Abstract
Viruses are the most prevalent infectious agents, populating almost every ecosystem on earth. Most viruses carry only a handful of genes supporting their replication and the production of capsids. It came as a great surprise in 2003 when the first giant virus was discovered and found to have a >1 Mbp genome encoding almost a thousand proteins. Following this first discovery, dozens of giant virus strains across several viral families have been reported. Here, we provide an updated quantitative and qualitative view on giant viruses and elaborate on their shared and variable features. We review the complexity of giant viral proteomes, which include functions traditionally associated only with cellular organisms. These unprecedented functions include components of the translation machinery, DNA maintenance, and metabolic enzymes. We discuss the possible underlying evolutionary processes and mechanisms that might have shaped the diversity of giant viruses and their genomes, highlighting their remarkable capacity to hijack genes and genomic sequences from their hosts and environments. This leads us to examine prominent theories regarding the origin of giant viruses. Finally, we present the emerging ecological view of giant viruses, found across widespread habitats and ecological systems, with respect to the environment and human health.
1. Giant Viruses and the Viral World
Viruses are cell infecting agents present in almost every ecosystem. Questions the regarding viral origin and early evolution alongside all living organisms (bacteria, archaea and eukarya) are still wide open, and relevant theories remain speculative [1,2,3,4,5]. As viruses are exceptionally diverse and undergo rapid changes, it is impossible to construct an ancestral lineage tree for the viral world [6,7,8,9,10]. Instead, virus families are categorized according to the nature of their genetic material, mode of replication, pathogenicity, and structural properties [11].
At present, the viral world is represented by over 8,000 reference genomes [12]. The International Committee on Taxonomy of Viruses (ICTV) provides a universal virus taxonomical classification proposal that covers ~150 families and ~850 genera, with many viruses yet unclassified [13]. This collection provides a comprehensive, compact set of virus representatives.
Inspection of viral genomes reveals that most known viruses have genomes encoding only a few proteins. Actually, 69% of all known viruses have less than 10 proteins encoded in their genomes (Figure 1). It is a common assumption that viruses demonstrate near-optimal genome packing and information compression, presumably in order to maximize their replication rate, number of progenies, and other parameters that increase infectivity [14,15]. However, a debate is still ongoing over the generality of these phenomena [16], and there is a non-negligible percentage of larger viruses (Figure 1). On the far end of the distribution, there are viruses with hundreds of genes, most of them are considered giant viruses. While only 0.3% of the known viral proteomes contain 500 or more proteins, they encode as much as 7.5% of the total number of viral proteins (Figure 1B).
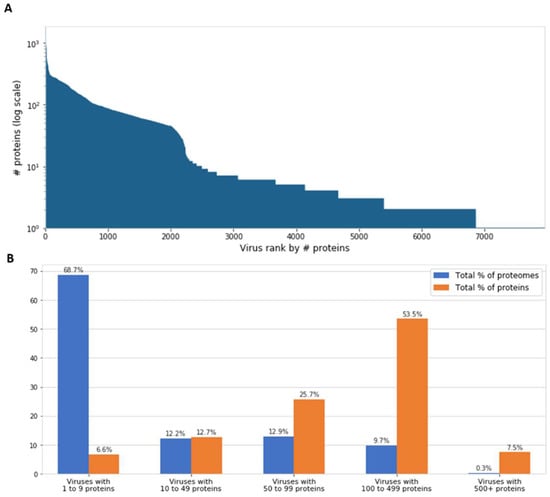
Figure 1.
Number of proteins encoded by viruses. (A) The number of encoded proteins (y-axis) in all 7,959 viral representatives, ranked in descending order. (B) Partitioning of the 7959 viral proteomes by the number of encoded proteins. The 0.3% viral proteomes with the highest number of proteins (over 500) encode 7.5% of the total number of viral proteins.
