Species-Specific Conservation of Linear Antigenic Sites on Vaccinia Virus A27 Protein Homologs of Orthopoxviruses
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
2.2. Polyclonal and Monoclonal Antibodies
2.3. Plaque Reduction Test
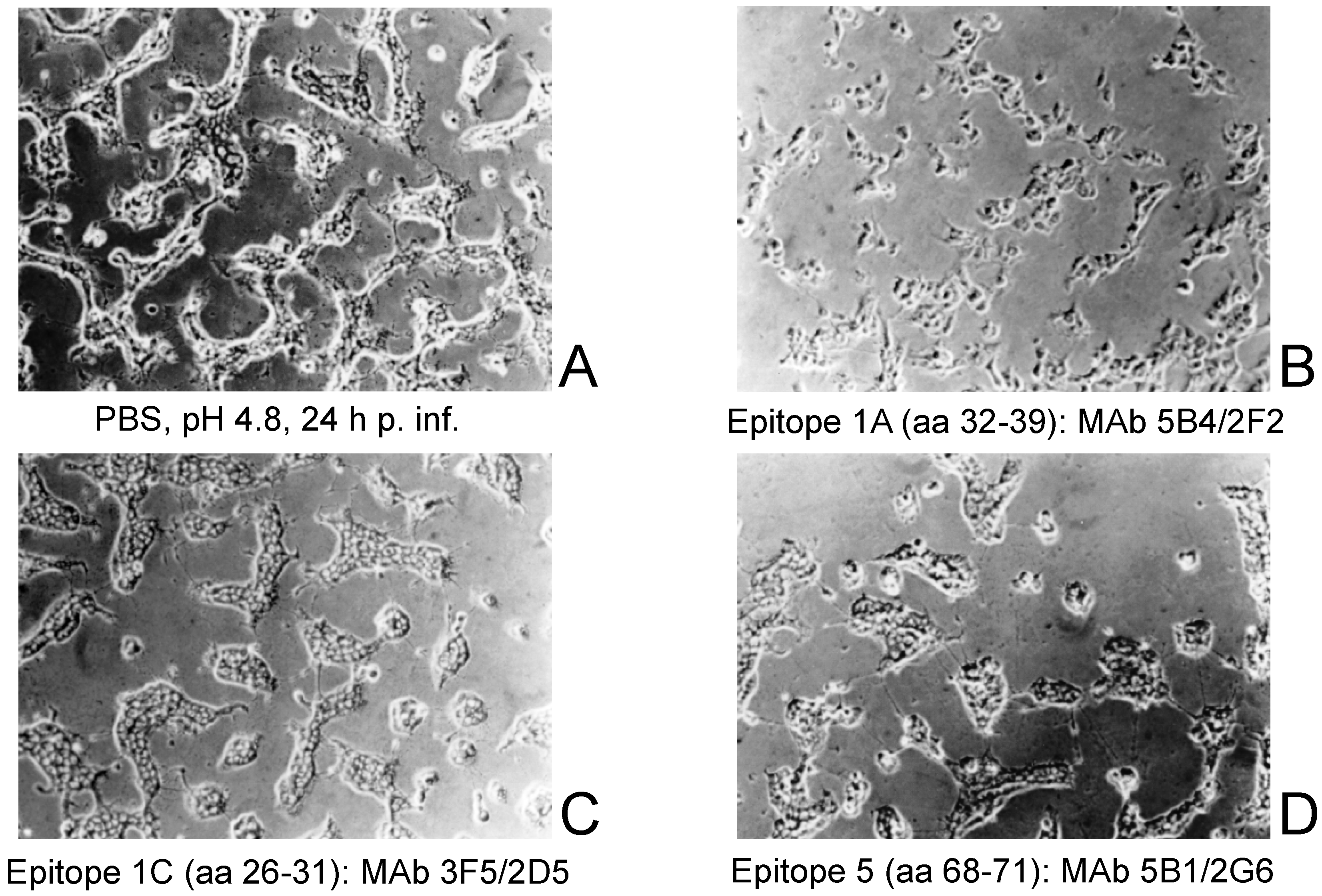
2.4. Inhibition of Cell Fusion and Syncytium Formation
2.5. Binding Affinities of the Monoclonal Antibodies (mAbs) in Indirect Enzyme-Linked Immunosorbent Assays (ELISAs)
2.6. Epitope Mapping by SPOT Synthesis on Cellulose Membranes
2.7. Epitope Mapping by Microarray Scanning Chips
2.8. DataBase Analysis of A27 Protein Sequences and Orthopoxvirus (OPXV) Phylogeny
3. Results
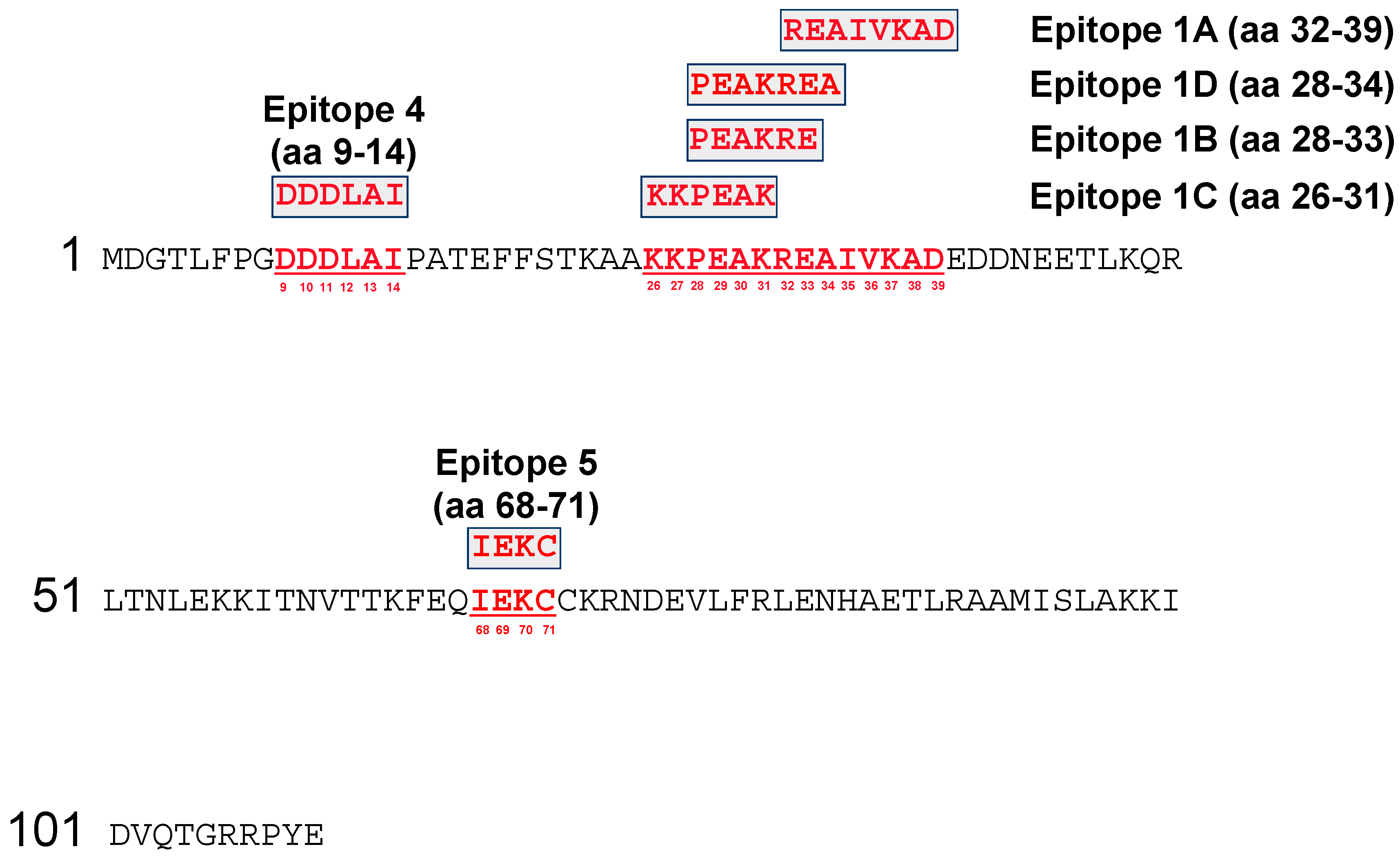
3.1. Fine Mapping of the Vaccinia Virus (VACV) A27 Epitopes by SPOTs Membrane
3.2. Fine Mapping of the VACV A27 Epitopes by Microarray Analysis
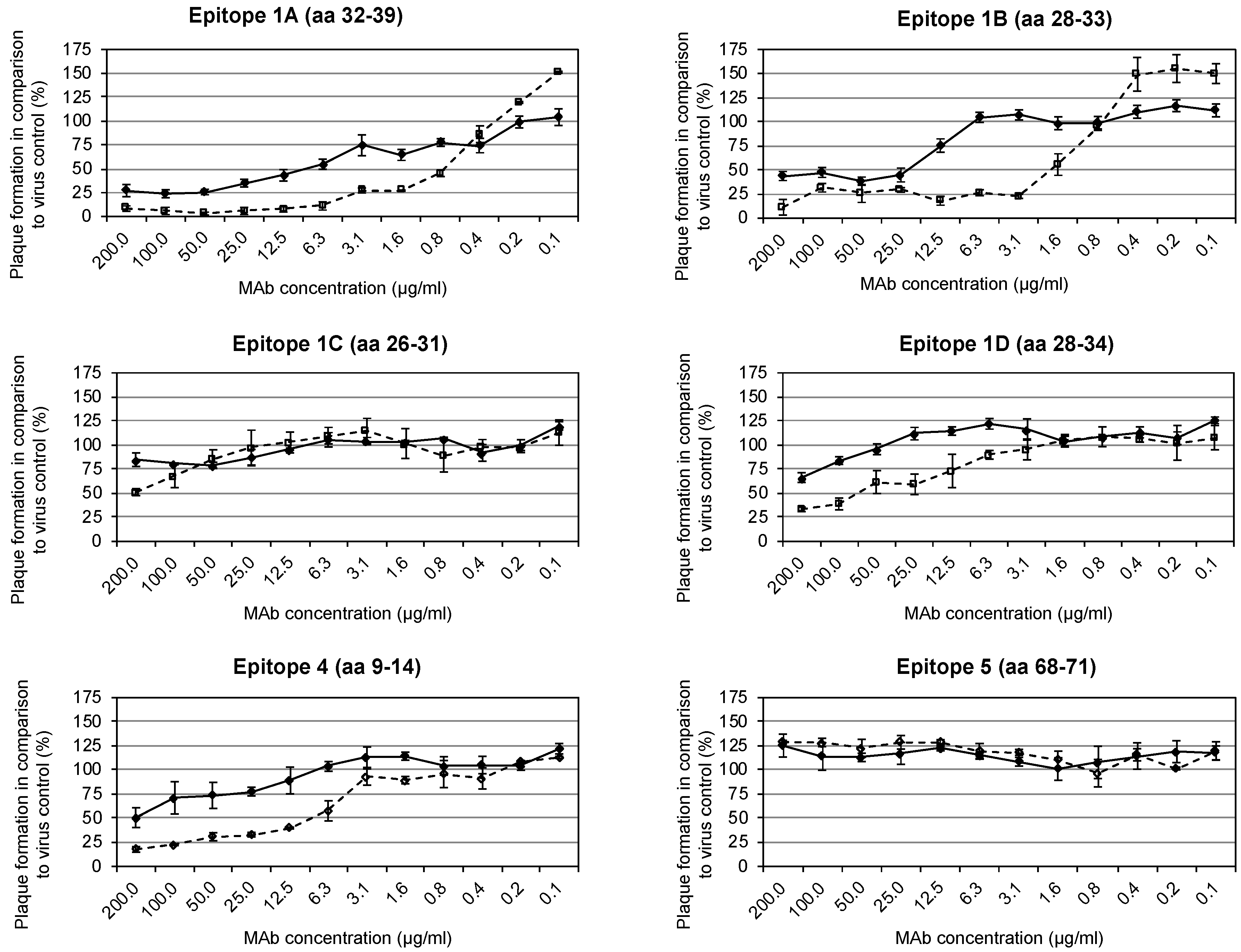
3.3. Identification of Neutralization-Mediating Epitopes with/without Complement
3.4. Inhibition of Cell Fusion
3.5. Binding Affinities of the mAbs to Various Variants of OPXVs
3.6. Species-Specific Epitope Conservation and Variation among the OPXV Members
3.7. Silent Mutations in the Epitope Sequences among the OPXV Members
4. Discussion
Supplementary Materials

Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Condit, R.C.; Moussatche, N.; Traktman, P. In a nutshell: Structure and assembly of the vaccinia virion. Adv. Virus Res. 2006, 66, 31–124. [Google Scholar] [CrossRef]
- Kurth, A.; Wibbelt, G.; Gerber, H.P.; Petschaelis, A.; Pauli, G.; Nitsche, A. Rat-to-elephant-to-human transmission of cowpox virus. Emerg. Infect. Dis. 2008, 14, 670–671. [Google Scholar] [CrossRef]
- Fenner, F.; Henderson, D.A.; Arita, I.; Jezek, Z.; Ladnyi, I.D. Smallpox and Its Eradication; World Health Organization: Geneva, Swithzerland, 1988; pp. 1–1460. [Google Scholar]
- Becker, C.; Kurth, A.; Hessler, F.; Kramp, H.; Gokel, M.; Hoffmann, R.; Kuczka, A.; Nitsche, A. Cowpox virus infection in pet rat owners: Not always immediately recognized. Dtsch. Arztebl. Int. 2009, 106, 329–334. [Google Scholar] [CrossRef]
- Novembre, F.J.; Raska, K., Jr.; Holowczak, J.A. The immune response to vaccinia virus infection in mice: Analysis of the role of antibody. Arch. Virol. 1989, 107, 273–289. [Google Scholar] [CrossRef]
- Henderson, D.A. The looming threat of bioterrorism. Science 1999, 283, 1279–1282. [Google Scholar] [CrossRef]
