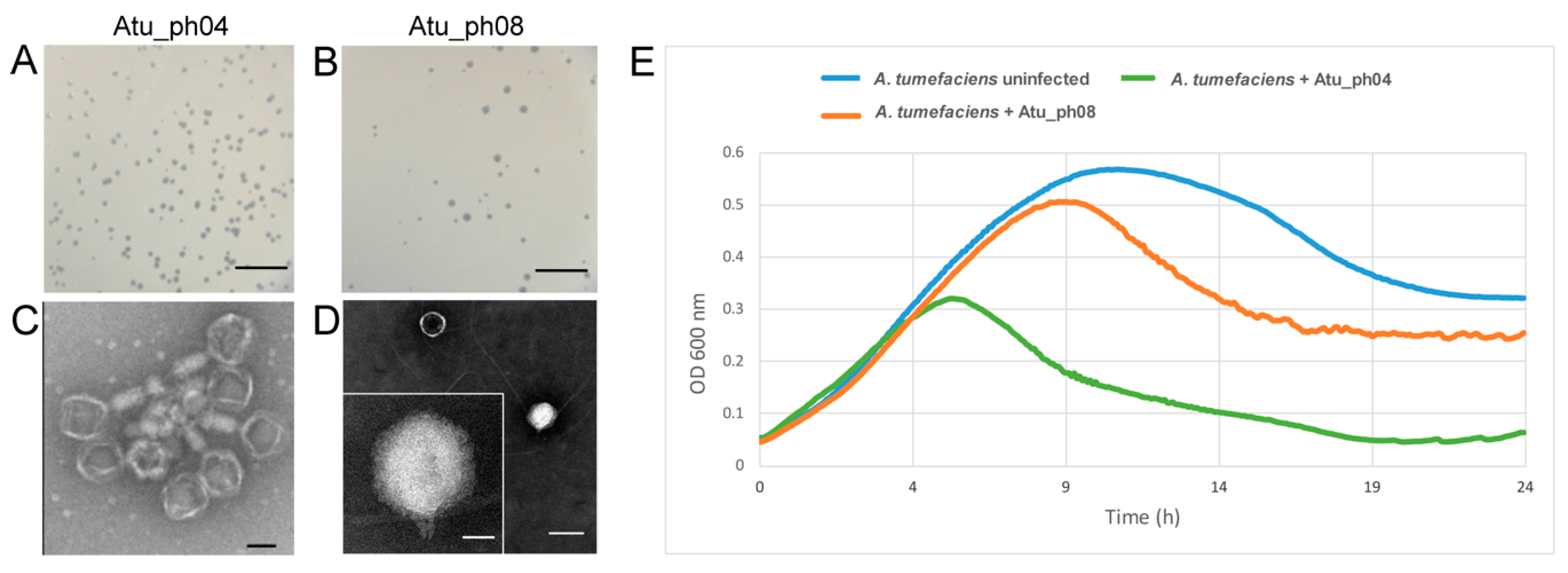
3.1. Phage Atu_ph08 has Higher Lytic Activity than Atu_ph04
3.2. Host Ranges of Atu_ph04 and Atu_ph08 are Limited to A. tumefaciens Strains
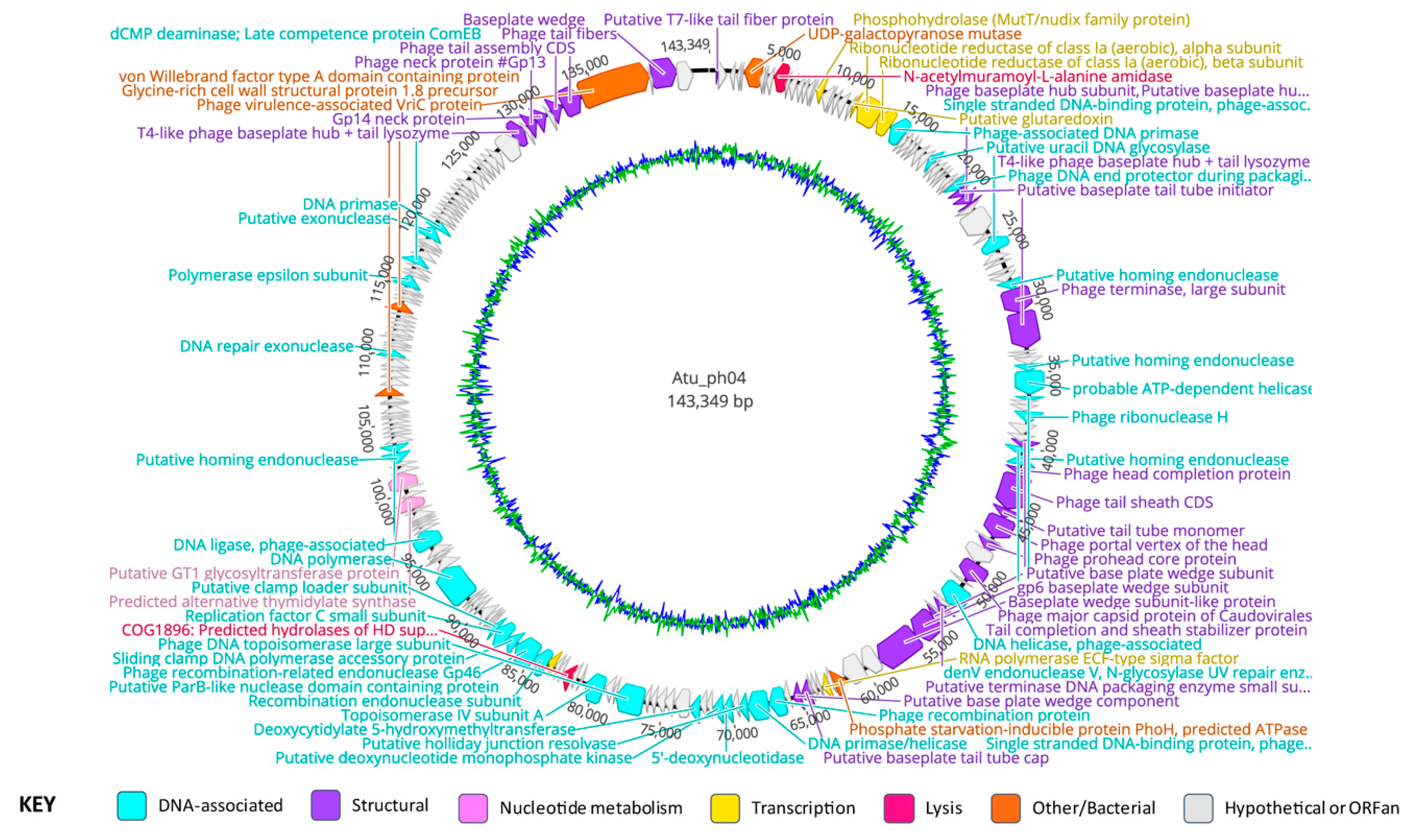
3.3. Genomic Characteristics of Atu_ph04
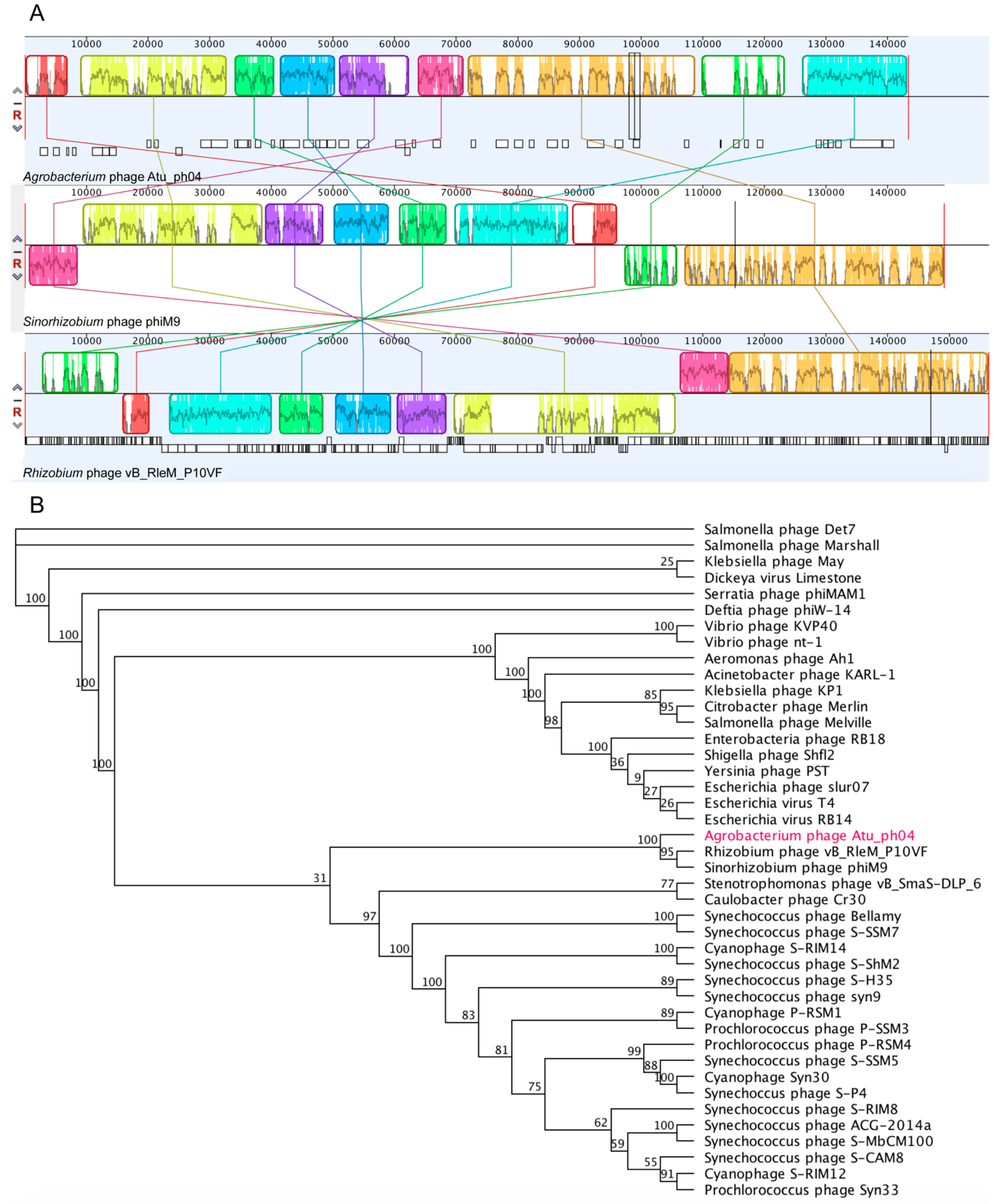
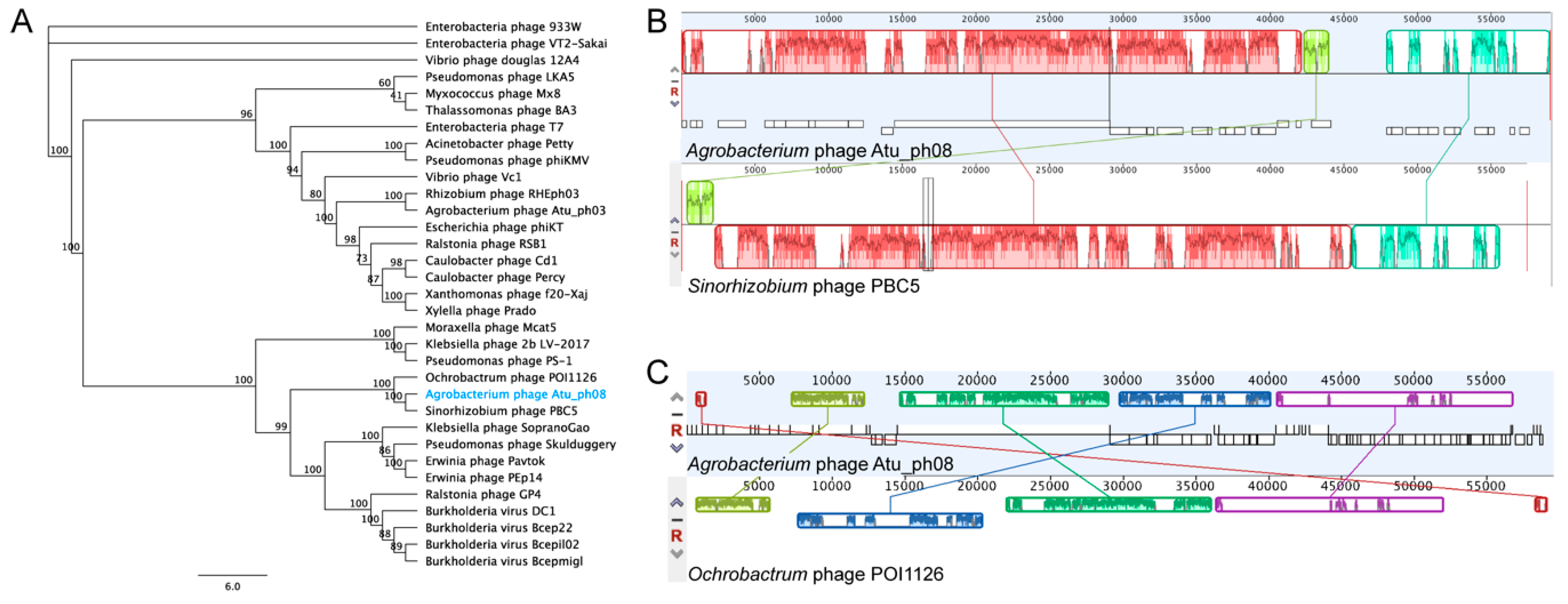
3.4. Phylogenetic Analysis Shows Atu_ph04 is Closely Related to T4-Like Sinorhizobium Phage phiM9 and Rhizobium Phage vB_RleM_P10VF
3.5. Atu_ph04 is a T4-like Phage but Lacks Several T4 Core Proteins
3.6. Major Gene Categories of Atu_ph04
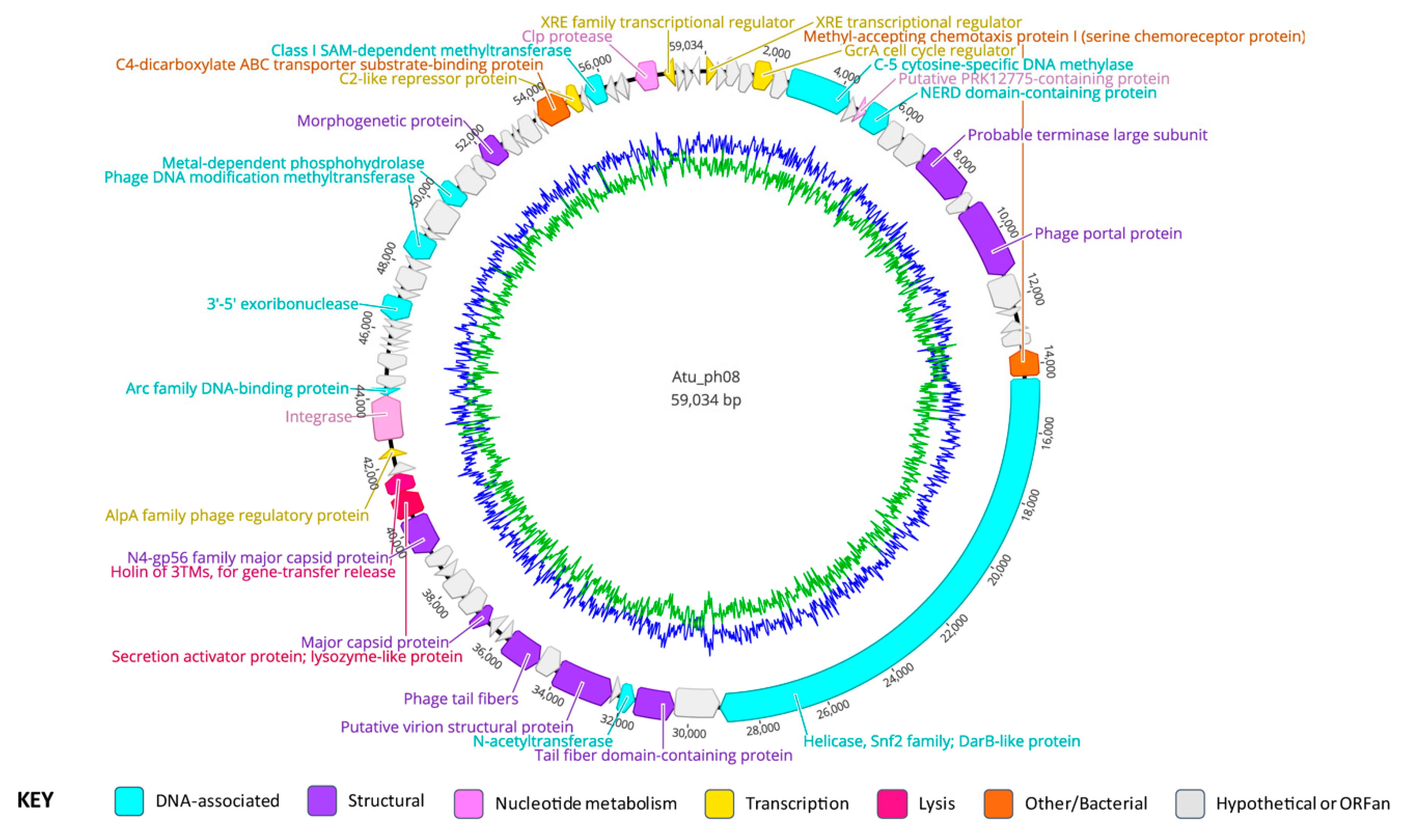
3.7. Atu_ph08 Genomic Summary
3.8. Gene Organization of Atu_ph08
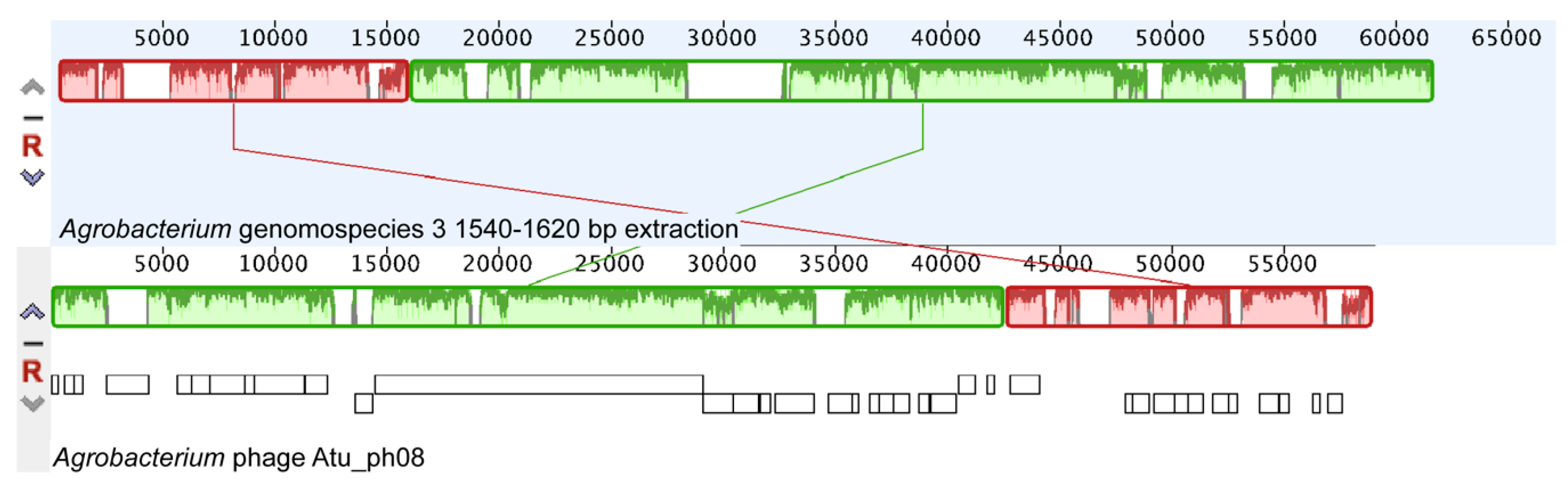
3.9. Atu_ph08 has Some Features of a Temperate Phage and Shares High Homology with A. tumefaciens genomospecies 3
3.10. The Atu_ph08 Genome is Highly Syntenic with the Genome of the T7-Like Sinorhizobium Phage PBC5
3.11. The Atu_ph08 Genome Encodes a DarB-like Protein, Commonly Found Among PBC5-Like Phages
3.12. The Atu_ph08 Genome Encodes a Putative Holin-Endolysin Cassette
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pulawska, J. Crown gall of stone fruits and nuts, economic significance and diversity of its causal agents: Tumorigenic Agrobacterium spp. J. Plant Pathol. 2010, 92, S87–S98. [Google Scholar]
- Sardesai, N.; Subramanyam, S. Agrobacterium: A genome-editing tool-delivery system. In Agrobacterium Biology: From Basic Science to Biotechnology; Gelvin, S.B., Ed.; Springer International Publishing: Cham, Switzerland, 2018; pp. 463–488. ISBN 978-3-030-03257-9. [Google Scholar]