2. The Discovery of Giant Viruses
The first giant virus, Acanthamoeba polyphaga mimivirus (APMV), was discovered in 2003 [17]. Its size was unprecedented, being on the scale of small bacteria or archaea cells [18]. Unlike any previously identified virus, APMV could be seen with a light microscope [19,20]. Initially it was mistaken for a bacterium and recognized as a virus only ten years after its isolation [21]. Up to this day, most of its proteins remain uncharacterized [22,23]. Notably, even more than a decade after the discovery of APMV, the identification of giant viruses still sometimes involves confusion, as illustrated in the discovery of the Pandoravirus inopinatum [24], which was initially described as an endoparasitic organism, and Pithovirus sibericum [25], which was also misinterpreted as an archaeal endosymbiont (see discussion in References [21,26]).
In the following years after the initial discovery of APMV, many additional giant viral species have been identified and their genomes fully sequenced. Most giant viral genomes have been obtained from large-scale metagenomic sequencing projects covering aquatic ecosystems (e.g., oceans, pools, lakes and cooling wastewater units) [27,28]; others have been sequenced from samples extracted from underexplored geographical and ecological niches (e.g., the Amazon River, deep seas and forest soils) [29,30,31,32]. Despite the accumulation of many more giant virus representatives, the fraction of uncharacterized proteins in their proteomes remains exceptionally high [33]. Many of these uncharacterized proteins were also considered orphan genes (ORFans), i.e., no significant match to any other sequence was identified. For example, 93% of the Pandoravirus salinus proteins, the first representative of this family [34] were reported as ORFans.
At present, there are over a hundred giant virus isolates, which reveal fascinating and unexpected characteristics. These extreme instances on the viral landscape challenge the current theories on genome size and compactness in viruses, and provide a new perspective on the very concept of a virus and viral origin [4,20,28,35,36,37].
3. Definition of Giant Viruses
Attempts to distinguish giant viruses from other large viruses remain somewhat fuzzy [38,39]. Any definition for giant viruses would necessarily involve some arbitrary threshold, as virus size, whether physical, genomic or proteomic, is clearly a continuum (Figure 2). Giant viruses were initially defined by their physical size as allowing visibility by a light microscope [33]. In this report, we prefer a proteomic definition, even if somewhat arbitrary. We consider giant viruses as eukaryote-infecting viruses with at least 500 protein-coding genes (Figure 2). Of the 7,959 curated viral genomes (extracted from NCBI Taxonomy complete genomes), 24 represented genomes meet this threshold. Of these, we consider the 19 eukaryote-infecting viruses to be the giant virus representatives (Table 1), excluding the five bacteria-infecting viruses.
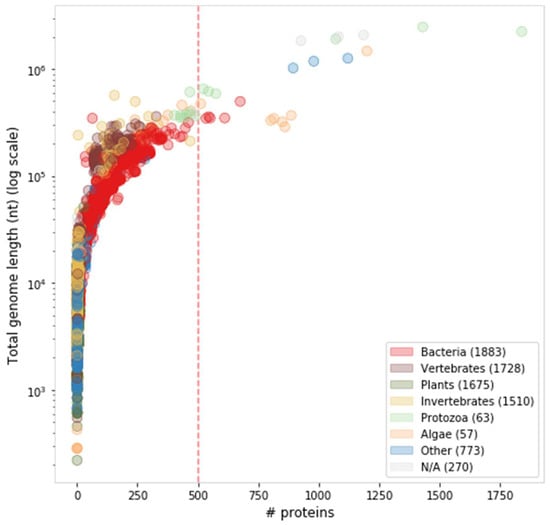
Figure 2.
Distribution of viral proteome and genome sizes, colored by host taxonomy. There are 24 represented genomes that meet the threshold of ≥500 proteins (dashed red line), comprising five bacteria-infecting and 19 eukaryote-infecting viruses.

Table 1.
Giant viruses.
Recall that reported proteome sizes are primarily based on automatic bioinformatics tools, which may differ from the experimental expression measurements (e.g., mimivirus APMV [40]). Moreover, physical dimensions are not in perfect correlation with the number of proteins or genome size. For example, Pithovirus sibericum, which was recovered from a 30,000-year-old permafrost sample [25], is one of the largest viruses by its physical dimensions (1.5 µm in length and 0.5 µm in diameter). However, it is excluded from this report, as its genome encodes only 467 proteins.
