Targeting Human Parainfluenza Virus Type-1 Haemagglutinin-Neuraminidase with Mechanism-Based Inhibitors
Abstract
1. Introduction
2. Material and Methods
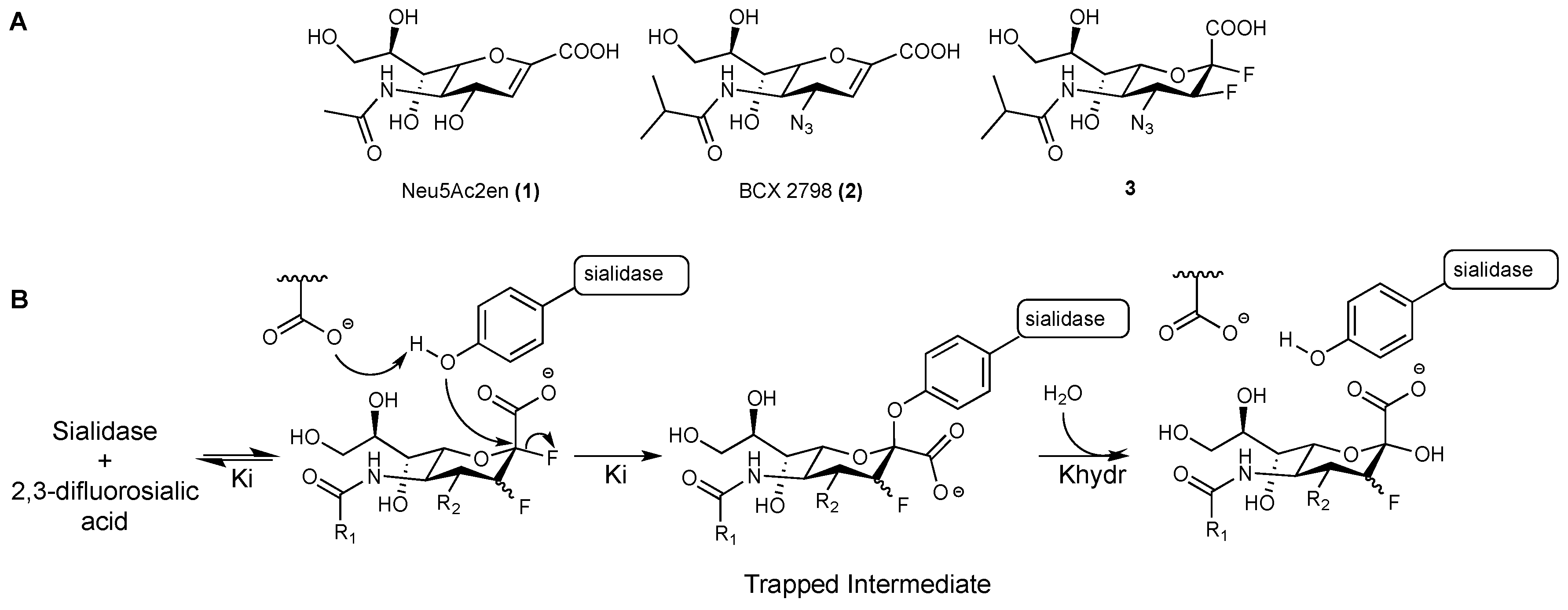
2.1. hPIV-1 HN Inhibitors
2.2. Cells and Viruses
2.3. Neuraminidase Inhibition Assay
2.4. Haemagglutination Inhibition Assay
2.5. In Situ ELISA
2.6. Recombinant hPIV-1 HN Expression and Purification
2.7. 1H NMR Experiments
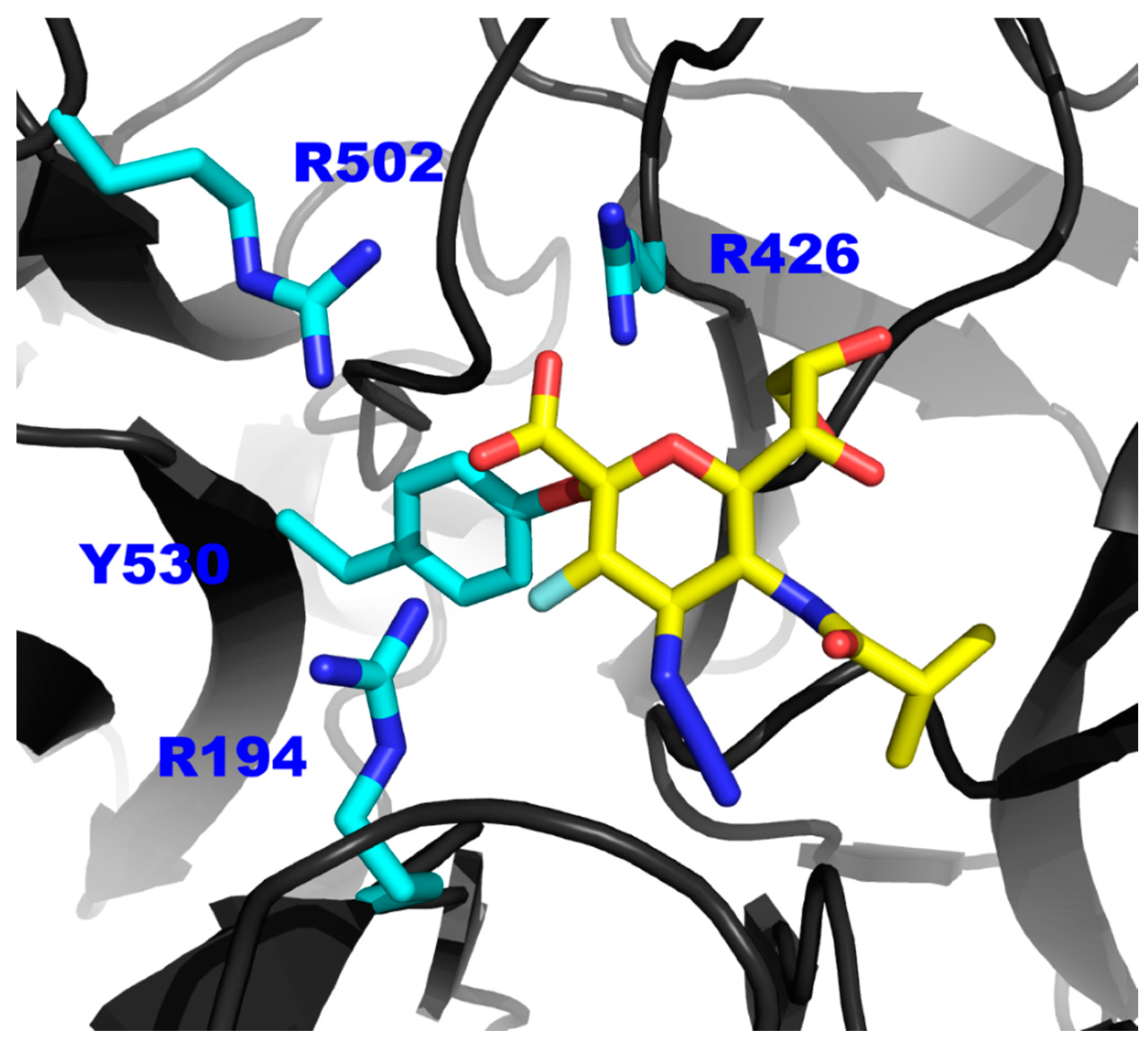
3. Results
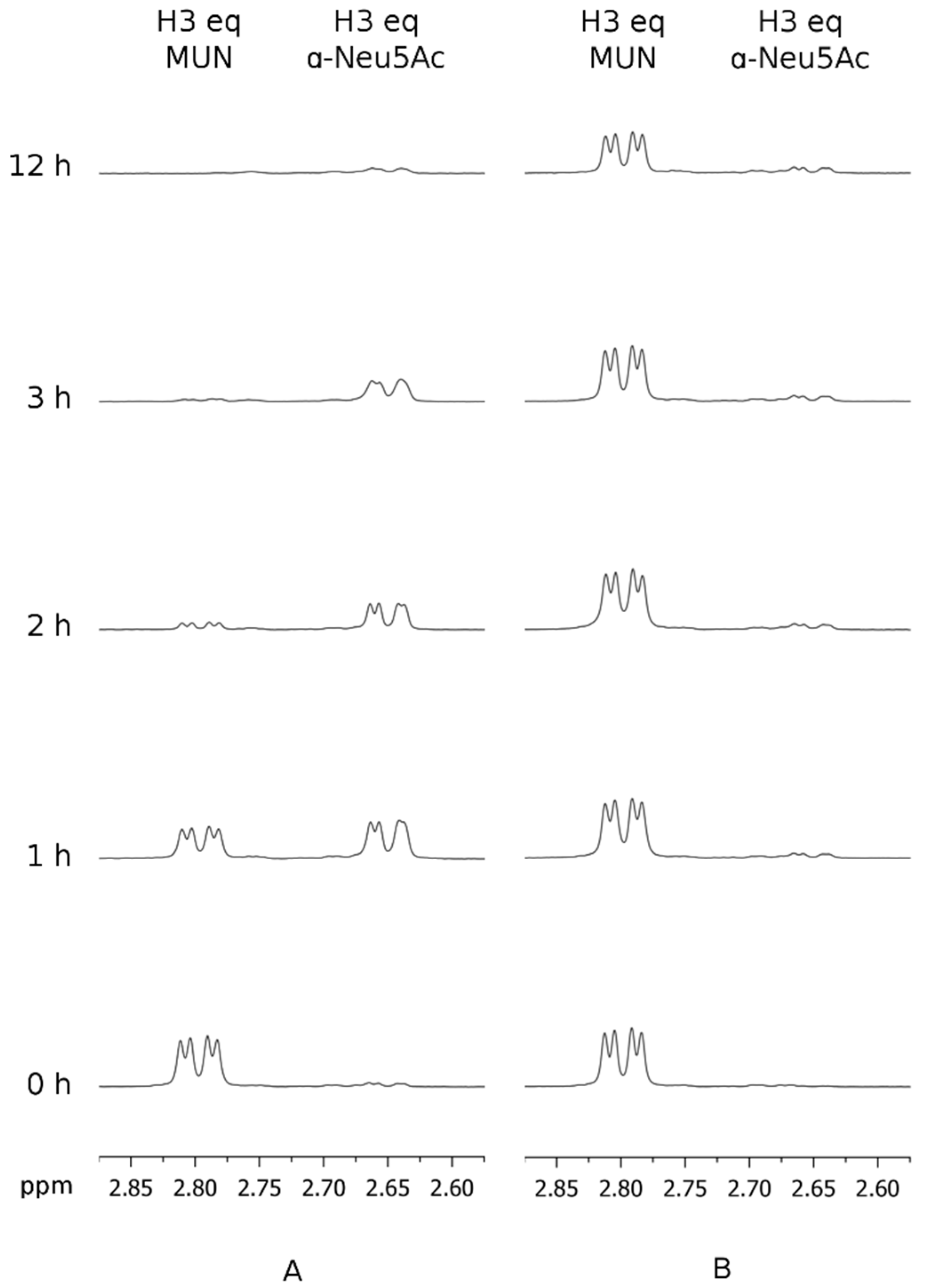
3.1. hPIV-1 HN Neuraminidase Activity Characterisation and Blockade
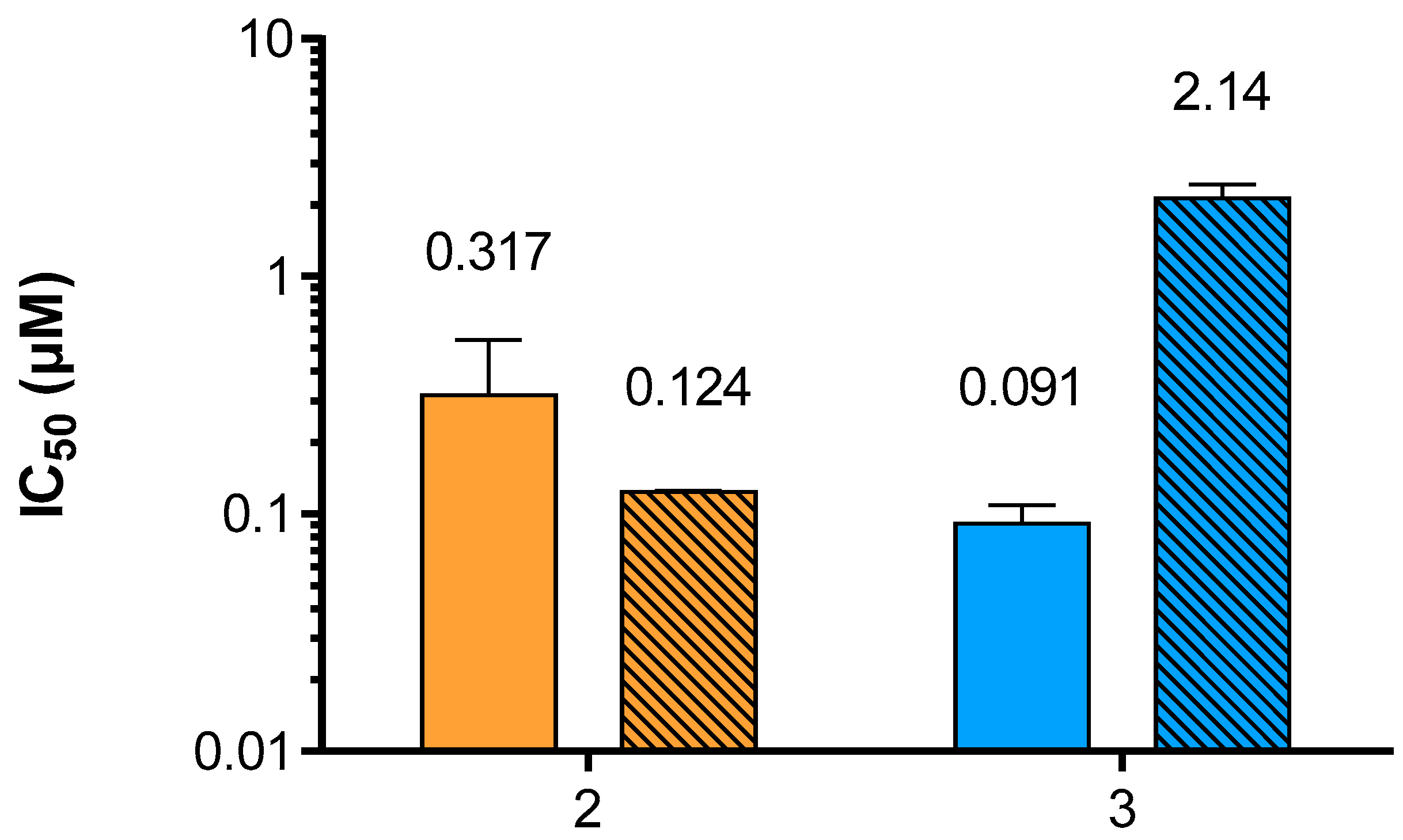
3.2. Inhibition of hPIV-1 HN Haemagglutination and Neuraminidase Functions
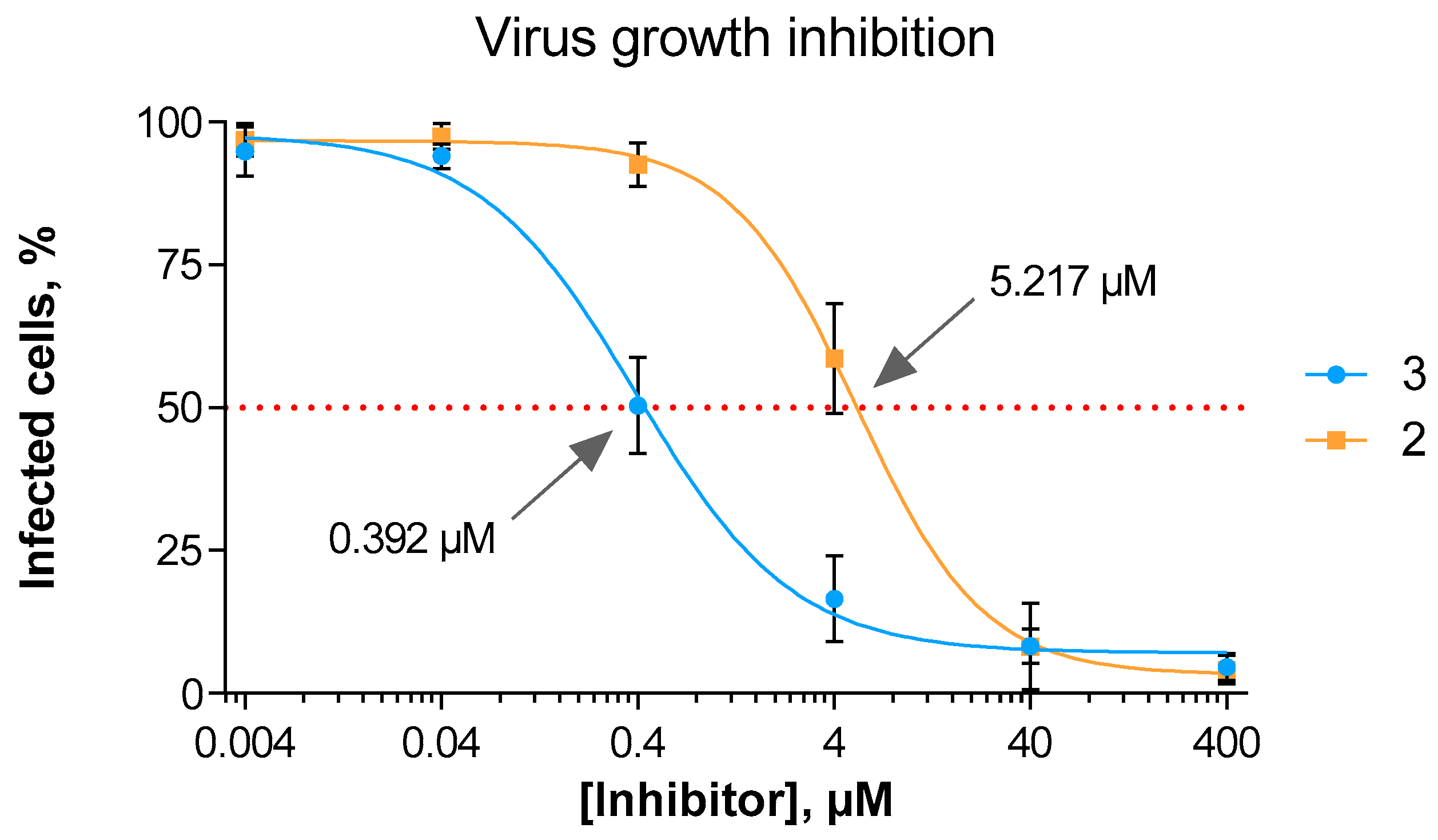
3.3. In Vitro Viral Replication Inhibition
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Pavia, A.T. Viral Infections of the Lower Respiratory Tract: Old Viruses, New Viruses, and the Role of Diagnosis. Clin. Infect. Dis. 2011, 52, S284–S289. [Google Scholar] [CrossRef] [PubMed]
- Karron, R.A.; Collins, P.L. Parainfluenza viruses. In Fields Virology: Sixth Edition; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; Volume 1. [Google Scholar]