- Rimoin, A.W.; Mulembakani, P.M.; Johnston, S.C.; Lloyd Smith, J.O.; Kisalu, N.K.; Kinkela, T.L.; Blumberg, S.; Thomassen, H.A.; Pike, B.L.; Fair, J.N.; et al. Major increase in human monkeypox incidence 30 years after smallpox vaccination campaigns cease in the Democratic Republic of Congo. Proc. Natl. Acad. Sci. USA 2010, 107, 16262–16267. [Google Scholar] [CrossRef]
- Ladnyj, I.D.; Ziegler, P.; Kima, E. A human infection caused by monkeypox virus in Basankusu Territory, Democratic Republic of the Congo. Bull. World Health Organ. 1972, 46, 593–597. [Google Scholar]
- Reed, K.D.; Melski, J.W.; Graham, M.B.; Regnery, R.L.; Sotir, M.J.; Wegner, M.V.; Kazmierczak, J.J.; Stratman, E.J.; Li, Y.; Fairley, J.A.; et al. The detection of monkeypox in humans in the Western Hemisphere. N. Engl. J. Med. 2004, 350, 342–350. [Google Scholar] [CrossRef]
- Vaughan, A.; Aarons, E.; Astbury, J.; Balasegaram, S.; Beadsworth, M.; Beck, C.R.; Chand, M.; O’Connor, C.; Dunning, J.; Ghebrehewet, S.; et al. Two cases of monkeypox imported to the United Kingdom, September 2018. Euro. Surveill. 2018, 23. [Google Scholar] [CrossRef]
- WHO. Weekly Bulletin On Outbreaks And Other Emergencies. Available online: http://apps.who.int/iris/bitstream/10665/259352/1/OEW42-1420102017.pdf?ua=1 (accessed on 28 May 2019).