4. Classification of Giant Viruses and the Question of Origin
All giant viruses belong to the superfamily of nucleocytoplasmic large DNA viruses (NCLDV), which was substantially expanded following the discoveries of giant viruses [41,42]. The NCLDV superfamily had traditionally been comprised of the following families: Phycodnaviridae, Iridoviridae, Poxviridae, Asfarviridae and Ascoviridae [43,44], for which a common ancestor had been proposed [45,46]. Following the inclusion of additional giant virus taxonomy groups (Mimiviradae, Pandoravirus and Marseillevirus) into the NCLDV superfamily, there remained only a handful of genes shared by the entire superfamily. Additional disparities in virion shapes and replication modes among NCLDV has led to the conclusion that the superfamily is not necessarily a taxonomic group, and that NCLDV families are more likely to have evolved separately [47,48,49].
Two models have been proposed for the evolution of giant viruses. According to the reductive model, an ancestral cellular genome became reduced in size, leading to the dependence of the resulting genome on host cells. The presence of genes carrying cellular functions in almost any giant virus (e.g., translation components) [50] is consistent with this model. An alternative and more accepted theory argues for an expansion model. According to this model, current giant viruses originated from smaller ancestral viruses carrying only a few dozens of genes, and through gene duplications and horizontal gene transfer (HGT), have rapidly expanded and diversified [48,51,52,53]. This model agrees with metagenomic studies and the wave of giant virus discoveries in recent years, suggesting massive gene exchange between giant viruses and a variety of organisms sharing the same ecosystems (e.g., Reference [32]).
Of special interest is the degree of similarity between giant viruses and their hosts. The amebae host in particular is often described as a melting pot for DNA exchange [54] that leads to chimeric genomes. The majority of genes in giant viruses and specifically Mimiviridae, have originated from the cells they parasitize (mostly amoeba and bacteria). Based on phylogenetic trees, it is likely that extensive HGT events have led to their chimeric genomes. It was also suggested that the spectrum of viral hosts may be larger than anticipated, including yet unknown species [55]. Therefore, comparative genomics of giant viruses infecting the same host is unlikely to unambiguously resolve questions of gene origin, namely, whether shared genes have originated from a common viral ancestor. Thus, the degree of similarity among giant viruses infecting different hosts is of special interest. For example, the phyletic relationship between Mimiviridae (which infect Acanthamoeba) and Phycodnaviridae (which infect algae) was investigated, and it was found that the algae-infecting Chrysochromulina ericina virus (CeV, Table 1) showed moderate resemblance to the amebae-infecting mimivirus [56]. As a result, suggestions were made to reclassify CeV as a new clade of Mimiviridae, rather than Phycodnaviridae. However, a later discovery of another algae-infecting Phycodnaviridae virus (Heterosigma akashiwo virus, HaV53) has provided a coherent phyletic relationship among Phycodnaviridae, thereby questioning this reclassification [52].
In summary, the taxonomy of giant viruses, like all viruses, is still unstable, and rapidly updated with new discoveries [31,57]. The origin and ancestry of giant viruses have remained controversial with questions of origin also unresolved [39]. Many newly discovered giant viruses are not compatible with the notion of a single common ancestor, as some giant viruses remain taxonomically isolated [4].
5. Common Features
Despite the ongoing debate on their origin, giant viruses still share some important features. All giant viruses belong to the dsDNA group, as do all NCLDV families. The total genome size of all the giant viruses listed in Table 1 is at least 288 Kbp (Figure 2). These giant viruses are classified into several families: Mimiviridae, Pithoviridae, Pandoraviridae, Phycodnaviridae and the Mollivirus genus [21,25].
All amoebae-infecting giant viruses rely on the non-specific phagocytosis by the amoebae host [55]. Interestingly, a necessary condition for phagocytosis is a minimal particle size (~0.6 μm [58]), as amebae (and related protozoa) naturally feed on bacteria. It is likely that this minimal size for inducing phagocytosis has become an evolutionary driving force for giant viruses. This fact, together with the largely uncharacterized genomic content of giant viruses, may suggest that much of the content in the genomes of giant viruses serves only for volume filling to increase their physical size.
Giant viruses share not only the cell entry process. When they exit the host cells during lysis, as many as 1000 virions are released from each lysed host via membrane fusion and active exocytosis [59], which are relatively rare exit mechanisms in viruses.