- Schomacker, H.; Schaap-Nutt, A.; Collins, P.L.; Schmidt, A.C. Pathogenesis of acute respiratory illness caused by human parainfluenza viruses. Curr. Opin. Virol. 2012, 2, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Abedi, G.R.; Prill, M.M.; Langley, G.E.; Wikswo, M.E.; Weinberg, G.A.; Curns, A.T.; Schneider, E. Estimates of Parainfluenza Virus-Associated Hospitalizations and Cost Among Children Aged Less Than 5 Years in the United States, 1998–2010. J. Pediatr. Infect. Dis. Soc. 2016, 5, 7–13. [Google Scholar] [CrossRef]
- Bayon, J.-C.L.; Lina, B.; Rosa-Calatrava, M.; Boivin, G. Recent developments with live-attenuated recombinant paramyxovirus vaccines. Rev. Med. Virol. 2013, 23, 15–34. [Google Scholar] [CrossRef]
- Schmidt, A.C.; Schaap-Nutt, A.; Bartlett, E.J.; Schomacker, H.; Boonyaratanakornkit, J.; Karron, R.A.; Collins, P.L. Progress in the development of human parainfluenza virus vaccines. Expert Rev. Respir. Med. 2011, 5, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Alymova, I.V.; Taylor, G.; Takimoto, T.; Lin, T.