- Anand, A.; Jones, T.J. Advancing Agrobacterium-based crop transformation and genome modification technology for agricultural biotechnology. In Current Topics in Microbiology and Immunology; Springer: Berlin, Germany, 2018. [Google Scholar]
- Buttimer, C.; McAuliffe, O.; Ross, R.P.; Hill, C.; O’Mahony, J.; Coffey, A. Bacteriophages and bacterial plant diseases. Front. Microbiol. 2017, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Dy, R.L.; Rigano, L.A.; Fineran, P.C. Phage-based biocontrol strategies and their application in agriculture and aquaculture. Biochem. Soc. Trans. 2018, 46, 1605–1613. [Google Scholar] [CrossRef] [PubMed]
- Pratama, A.A.; van Elsas, J.D. The ‘neglected’ soil virome–potential role and impact. Trends Microbiol. 2018, 26, 649–662. [Google Scholar] [CrossRef] [PubMed]
- Domingo-Calap, P.; Delgado-Martínez, J. Bacteriophages: Protagonists of a post-antibiotic era. Antibiotics 2018, 7, 66. [Google Scholar] [CrossRef]
- Williamson, K.E.; Fuhrmann, J.J.; Wommack, K.E.; Radosevich, M. Viruses in soil ecosystems: An unknown quantity within an unexplored territory. Annu. Rev. Virol. 2017, 4, 201–219. [Google Scholar] [CrossRef] [PubMed]
- Kropinski, A.M.; Van Den Bossche, A.; Lavigne, R.; Noben, J.P.; Babinger, P.; Schmitt, R. Genome and proteome analysis of 7-7-1, a flagellotropic phage infecting Agrobacterium sp H13-3. Virol. J. 2012, 9, 102. [Google Scholar] [CrossRef] [PubMed]
- Attai, H.; Rimbey, J.; Smith, G.P.; Brown, P.J.B. Expression of a peptidoglycan hydrolase from lytic bacteriophages Atu_ph02 and Atu_ph03 triggers lysis of Agrobacterium tumefaciens. Appl. Environ. Microbiol. 2017, 83, e01498-17. [Google Scholar] [CrossRef] [PubMed]