Other than these genomic and cell-biology similarities, other features of giant viruses are mostly family-specific. For example, virion shapes and symmetries, nuclear involvement, duration of the infection cycle, and the stages of virion assembly—all substantially vary among giant viruses from different families [21,60,61].
6. Proteome Complexity and Functional Diversity
The majority of the giant virus proteomes remain with no known function (Figure 3). Actually, the fraction of uncharacterized proteins reaches 65–85% of all reported proteins in giant viral proteomes, many of them are ORFans. However, when proteomes of closely related species are considered, the fraction of ORFans obviously drops (by definition). For example, 93% of the proteins were reported as ORFans for the first representative of the Pandoraviridae family (P. salinus) [34]. But later, following the completion of five additional Pandoravirus proteomes (of the species inopinatum, macleodensis, neocaledonia, dulcis, and quercus), the number of P. salinus ORFans dropped to 29% (i.e., 71% of its genes now had a significant similarity to at least one other Pandoravirus protein sequence). Still, the vast majority of Pandoravirus proteins remain uncharacterized.
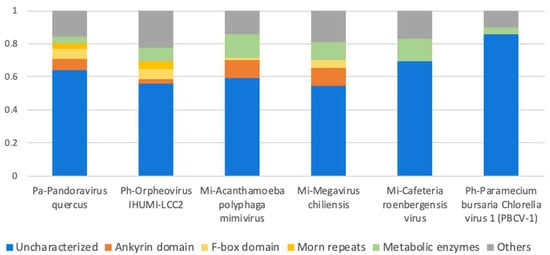
Figure 3.
Protein function categories in six giant virus representatives from three families: Mimiviridae (Mi), Pandoviridae (Pa) and Phycodnaviridae (Ph). In all proteomes, the majority of proteins are uncharacterized. Short repeated domains such as ankyrin, F-box and MORM repeats are abundant in the proteomes of amebae-infecting giant viruses [71].
The most striking finding regarding the proteomes of giant viruses is the presence of protein functions that are among the hallmarks of cellular organisms, and are never detected in other viruses. To exemplify the complexity of proteome functions in giant viruses, we examined the proteome of the Cafeteria roenbergensis virus (CroV), which infects the marine plankton community in the Gulf of Mexico. CroV was sequenced in 2010 as the first algae-infecting virus in the Mimiviridae family. Unexpectedly, despite its affiliation with a recognized viral family, the majority of its proteins showed no significant similarity to any other known protein sequence. Of the remaining proteins that show significant basic local alignment search tool (BLAST) hits to other proteins from all domains of life, 45% are eukaryotic sequences, 22% are from bacteria, and the rest are mostly from other viruses, including other mimivirus strains. A similar partition of protein origin applies across other members of the Mimiviridae family [33].
The CroV proteome includes a rich set of genes involved in protein translation [62]. These genes include multiple translation factors, a dozen of ribosomal proteins, tRNA synthetases, and 22 sequences encoding five different tRNAs [62]. As a lack of translation potential is considered a hallmark of the virosphere, the presence of translation machinery components raised a debate on the very definition of viruses [63,64]. Similar findings were replicated in two other giant virus strains of the Tupanvirus genus in the same Mimiviridae family, which were recently isolated in Brazil [65]. The two strains have 20 open reading frames (ORFs) related to tRNA aminoacylation (aaRS), ~70 tRNA sequences decoding the majority of the codons, eight translation initiation factors, and elongation and release factors. The theory that translation optimization is an evolutionary driving force in viruses [66] may in part explain the curious presence of translation machinery in giant viruses.
In addition to translation, numerous CroV proteins are associated with the transcription machinery. Specifically, the CroV proteome contains several subunits of the DNA-dependent RNA polymerase II, initiation, elongation, and termination factors, the mRNA capping enzyme, and a poly(A) polymerase. Presumably, the virus can activate its own transcription in the viral factory foci in the cytoplasm of its host cell [47].
Another unexpected function detected in CroV is DNA repair, specifically of UV radiation damage and base-excision repair. Other DNA-maintenance functions found in CroV include helicase and topoisomerases (type I and II), suggesting regulation of DNA replication, recombination and chromatin remodeling.