- Vorou, R.M.; Papavassiliou, V.G.; Pierroutsakos, I.N. Cowpox virus infection: An emerging health threat. Curr. Opin. Infect. Dis. 2008, 21, 153–156. [Google Scholar] [CrossRef]
- Campe, H.; Zimmermann, P.; Glos, K.; Bayer, M.; Bergemann, H.; Dreweck, C.; Graf, P.; Weber, B.K.; Meyer, H.; Buttner, M.; et al. Cowpox virus transmission from pet rats to humans, Germany. Emerg. Infect. Dis. 2009, 15, 777–780. [Google Scholar] [CrossRef]
- Howard, A.R.; Senkevich, T.G.; Moss, B. Vaccinia virus A26 and A27 proteins form a stable complex tethered to mature virions by association with the A17 transmembrane protein. J. Virol. 2008, 82, 12384–12391. [Google Scholar] [CrossRef]
- Vogel, S.; Sardy, M.; Glos, K.; Korting, H.C.; Ruzicka, T.; Wollenberg, A. The Munich outbreak of cutaneous cowpox infection: Transmission by infected pet rats. Acta. Derm. Venereol. 2012, 92, 126–131. [Google Scholar] [CrossRef]
- Redfield, R.R.; Wright, D.C.; James, W.D.; Jones, T.S.; Brown, C.; Burke, D.S. Disseminated vaccinia in a military recruit with human immunodeficiency virus (HIV) disease. N. Engl. J. Med. 1987, 316, 673–676. [Google Scholar] [CrossRef] [PubMed]
- Eis-Hubinger, A.M.; Gerritzen, A.; Schneweis, K.E.; Pfeiff, B.; Pullmann, H.; Mayr, A.; Czerny, C.P. Fatal cowpox-like virus infection transmitted by cat. Lancet 1990, 336, 880. [Google Scholar] [CrossRef]
- Czerny, C.P.; Eis-Hubinger, A.M.; Mayr, A.; Schneweis, K.E.; Pfeiff, B. Animal poxviruses transmitted from cat to man: Current event with lethal end. Zent. Vet. B 1991, 38, 421–431. [Google Scholar] [CrossRef]
- Fassbender, P.; Zange, S.; Ibrahim, S.; Zoeller, G.; Herbstreit, F.; Meyer, H. Generalized Cowpox Virus Infection in a Patient with HIV, Germany, 2012. Emerg. Infect. Dis. 2016, 22, 553–555. [Google Scholar] [CrossRef]
- Kinnunen, P.M.; Holopainen, J.M.; Hemmila, H.; Piiparinen, H.; Sironen, T.; Kivela, T.; Virtanen, J.; Niemimaa, J.; Nikkari, S.; Jarvinen, A.; et al. Severe Ocular Cowpox in a Human, Finland. Emerg. Infect. Dis. 2015, 21, 2261–2263. [Google Scholar] [CrossRef]
- Moss, B. Poxvirus entry and membrane fusion. Virology 2006, 344, 48–54. [Google Scholar] [CrossRef]
- Smith, G.L.; Vanderplasschen, A.; Law, M. The formation and function of extracellular enveloped vaccinia virus. J. Gen. Virol. 2002, 83, 2915–2931. [Google Scholar] [CrossRef]
- Ulaeto, D.; Grosenbach, D.; Hruby, D.E. The vaccinia virus 4c and A-type inclusion proteins are specific markers for the intracellular mature virus particle. J. Virol. 1996, 70, 3372–3377. [Google Scholar]
- Blasco, R.; Moss, B. Role of cell-associated enveloped vaccinia virus in cell-to-cell spread. J. Virol. 1992, 66, 4170–4179. [Google Scholar]
- Smith, G.L.; Murphy, B.J.; Law, M. Vaccinia virus motility. Annu. Rev. Microbiol. 2003, 57, 323–342. [Google Scholar] [CrossRef]
- Appleyard, G.; Hapel, A.J.; Boulter, E.A. An antigenic difference between intracellular and extracellular rabbitpox virus. J. Gen. Virol. 1971, 13, 9–17. [Google Scholar] [CrossRef]
- Boulter, E.A.; Appleyard, G. Differences between extracellular and intracellular forms of poxvirus and their implications. Prog. Med. Virol. 1973, 16, 86–108. [Google Scholar]
- Payne, L. Polypeptide composition of extracellular enveloped vaccinia virus. J. Virol. 1978, 27, 28–37. [Google Scholar]
- Payne, L.G. Significance of extracellular enveloped virus in the in vitro and in vivo dissemination of vaccinia. J. Gen. Virol. 1980, 50, 89–100. [Google Scholar] [CrossRef]
- Isaacs, S.N.; Wolffe, E.J.; Payne, L.G.; Moss, B. Characterization of a vaccinia virus-encoded 42-kilodalton class I membrane glycoprotein component of the extracellular virus envelope. J. Virol. 1992, 66, 7217–7224. [Google Scholar]
- Roper, R.L.; Wolffe, E.J.; Weisberg, A.; Moss, B. The envelope protein encoded by the A33R gene is required for formation of actin-containing microvilli and efficient cell-to-cell spread of vaccinia virus. J. Virol. 1998, 72, 4192–4204. [Google Scholar]
- Rodriguez, J.F.; Janeczko, R.; Esteban, M. Isolation and characterization of neutralizing monoclonal antibodies to vaccinia virus. J. Virol. 1985, 56, 482–488. [Google Scholar]