- Attai, H.; Boon, M.; Phillips, K.; Noben, J.-P.; Lavigne, R.; Brown, P.J.B. Larger than life: Isolation and genomic characterization of a jumbo phage that infects the bacterial plant pathogen, Agrobacterium tumefaciens. Front. Microbiol. 2018, 9, 1861. [Google Scholar] [CrossRef] [PubMed]
- Miller, E.S.; Kutter, E.; Mosig, G.; Arisaka, F.; Kunisawa, T.; Ruger, W. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 2003, 67, 86–156. [Google Scholar] [CrossRef] [PubMed]
- Kutter, E.; Bryan, D.; Ray, G.; Brewster, E.; Blasdel, B.; Guttman, B. From host to phage metabolism: Hot tales of phage T4′s takeover of E. coli. Viruses 2018, 10, 387. [Google Scholar] [CrossRef] [PubMed]
- Poindexter, J.S. Biological properties and classification of the Caulobacter group. Bacteriol. Rev. 1964, 28, 231–295. [Google Scholar] [PubMed]
- Watson, B.; Currier, T.C.; Gordon, M.P.; Chilton, M.-D.; Nester, E.W. Plasmid required for virulance of Agrobacterium tumefaciens. J. Bacteriol. 1975, 123, 255–264. [Google Scholar] [PubMed]
- Luo, Z.Q.; Clemente, T.E.; Farrand, S.K. Construction of a derivative of Agrobacterium tumefaciens C58 that does not mutate to tetracycline resistance. Mol. Plant-Microbe Interact. 2001, 14, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Bush, A.L.; Pueppke, S.G. Characterization of an unusual new Agrobacterium tumefaciens strain from Chrysanthemum morifolium ram. Appl. Environ. Microbiol. 1991, 57, 2468–2472. [Google Scholar] [PubMed]
- Slater, S.C.; Goldman, B.S.; Goodner, B.; Setubal, J.C.; Farrand, S.K.; Nester, E.W.; Burr, T.J.; Banta, L.; Dickerman, A.W.; Paulsen, I.; et al. Genome sequences of three Agrobacterium biovars help elucidate the evolution of multichromosome genomes in bacteria. J. Bacteriol. 2009, 191, 2501–2511. [Google Scholar] [CrossRef] [PubMed]