Another rich set of functions related to protein maintenance include chaperons [67] and the ubiquitin-proteasome system [68]. Interestingly, some of these genes seem to be acquired from bacteria (e.g., a homolog of the Escherichia coli heat-shock chaperon). In addition, a rich collection of sugar-, lipid- and amino acid-related metabolic enzymes were also found [18,69], which occupy 13% of the CroV proteome (Figure 3).
It appears that the CroV proteome covers most functions traditionally attributed to cellular organisms, including: Protein translation, RNA maturation, DNA maintenance, proteostasis and metabolism. Although CroV exemplifies many widespread functions in giant viruses, each strain has its own unique functional composition. For example, the most abundant group of giant viruses in ocean metagenomes, the Bodo saltans virus (BsV), was recently identified and classified into the same microzooplankton-infecting Mimiviridae family [70]. Unlike the other family members, BsV does not have an elaborate translation apparatus or tRNA genes, but it carries proteins active in cell membrane trafficking and phagocytosis, yet more unprecedented functions discovered in viruses.
7. Virophages and Defense Mechanism in Giant Viruses
Additional important players of genome dynamics in giant viruses are the virophages [72]. These are small double-stranded DNA (dsDNA) viruses that hitchhike the replication system of giant viruses following coinfection of the host, and are considered parasites of the coinfecting giant viruses [73]. Virophages (e.g., Sputnik 1–3, Zamilion) are associated with Mimiviridae representatives and their specific viral strain infectivity [21,74]. Additionally, short mobile genetic agents, called transpovirions (combining features of a transposon and a virion) [73], together with other mobile elements display complex ecological interactions with their hosts. Indeed, similar to eukaryotic transposons and endogenous viruses, sequences of a virophage (Mavirus) of Cafeteria roenbergensis virus (CroV), that were integrated to the genome of the protozoan host, serves an antiviral defense mechanism, which is activated by giant virus infection [75].
An even more unexpected finding is the discovery of a nucleic acid-based immunity in mimiviruses, resembling the adaptive (clustered regularly interspaced short palindromic repeats) CRISPR-Cas system in bacteria and archaea. Despite the differences to the canonical CRISPR-Cas system, an operon-like cluster of sequences derived from the Zamilion virophage was identified in mimivirus and experimentally validated to govern virophage coinfection. This cluster, coined MIMIVIRE, acts as a mimivirus virophage resistance element system with an exonuclease, helicase and RNase III identified in its vicinity [76]. The homology between the MIMIVIRE-associated exonuclease to the bacterial Cas-4 exonuclease was revealed by 3D protein structure analysis [77]. This CRISPR-Cas related function in mimiviruses are assumed to degrade foreign DNA, thereby constituting an antiviral innate immune system. It is likely that the CRISPR-Cas immune system in mimiviruses contributes to its sequence diversification as well by removing unessential host sequences. Alternative mechanisms that govern viral-host infection specificity and immunity may be discovered as our knowledge on virophages and other mobile elements is expending [72,78].
Altogether, a rich network of mobile genetic elements contributes to the host-virus coevolution and interviral gene transfer [78]. Virophages and other mobile elements could facilitate gene transfer, thereby having the potential to shape the genomes of giant viruses and impact their diversity [21,79,80].
8. The Emerging Ecological View
Viruses are the most abundant entities in nature. In marine and fresh water habitats, there are millions of viruses in each milliliter of water [81]. However, the collection of virus isolates is often sporadic, especially for those without clinical or agricultural relevance. The accelerated pace in the discovery of giant viruses reflects the increasing number of sequencing projects of exotic environments, including metagenomic projects [32,82].
Giant viruses have been isolated from numerous environmental niches and distant geographic locations, revealing their global distribution and diversity. Current evidence suggests that the representation of giant viruses is underexplored, especially in soil ecosystems [31] and unique ecological niches [83,84]. In fact, ~60% of the giant viral genomes were completed after 2013 (Table 1). Many more virus–host systems, most of them reported in the last five years, still await isolation, characterization and classification [84,85].