- Aldaz-Carroll, L.; Whitbeck, J.C.; Ponce de Leon, M.; Lou, H.; Hirao, L.; Isaacs, S.N.; Moss, B.; Eisenberg, R.J.; Cohen, G.H. Epitope-mapping studies define two major neutralization sites on the vaccinia virus extracellular enveloped virus glycoprotein B5R. J. Virol. 2005, 79, 6260–6271. [Google Scholar] [CrossRef]
- Hsiao, J.C.; Chung, C.S.; Chang, W. Cell surface proteoglycans are necessary for A27L protein-mediated cell fusion: Identification of the N-terminal region of A27L protein as the glycosaminoglycan-binding domain. J. Virol. 1998, 72, 8374–8379. [Google Scholar]
- Hsiao, J.C.; Chung, C.S.; Chang, W. Vaccinia virus envelope D8L protein binds to cell surface chondroitin sulfate and mediates the adsorption of intracellular mature virions to cells. J. Virol. 1999, 73, 8750–8761. [Google Scholar]
- Moss, B. Membrane fusion during poxvirus entry. Semin. Cell. Dev. Biol. 2016, 60, 89–96. [Google Scholar] [CrossRef]
- Moss, B. Poxvirus cell entry: How many proteins does it take? Viruses 2012, 4, 688–707. [Google Scholar] [CrossRef]
- Matho, M.H.; Schlossman, A.; Meng, X.; Benhnia, M.R.; Kaever, T.; Buller, M.; Doronin, K.; Parker, S.; Peters, B.; Crotty, S.; et al. Structural and Functional Characterization of Anti-A33 Antibodies Reveal a Potent Cross-Species Orthopoxviruses Neutralizer. PLoS Pathog. 2015, 11, e1005148. [Google Scholar] [CrossRef]
- Czerny, C.P.; Mahnel, H. Structural and functional analysis of orthopoxvirus epitopes with neutralizing monoclonal antibodies. J. Gen. Virol. 1990, 71 (Pt 10), 2341–2352. [Google Scholar] [CrossRef]
- Czerny, C.P.; Johann, S.; Holzle, L.; Meyer, H. Epitope detection in the envelope of intracellular naked orthopox viruses and identification of encoding genes. Virology 1994, 200, 764–777. [Google Scholar] [CrossRef]
- Matho, M.H.; Maybeno, M.; Benhnia, M.R.; Becker, D.; Meng, X.; Xiang, Y.; Crotty, S.; Peters, B.; Zajonc, D.M. Structural and biochemical characterization of the vaccinia virus envelope protein D8 and its recognition by the antibody LA5. J. Virol. 2012, 86, 8050–8058. [Google Scholar] [CrossRef]
- Kaever, T.; Matho, M.H.; Meng, X.; Crickard, L.; Schlossman, A.; Xiang, Y.; Crotty, S.; Peters, B.; Zajonc, D.M. Linear Epitopes in Vaccinia Virus A27 Are Targets of Protective Antibodies Induced by Vaccination against Smallpox. J. Virol. 2016, 90, 4334–4345. [Google Scholar] [CrossRef]
- Hooper, J.W.; Custer, D.M.; Schmaljohn, C.S.; Schmaljohn, A.L. DNA vaccination with vaccinia virus L1R and A33R genes protects mice against a lethal poxvirus challenge. Virology 2000, 266, 329–339. [Google Scholar] [CrossRef]
- Galmiche, M.C.; Goenaga, J.; Wittek, R.; Rindisbacher, L. Neutralizing and protective antibodies directed against vaccinia virus envelope antigens. Virology 1999, 254, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Benhnia, M.R.; McCausland, M.M.; Moyron, J.; Laudenslager, J.; Granger, S.; Rickert, S.; Koriazova, L.; Kubo, R.; Kato, S.; Crotty, S. Vaccinia virus extracellular enveloped virion neutralization in vitro and protection in vivo depend on complement. J. Virol. 2009, 83, 1201–1215. [Google Scholar] [CrossRef] [PubMed]
- McCausland, M.M.; Benhnia, M.R.; Crickard, L.; Laudenslager, J.; Granger, S.W.; Tahara, T.; Kubo, R.; Koriazova, L.; Kato, S.; Crotty, S. Combination therapy of vaccinia virus infection with human anti-H3 and anti-B5 monoclonal antibodies in a small animal model. Antivir. Ther. 2010, 15, 661–675. [Google Scholar] [CrossRef]
- Ramirez, J.C.; Tapia, E.; Esteban, M. Administration to mice of a monoclonal antibody that neutralizes the intracellular mature virus form of vaccinia virus limits virus replication efficiently under prophylactic and therapeutic conditions. J. Gen. Virol. 2002, 83, 1059–1067. [Google Scholar] [CrossRef]
- Lai, C.F.; Gong, S.C.; Esteban, M. The purified 14-kilodalton envelope protein of vaccinia virus produced in Escherichia coli induces virus immunity in animals. J. Virol. 1991, 65, 5631–5635. [Google Scholar]
- Ichihashi, Y.; Oie, M. Neutralizing epitope on penetration protein of vaccinia virus. Virology 1996, 220, 491–494. [Google Scholar] [CrossRef]