- Nierman, W.C.; Feldblyum, T.V.; Laub, M.T.; Paulsen, I.T.; Nelson, K.E.; Eisen, J.A.; Heidelberg, J.F.; Alley, M.R.; Ohta, N.; Maddock, J.R.; et al. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 2001, 98, 4136–4141. [Google Scholar] [CrossRef]
- Santamaría, R.I.; Bustos, P.; Sepúlveda-Robles, O.; Lozano, L.; Rodríguez, C.; Fernández, J.L.; Juárez, S.; Kameyama, L.; Guarneros, G.; Dávila, G.; et al. Narrow-host-range bacteriophages that infect Rhizobium etli associate with distinct genomic types. Appl. Environ. Microbiol. 2014, 80, 446–454. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.C.E.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, H.W. Phage classification and characterization. Methods Mol. Biol. 2009, 501, 127–140. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.C.; Tatum, K.B.; Lynn, J.S.; Brewer, T.E.; Lu, S.; Washburn, B.K.; Stroupe, M.E.; Jones, K.M. Sinorhizobium meliloti phage phiM9 defines a new group of T4 superfamily phages with unusual genomic features but a common T=16 capsid. J. Virol. 2015, 89, 10945–10958. [Google Scholar] [CrossRef]
- Habann, M.; Leiman, P.G.; Vandersteegen, K.; Van den Bossche, A.; Lavigne, R.; Shneider, M.M.; Bielmann, R.; Eugster, M.R.; Loessner, M.J.; Klumpp, J. Listeria phage A511, a model for the contractile tail machineries of SPO1-related bacteriophages. Mol. Microbiol. 2014, 92, 84–99. [Google Scholar] [CrossRef]
- Nováček, J.; Šiborová, M.; Benešík, M.; Pantůček, R.; Doškař, J.; Plevka, P. Structure and genome release of Twort-like Myoviridae phage with a double-layered baseplate. Proc. Natl. Acad. Sci. USA 2016, 113, 9351–9356. [Google Scholar] [CrossRef] [PubMed]