-H.; Chand, P.; Babu, Y.S.; Li, C.; Xiong, X.; Portner, A. Efficacy of Novel Hemagglutinin-Neuraminidase Inhibitors BCX 2798 and BCX 2855 against Human Parainfluenza Viruses In Vitro and In Vivo. Antimicrob. Agents Chemother. 2004, 48, 1495–1502. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Alymova, I.V.; Watanabe, M.; Boyd, K.L.; Chand, P.; Babu, Y.S.; Portner, A. Efficacy of the Novel Parainfluenza Virus Hemagglutinin-Neuraminidase Inhibitor BCX 2798 in Mice--Further Evaluation. Antivir. Ther. 2009, 14, 891–898. [Google Scholar] [CrossRef]
- Suzuki, T.; Ikeda, K.; Koyama, N.; Hosokawa, C.; Kogure, T.; Takahashi, T.; Jwa Hidari, K.-P.; Miyamoto, D.; Tanaka, K.; Suzuki, Y. Inhibition of human parainfluenza virus type 1 sialidase by analogs of 2-deoxy-2,3-didehydro-N-acetylneuraminic acid. Glycoconj. J. 2001, 18, 331–337. [Google Scholar] [CrossRef]
- Ikeda, K.; Sato, K.; Kitani, S.; Suzuki, T.; Maki, N.; Suzuki, Y.; Sato, M. 2-Deoxy-2,3-didehydro-N-acetylneuraminic acid analogues structurally modified at the C-4 position: Synthesis and biological evaluation as inhibitors of human parainfluenza virus type 1. Bioorg. Med. Chem. 2006, 14, 7893–7897. [Google Scholar] [CrossRef]