- Pulford, D.J.; Gates, A.; Bridge, S.H.; Robinson, J.H.; Ulaeto, D. Differential efficacy of vaccinia virus envelope proteins administered by DNA immunisation in protection of BALB/c mice from a lethal intranasal poxvirus challenge. Vaccine 2004, 22, 3358–3366. [Google Scholar] [CrossRef]
- Reeman, S.; Gates, A.J.; Pulford, D.J.; Krieg, A.; Ulaeto, D.O. Protection of Mice from Lethal Vaccinia Virus Infection by Vaccinia Virus Protein Subunits with a CpG Adjuvant. Viruses 2017, 9, 378. [Google Scholar] [CrossRef]
- Rodriguez, J.F.; Esteban, M. Mapping and nucleotide sequence of the vaccinia virus gene that encodes a 14-kilodalton fusion protein. J. Virol. 1987, 61, 3550–3554. [Google Scholar]
- Gong, S.C.; Lai, C.F.; Dallo, S.; Esteban, M. A single point mutation of Ala-25 to Asp in the 14,000-Mr envelope protein of vaccinia virus induces a size change that leads to the small plaque size phenotype of the virus. J. Virol. 1989, 63, 4507–4514. [Google Scholar]
- Gong, S.C.; Lai, C.F.; Esteban, M. Vaccinia virus induces cell fusion at acid pH and this activity is mediated by the N-terminus of the 14-kDa virus envelope protein. Virology 1990, 178, 81–91. [Google Scholar] [CrossRef]
- Lai, C.F.; Gong, S.C.; Esteban, M. Structural and functional properties of the 14-kDa envelope protein of vaccinia virus synthesized in Escherichia coli. J. Biol. Chem. 1990, 265, 22174–22180. [Google Scholar]
- Goebel, S.J.; Johnson, G.P.; Perkus, M.E.; Davis, S.W.; Winslow, J.P.; Paoletti, E. The complete DNA sequence of vaccinia virus. Virology 1990, 179, 247–266, 517–563. [Google Scholar] [CrossRef]
- Shih, P.C.; Yang, M.S.; Lin, S.C.; Ho, Y.; Hsiao, J.C.; Wang, D.R.; Yu, S.S.; Chang, W.; Tzou, D.L. A turn-like structure "KKPE" segment mediates the specific binding of viral protein A27 to heparin and heparan sulfate on cell surfaces. J. Biol. Chem. 2009, 284, 36535–36546. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, D.; Rodriguez, J.R.; Esteban, M. The vaccinia virus 14-kilodalton fusion protein forms a stable complex with the processed protein encoded by the vaccinia virus A17L gene. J. Virol. 1993, 67, 3435–3440. [Google Scholar]
- Vazquez, M.I.; Rivas, G.; Cregut, D.; Serrano, L.; Esteban, M. The vaccinia virus 14-kilodalton (A27L) fusion protein forms a triple coiled-coil structure and interacts with the 21-kilodalton (A17L) virus membrane protein through a C-terminal alpha-helix. J. Virol. 1998, 72, 10126–10137. [Google Scholar]
- Unger, B.; Mercer, J.; Boyle, K.A.; Traktman, P. Biogenesis of the vaccinia virus membrane: Genetic and ultrastructural analysis of the contributions of the A14 and A17 proteins. J. Virol. 2013, 87, 1083–1097. [Google Scholar] [CrossRef]
- Chang, T.H.; Chang, S.J.; Hsieh, F.L.; Ko, T.P.; Lin, C.T.; Ho, M.R.; Wang, I.; Hsu, S.T.; Guo, R.T.; Chang, W.; et al. Crystal structure of vaccinia viral A27 protein reveals a novel structure critical for its function and complex formation with A26 protein. PLoS Pathog. 2013, 9, e1003563. [Google Scholar] [CrossRef]
- Chung, C.S.; Hsiao, J.C.; Chang, Y.S.; Chang, W. A27L protein mediates vaccinia virus interaction with cell surface heparan sulfate. J. Virol. 1998, 72, 1577–1585. [Google Scholar] [PubMed]
- Ching, Y.C.; Chung, C.S.; Huang, C.Y.; Hsia, Y.; Tang, Y.L.; Chang, W. Disulfide bond formation at the C termini of vaccinia virus A26 and A27 proteins does not require viral redox enzymes and suppresses glycosaminoglycan-mediated cell fusion. J. Virol. 2009, 83, 6464–6476. [Google Scholar] [CrossRef]
- Wang, D.R.; Hsiao, J.C.; Wong, C.H.; Li, G.C.; Lin, S.C.; Yu, S.S.; Chen, W.; Chang, W.; Tzou, D.L. Vaccinia viral protein A27 is anchored to the viral membrane via a cooperative interaction with viral membrane protein A17. J. Biol. Chem. 2014, 289, 6639–6655. [Google Scholar] [CrossRef] [PubMed]
- Kochan, G.; Escors, D.; Gonzalez, J.M.; Casasnovas, J.M.; Esteban, M. Membrane cell fusion activity of the vaccinia virus A17-A27 protein complex. Cell Microbiol. 2008, 10, 149–164. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rodriguez, J.F.; Smith, G.L. IPTG-dependent vaccinia virus: Identification of a virus protein enabling virion envelopment by Golgi membrane and egress. Nucleic Acids Res. 1990, 18, 5347–5351. [Google Scholar] [CrossRef]