- Mueser, T.C.; Nossal, N.G.; Hyde, C.C. Structure of bacteriophage T4 RNase H, a 5′ to 3′ RNA-DNA and DNA-DNA exonuclease with sequence similarity to the RAD2 family of eukaryotic proteins. Cell 1996, 85, 1101–1112. [Google Scholar] [CrossRef]
- Kala, S.; Cumby, N.; Sadowski, P.D.; Hyder, B.Z.; Kanelis, V.; Davidson, A.R.; Maxwell, K.L. HNH proteins are a widespread component of phage DNA packaging machines. Proc. Natl. Acad. Sci. USA 2014, 111, 6022–6027. [Google Scholar] [CrossRef] [PubMed]
- McMillan, S.; Edenberg, H.J.; Radany, E.H.; Friedberg, R.C.; Friedberg, E.C. denV gene of bacteriophage T4 codes for both pyrimidine dimer-DNA glycosylase and apyrimidinic endonuclease activities. J. Virol. 1981, 40, 211–223. [Google Scholar] [PubMed]
- Sullivan, M.B.; Coleman, M.L.; Weigele, P.; Rohwer, F.; Chisholm, S.W. Three Prochlorococcus cyanophage genomes: Signature features and ecological interpretations. PLoS Biol. 2005, 3, e144. [Google Scholar] [CrossRef] [PubMed]
- Tanner, J.J.; Boechi, L.; Andrew McCammon, J.; Sobrado, P. Structure, mechanism, and dynamics of UDP-galactopyranose mutase. Arch. Biochem. Biophys. 2014, 544, 128–141. [Google Scholar] [CrossRef]
- Grynberg, M.; Godzik, A. NERD: A DNA processing-related domain present in the anthrax virulence plasmid, pXO1. Trends Biochem. Sci. 2004, 29, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Haakonsen, D.L.; Yuan, A.H.; Laub, M.T. The bacterial cell cycle regulator GcrA is a σ70 cofactor that drives gene expression from a subset of methylated promoters. Genes Dev. 2015, 29, 2272–2286. [Google Scholar] [CrossRef]
- Fioravanti, A.; Fumeaux, C.; Mohapatra, S.S.; Bompard, C.; Brilli, M.; Frandi, A.; Castric, V.; Villeret, V.; Viollier, P.H.; Biondi, E.G. DNA Binding of the cell cycle transcriptional regulator GcrA depends on N6-adenosine methylation in Caulobacter crescentus and other Alphaproteobacteria. PLoS Genet. 2013, 9, e1003541. [Google Scholar] [CrossRef]