- Ikeda, K.; Sato, K.; Nishino, R.; Aoyama, S.; Suzuki, T.; Sato, M. 2-Deoxy-2,3-didehydro-N-acetylneuraminic acid analogs structurally modified by thiocarbamoylalkyl groups at the C-4 position: Synthesis and biological evaluation as inhibitors of human parainfluenza virus type 1. Bioorg. Med. Chem. 2008, 16, 6783–6788. [Google Scholar] [CrossRef] [PubMed]
- Itoh, Y.; Sando, A.; Ikeda, K.; Suzuki, T.; Tokiwa, H. Origin of the inhibitory activity of 4-O-substituted sialic derivatives of human parainfluenza virus. Glycoconj. J. 2012, 29, 231–237. [Google Scholar] [CrossRef]
- El-Deeb, I.M.; Guillon, P.; Winger, M.; Eveno, T.; Haselhorst, T.; Dyason, J.C.; von Itzstein, M. Exploring Human Parainfluenza Virus Type-1 Hemagglutinin–Neuraminidase as a Target for Inhibitor Discovery. J. Med. Chem. 2014, 57, 7613–7623. [Google Scholar] [CrossRef]
- Buchini, S.; Gallat, F.-X.; Greig, I.R.; Kim, J.-H.; Wakatsuki, S.; Chavas, L.M.G.; Withers, S.G. Tuning Mechanism-Based Inactivators of Neuraminidases: Mechanistic and Structural Insights. Angew. Chem. Int. Ed. 2014, 53, 3382–3386. [Google Scholar] [CrossRef] [PubMed]