- Roper, R.L. Characterization of the vaccinia virus A35R protein and its role in virulence. J. Virol. 2006, 80, 306–313. [Google Scholar] [CrossRef]
- Joklik, W.K. The purification fo four strains of poxvirus. Virology 1962, 18, 9–18. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [PubMed]
- Wolffe, E.J.; Isaacs, S.N.; Moss, B. Deletion of the vaccinia virus B5R gene encoding a 42-kilodalton membrane glycoprotein inhibits extracellular virus envelope formation and dissemination. J. Virol. 1993, 67, 4732–4741. [Google Scholar]
- Michaelis, L.; Menten, M.L. Die Kinetik der Invertinwirkung. Biochem. Z. 1913, 49, 333–369. [Google Scholar]
- Michaelis, L.; Menten, M.L.; Johnson, K.A.; Goody, R.S. The original Michaelis constant: Translation of the 1913 Michaelis-Menten paper. Biochemistry 2011, 50, 8264–8269. [Google Scholar] [CrossRef]
- Frank, R. Spot-Synthesis—an Easy Technique for the Positionally Addressable, Parallel Chemical Synthesis on a Membrane Support. Tetrahedron 1992, 48, 9217–9232. [Google Scholar] [CrossRef]
- Dikmans, A.; Beutling, U.; Schmeisser, E.; Thiele, S.; Frank, R. SC2: A novel process for manufacturing multipurpose high-density chemical microarrays. Qsar Comb Sci. 2006, 25, 1069–1080. [Google Scholar] [CrossRef]
- Hotop, S.K.; Abd El Wahed, A.; Beutling, U.; Jentsch, D.; Motzkus, D.; Frank, R.; Hunsmann, G.; Stahl-Hennig, C.; Fritz, H.J. Multiple antibody targets on herpes B glycoproteins B and D identified by screening sera of infected rhesus macaques with peptide microarrays. PLoS ONE 2014, 9, e86857. [Google Scholar] [CrossRef][Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007, 35, D61–65. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Rodriguez, J.F.; Paez, E.; Esteban, M. A 14,000-Mr envelope protein of vaccinia virus is involved in cell fusion and forms covalently linked trimers. J. Virol. 1987, 61, 395–404. [Google Scholar]
- Moss, B. Smallpox vaccines: Targets of protective immunity. Immunol. Rev. 2011, 239, 8–26. [Google Scholar] [CrossRef]
- Diesterbeck, U.S.; Gittis, A.G.; Garboczi, D.N.; Moss, B. The 2.1 A structure of protein F9 and its comparison to L1, two components of the conserved poxvirus entry-fusion complex. Sci. Rep. 2018, 8, 16807. [Google Scholar] [CrossRef] [PubMed]
- Senkevich, T.G.; Ojeda, S.; Townsley, A.; Nelson, G.E.; Moss, B. Poxvirus multiprotein entry-fusion complex. Proc. Natl. Acad. Sci. USA 2005, 102, 18572–18577. [Google Scholar] [CrossRef]
- Abd El Wahed, A. Use of peptide microarrays for mapping viral b cell epitopes. Ph.D. Thesis, Georg-August-Universität Göttingen, Göttingen, Germany, March 2011. [Google Scholar]
- Garnier, J.; Osguthorpe, D.J.; Robson, B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J. Mol. Biol. 1978, 120, 97–120. [Google Scholar] [CrossRef]
- Chou, P.Y.; Fasman, G.D. Prediction of beta-turns. Biophys. J. 1979, 26, 367–383. [Google Scholar] [CrossRef]
- Fanning, D.W.; Smith, J.A.; Rose, G.D. Molecular cartography of globular proteins with application to antigenic sites. Biopolymers 1986, 25, 863–883. [Google Scholar] [CrossRef] [PubMed]
- Novotny, J.; Handschumacher, M.; Haber, E. Location of antigenic epitopes on antibody molecules. J. Mol. Biol. 1986, 189, 715–721. [Google Scholar] [CrossRef]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Emini, E.A.; Hughes, J.V.; Perlow, D.S.; Boger, J. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J. Virol. 1985, 55, 836–839. [Google Scholar] [PubMed]
- Vazquez, M.I.; Esteban, M. Identification of functional domains in the 14-kilodalton envelope protein (A27L) of vaccinia virus. J. Virol. 1999, 73, 9098–9109. [Google Scholar]
- Meyer, H.; Osterrieder, N.; Czerny, C.P. Identification of binding sites for neutralizing monoclonal antibodies on the 14-kDa fusion protein of orthopox viruses. Virology 1994, 200, 778–783. [Google Scholar] [CrossRef]