- Gill, J.J.; Berry, J.D.; Russell, W.K.; Lessor, L.; Escobar-Garcia, D.A.; Hernandez, D.; Kane, A.; Keene, J.; Maddox, M.; Martin, R.; et al. The Caulobacter crescentus phage phiCbK: Genomics of a canonical phage. BMC Genom. 2012, 13, 542. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed]
- Barragán, M.J.L.; Bláquez, B.; Zamarro, M.T.; Mancheño, J.M.; García, J.L.; Díaz, E.; Carmona, M. BzdR, a repressor that controls the anaerobic catabolism of benzoate in Azoarcus sp. CIB, is the first member of a new subfamily of transcriptional regulators. J. Biol. Chem. 2005, 280, 10683–10694. [Google Scholar] [CrossRef] [PubMed]
- Knight, K.L.; Bowie, J.U.; Vershon, A.K.; Kelley, R.D.; Sauer, R.T.; Vershong, A.K.; Sauer, R.T.; Vershon, A.K.; Kelley, R.D.; Sauer, R.T.; et al. The Arc and Mnt repressors. J. Biol. Chem. 1989, 264, 3639–3642. [Google Scholar] [PubMed]
- Trempy, J.E.; Kirby, J.E.; Gottesman, S. Alp suppression of Lon: Dependence on the slpA gene. J. Bacteriol. 1994, 176, 2061–2067. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hatfull, G.F.; Hendrix, R.W. Bacteriophages and their genomes. Curr. Opin. Virol. 2011, 1, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Gill, J.J.; Summer, E.J.; Russell, W.K.; Cologna, S.M.; Carlile, T.M.; Fuller, A.C.; Kitsopoulos, K.; Mebane, L.M.; Parkinson, B.N.; Sullivan, D.; et al. Genomes and characterization of phages Bcep22 and BcepIL02, founders of a novel phage type in Burkholderia cenocepacia. J. Bacteriol. 2011, 193, 5300–5313. [Google Scholar] [CrossRef] [PubMed]