- Watts, A.G.; Damager, I.; Amaya, M.L.; Buschiazzo, A.; Alzari, P.; Frasch, A.C.; Withers, S.G. Trypanosoma cruzi Trans-sialidase Operates through a Covalent Sialyl−Enzyme Intermediate: Tyrosine Is the Catalytic Nucleophile. J. Am. Chem. Soc. 2003, 125, 7532–7533. [Google Scholar] [CrossRef]
- Kim, J.-H.; Resende, R.; Wennekes, T.; Chen, H.-M.; Bance, N.; Buchini, S.; Watts, A.G.; Pilling, P.; Streltsov, V.A.; Petric, M.; et al. Mechanism-Based Covalent Neuraminidase Inhibitors with Broad-Spectrum Influenza Antiviral Activity. Science 2013, 340, 71–75. [Google Scholar] [CrossRef]
- Dirr, L.; El-Deeb, I.M.; Guillon, P.; Carroux, C.J.; Chavas, L.M.G.; von Itzstein, M. The Catalytic Mechanism of Human Parainfluenza Virus Type 3 Haemagglutinin-Neuraminidase Revealed. Angew. Chem. Int. Ed. 2015, 54, 2936–2940. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, K.; Kitani, S.; Sato, K.; Suzuki, T.; Hosokawa, C.; Suzuki, Y.; Tanaka, K.; Sato, M. 2β,3β-Difluorosialic acid derivatives structurally modified at the C-4 position: Synthesis and biological evaluation as inhibitors of human parainfluenza virus type 1. Carbohydr. Res. 2004, 339, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Guillon, P.; Dirr, L.; El-Deeb, I.M.; Winger, M.; Bailly, B.; Haselhorst, T.; Dyason, J.C.; von Itzstein, M. Structure-guided discovery of potent and dual-acting human parainfluenza virus haemagglutinin–neuraminidase inhibitors. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Potier, M.; Mameli, L.; Bélisle, M.; Dallaire, L.; Melançon, S.B. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-α-D-N-acetylneuraminate) substrate. Anal. Biochem. 1979, 94, 287–296. [Google Scholar] [CrossRef]
- Dirr, L.; El-Deeb, I.M.; Chavas, L.M.G.; Guillon, P.; von Itzstein, M. The impact of the butterfly effect on human parainfluenza virus haemagglutinin-neuraminidase inhibitor design. Sci. Rep. 2017, 7, 4507. [Google Scholar] [CrossRef]
- Zechel, D.L.; Withers, S.G. Dissection of nucleophilic and acid–base catalysis in glycosidases. Curr. Opin. Chem. Biol. 2001, 5, 643–649. [Google Scholar] [CrossRef]
- Villar, E.; Barroso, I.M. Role of sialic acid-containing molecules in paramyxovirus entry into the host cell: A minireview. Glycoconj. J. 2006, 23, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Pascolutti, M.; Dirr, L.; Guillon, P.; van den Bergh, A.; Ve, T.; Thomson, R.J.; von Itzstein, M. Structural Insights into Human Parainfluenza Virus 3 Hemagglutinin–Neuraminidase Using Unsaturated 3-N-Substituted Sialic Acids as Probes. ACS Chem. Biol. 2018, 13, 1544–1550. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eveno, T.; Dirr, L.; El-Deeb, I.M.; Guillon, P.; von Itzstein, M. Targeting Human Parainfluenza Virus Type-1 Haemagglutinin-Neuraminidase with Mechanism-Based Inhibitors. Viruses 2019, 11, 417. https://doi.org/10.3390/v11050417
Eveno T, Dirr L, El-Deeb IM, Guillon P, von Itzstein M. Targeting Human Parainfluenza Virus Type-1 Haemagglutinin-Neuraminidase with Mechanism-Based Inhibitors. Viruses. 2019; 11(5):417. https://doi.org/10.3390/v11050417
Chicago/Turabian StyleEveno, Tanguy, Larissa Dirr, Ibrahim M. El-Deeb, Patrice Guillon, and Mark von Itzstein. 2019. "Targeting Human Parainfluenza Virus Type-1 Haemagglutinin-Neuraminidase with Mechanism-Based Inhibitors" Viruses 11, no. 5: 417. https://doi.org/10.3390/v11050417
APA StyleEveno, T., Dirr, L., El-Deeb, I. M., Guillon, P., & von Itzstein, M. (2019). Targeting Human Parainfluenza Virus Type-1 Haemagglutinin-Neuraminidase with Mechanism-Based Inhibitors. Viruses, 11(5), 417. https://doi.org/10.3390/v11050417