- Massung, R.F.; Liu, L.I.; Qi, J.; Knight, J.C.; Yuran, T.E.; Kerlavage, A.R.; Parsons, J.M.; Venter, J.C.; Esposito, J.J. Analysis of the complete genome of smallpox variola major virus strain Bangladesh-1975. Virology 1994, 201, 215–240. [Google Scholar] [CrossRef]
- Franke, A.; Pfaff, F.; Jenckel, M.; Hoffmann, B.; Hoper, D.; Antwerpen, M.; Meyer, H.; Beer, M.; Hoffmann, D. Classification of Cowpox Viruses into Several Distinct Clades and Identification of a Novel Lineage. Viruses 2017, 9, 142. [Google Scholar] [CrossRef]
- Dabrowski, P.W.; Radonic, A.; Kurth, A.; Nitsche, A. Genome-wide comparison of cowpox viruses reveals a new clade related to Variola virus. PLoS ONE 2013, 8, e79953. [Google Scholar] [CrossRef]
- Carroll, D.S.; Emerson, G.L.; Li, Y.; Sammons, S.; Olson, V.; Frace, M.; Nakazawa, Y.; Czerny, C.P.; Tryland, M.; Kolodziejek, J.; et al. Chasing Jenner’s vaccine: Revisiting cowpox virus classification. PLoS ONE 2011, 6, e23086. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wharton, S.A.; Weissenhorn, W.; Calder, L.J.; Hughson, F.M.; Skehel, J.J.; Wiley, D.C. A soluble domain of the membrane-anchoring chain of influenza virus hemagglutinin (HA2) folds in Escherichia coli into the low-pH-induced conformation. Proc. Natl. Acad. Sci. USA 1995, 92, 12205–12209. [Google Scholar] [CrossRef] [PubMed]
- Dessen, A.; Volchkov, V.; Dolnik, O.; Klenk, H.D.; Weissenhorn, W. Crystal structure of the matrix protein VP40 from Ebola virus. EMBO J. 2000, 19, 4228–4236. [Google Scholar] [CrossRef]
- Weissenhorn, W.; Calder, L.J.; Wharton, S.A.; Skehel, J.J.; Wiley, D.C. The central structural feature of the membrane fusion protein subunit from the Ebola virus glycoprotein is a long triple-stranded coiled coil. Proc. Natl. Acad. Sci. USA 1998, 95, 6032–6036. [Google Scholar] [CrossRef]
- Skehel, J.J.; Wiley, D.C. Coiled coils in both intracellular vesicle and viral membrane fusion. Cell 1998, 95, 871–874. [Google Scholar] [CrossRef]
- Doms, R.W.; Blumenthal, R.; Moss, B. Fusion of intra- and extracellular forms of vaccinia virus with the cell membrane. J. Virol. 1990, 64, 4884–4892. [Google Scholar] [PubMed]
| Epitope | Position (aa) | MAb | Virus Strain Used for mAb Production | Isotype | Neutralization without Complement (µg/mL) | Neutralization with 1% Complement (µg/mL) |
|---|---|---|---|---|---|---|
| 1A | 32–39 | 5B4/2F2 | VACV MVA | IgG2a | 12.5 | 1.6 |
| 1B | 28–33 | 2C11/1B4 | VACV MVA | IgG2b | 25.0 | 3.1 |
| 1C | 26–31 | 3F5/2D5 | CPXV KR2 Brighton | IgG1 | - | 200.0 |
| 1D | 28–34 | 1D5/2D11 | CPXV KR2 Brighton | IgG1 | - | 100.0 |
| 4 | 9–14 | 2G8/1E4 | ECTV Munich 1 | IgG3 | 200.0 | 12.5 |
| 5 | 68–71 | 5B1/2G6 | ECTV Munich 1 | IgG2a | - | - |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahsendorf, H.P.; Gan, L.L.; Eltom, K.H.; Abd El Wahed, A.; Hotop, S.-K.; Roper, R.L.; Beutling, U.; Broenstrup, M.; Stahl-Hennig, C.; Hoelzle, L.E.; et al. Species-Specific Conservation of Linear Antigenic Sites on Vaccinia Virus A27 Protein Homologs of Orthopoxviruses. Viruses 2019, 11, 493. https://doi.org/10.3390/v11060493
Ahsendorf HP, Gan LL, Eltom KH, Abd El Wahed A, Hotop S-K, Roper RL, Beutling U, Broenstrup M, Stahl-Hennig C, Hoelzle LE, et al. Species-Specific Conservation of Linear Antigenic Sites on Vaccinia Virus A27 Protein Homologs of Orthopoxviruses. Viruses. 2019; 11(6):493. https://doi.org/10.3390/v11060493
Chicago/Turabian StyleAhsendorf, Henrike P., Li L. Gan, Kamal H. Eltom, Ahmed Abd El Wahed, Sven-Kevin Hotop, Rachel L. Roper, Ulrike Beutling, Mark Broenstrup, Christiane Stahl-Hennig, Ludwig E. Hoelzle, and et al. 2019. "Species-Specific Conservation of Linear Antigenic Sites on Vaccinia Virus A27 Protein Homologs of Orthopoxviruses" Viruses 11, no. 6: 493. https://doi.org/10.3390/v11060493
APA StyleAhsendorf, H. P., Gan, L. L., Eltom, K. H., Abd El Wahed, A., Hotop, S.-K., Roper, R. L., Beutling, U., Broenstrup, M., Stahl-Hennig, C., Hoelzle, L. E., & Czerny, C.-P. (2019). Species-Specific Conservation of Linear Antigenic Sites on Vaccinia Virus A27 Protein Homologs of Orthopoxviruses. Viruses, 11(6), 493. https://doi.org/10.3390/v11060493