- Iida, S.; Streiff, M.B.; Bickle, T.A.; Arber, W. Two DNA antirestriction systems of bacteriophage P1, darA, and darB: Characterization of darA-phages. Virology 1987, 157, 156–166. [Google Scholar] [CrossRef]
- Łobocka, M.B.; Rose, D.J.; Plunkett, G.; Rusin, M.; Samojedny, A.; Lehnherr, H.; Yarmolinsky, M.B.; Blattner, F.R. Genome of bacteriophage P1. J. Bacteriol. 2004, 186, 7032–7068. [Google Scholar] [CrossRef] [PubMed]
- Piya, D.; Vara, L.; Russell, W.K.; Young, R.; Gill, J.J. The multicomponent antirestriction system of phage P1 is linked to capsid morphogenesis. Mol. Microbiol. 2017, 105, 399–412. [Google Scholar] [CrossRef] [PubMed]
| Strain or Plasmid | Relevant Characteristics | Growth Medium | Reference or Source |
|---|---|---|---|
| A. tumefaciens strains | |||
| C58 | Nopaline type strain; pTiC58; pAtC58 | LB | [15] |
| EHA105 | C58 derived, succinamopine strain, T-DNA deletion derivative of pTiBo542 | LB | MU plant transformation core |
| EHA101 | C58 derived, nopaline strain, T-DNA deletion derivative of pTiBo542 | LB | MU plant transformation core |
| GV3101 | C58 derived, nopaline strain | LB | MU plant transformation core |
| NTL4 | C58 derived, nopaline-agrocinopine strain, ∆tetRA | LB | [16] |
| AGL-1 | C58 derived, succinamopine strain, T-DNA deletion derivative of pTiBo542 ∆recA | LB | MU plant transformation core |
| LBA4404 | Ach5 derived, octopine strain, T-DNA deletion derivative of pTiAch5 | YM | MU plant transformation core |
| Chry5 | Succinamopine strain, pTiChry5 | LB | [17] |
| Other bacterial strains | |||
| A. vitis S4 | Vitopine strain, pTiS4, pSymA, pSymB | Potato dextrose | [18] |
| Caulobacter crescentus CB15 | Alphaproteobacterium | PYE | [19] |
| Escherichia coli DH5α | Gammaproteobacterium | LB | Life Technologies |
| Strain | Susceptibility to Phage 1 | |
|---|---|---|
| Atu_ph04 | Atu_ph08 | |
| A. tumefaciens C58 | S | S |
| A. tumefaciens EHA105 | S | S |
| A. tumefaciens EHA101 | S | S |
| A. tumefaciens GV3101 | S | S |
| A. tumefaciens NTL4 | S | S |
| A. tumefaciens AGL-1 | R | I |
| A. tumefaciens LBA4404 | I | I |
| A. tumefaciens Chry5 | R | R |
| A. vitis S4 | R | R |
| C. crescentus CB15 | R | R |
| E. coli DH5α | R | R |
| Phage | Genome Length (bp) | G + C content (%) | Number of ORFs | Number of Hypothetical Proteins | Number of ORFs with Predicted Functions | Number of ORFans | Number of tRNAs |
|---|---|---|---|---|---|---|---|
| Atu_ph04 | 143,349 | 49.4 | 223 | 67 | 73 | 83 | 1 |
| Atu_ph08 | 59,034 | 59.7 | 75 | 43 | 32 | 3 | 0 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Attai, H.; Brown, P.J.B. Isolation and Characterization T4- and T7-Like Phages that Infect the Bacterial Plant Pathogen Agrobacterium tumefaciens. Viruses 2019, 11, 528. https://doi.org/10.3390/v11060528
Attai H, Brown PJB. Isolation and Characterization T4- and T7-Like Phages that Infect the Bacterial Plant Pathogen Agrobacterium tumefaciens. Viruses. 2019; 11(6):528. https://doi.org/10.3390/v11060528
Chicago/Turabian StyleAttai, Hedieh, and Pamela J.B. Brown. 2019. "Isolation and Characterization T4- and T7-Like Phages that Infect the Bacterial Plant Pathogen Agrobacterium tumefaciens" Viruses 11, no. 6: 528. https://doi.org/10.3390/v11060528
APA StyleAttai, H., & Brown, P. J. B. (2019). Isolation and Characterization T4- and T7-Like Phages that Infect the Bacterial Plant Pathogen Agrobacterium tumefaciens. Viruses, 11(6), 528. https://doi.org/10.3390/v11060528