The hosts of contemporary isolates include mainly protozoa, specifically amoeba (Table 1). However, the prevalence of amoeba as hosts may in part be attributed to sampling bias, specifically to the widespread use of amoebal coculture methods for testing ecological environments [28,85].
Despite their prevalence, the impact of giant viruses on human health deserves further investigation [22]. Some initial reports show that APMV giant virus is able to replicate in human peripheral blood cells and to induce the interferon system [86]. Sequences of numerous giant viruses were identified as part of large-scale human gut microbiome sequencing projects [87], but their abundance, compositions and ecological roles are yet to be determined [88]. Reports are accumulating on the presence of giant viral sequences in human blood, as well as antibodies against giant viral proteins. Some reports associate mimirus and marseillevirus with a broad collection of human diseases (e.g., rheumatoid arthritis, adenitis, unexplained pneumonia, lymphoma), yet causal relationship is mostly missing [89]. The presence of giant viruses in almost any environment, including extreme niches and manmade sites (e.g., sewage and wastewater plants), suggests that the ecological role of these fascinating entities and their impact on human health are yet to be fully explored.
Author Contributions
The authors contributed to writing and manuscript design.
Funding
This research received no external funding.
Acknowledgments
The authors wish to thank the support to M.L. from Yad Hanadiv. We are grateful to Elixir infrastructure tools and resources.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Holmes, E.C. What does virus evolution tell us about virus origins? J. Virol. 2011, 85, 5247–5251. [Google Scholar] [CrossRef] [PubMed]
- Forterre, P. The origin of viruses and their possible roles in major evolutionary transitions. Virus Res. 2006, 117, 5–16. [Google Scholar] [CrossRef]
- Yutin, N.; Koonin, E.V. Evolution of DNA ligases of nucleo-cytoplasmic large DNA viruses of eukaryotes: A case of hidden complexity. Biol. Direct. 2009, 4, 51. [Google Scholar] [CrossRef] [PubMed]
- Forterre, P. Viruses in the 21st Century: From the Curiosity-Driven Discovery of Giant Viruses to New Concepts and Definition of Life. Clin. Infect. Dis. 2017, 65, S74–S79. [Google Scholar] [CrossRef] [PubMed]
- Moelling, K.; Broecker, F. Viruses and Evolution - Viruses First? A Personal Perspective. Front. Microbiol. 2019, 10, 523. [Google Scholar] [CrossRef] [PubMed]
- Moreira, D.; Lopez-Garcia, P. Ten reasons to exclude viruses from the tree of life. Nat. Rev. Microbiol. 2009, 7, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Brüssow, H. The not so universal tree of life or the place of viruses in the living world. Philos. Trans. R. Soc. B Biol. Sci. 2009, 364, 2263–2274. [Google Scholar]
- Bamford, D.H.; Grimes, J.M.; Stuart, D.I. What does structure tell us about virus evolution? Curr. Opin. Struct. Biol. 2005, 15, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Domingo, E.; Escarmis, C.; Sevilla, N.; Moya, A.; Elena, S.; Quer, J.; Novella, I.; Holland, J. Basic concepts in RNA virus evolution. FASEB J. 1996, 10, 859–864. [Google Scholar] [CrossRef]
- Koonin, E.V.; Yutin, N. Origin and evolution of eukaryotic large nucleo-cytoplasmic DNA viruses. Intervirology 2010, 53, 284–292. [Google Scholar] [CrossRef]
- Murphy, F.A.; Fauquet, C.M.; Bishop, D.H.; Ghabrial, S.A.; Jarvis, A.W.; Martelli, G.P.; Mayo, M.A.; Summers, M.D. Virus taxonomy: Classification and nomenclature of viruses; Springer Science & Business Media: Berlin, Germany, 2012; Volume 10. [Google Scholar]
- Stano, M.; Beke, G.; Klucar, L. viruSITE-integrated database for viral genomics. Database (Oxford) 2016, 2016. [Google Scholar] [CrossRef]
- Lefkowitz, E.J.; Dempsey, D.M.; Hendrickson, R.C.; Orton, R.J.; Siddell, S.G.; Smith, D.B. Virus taxonomy: The database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids. Res. 2018, 46, D708–D717. [Google Scholar] [CrossRef] [PubMed]
- Pybus, O.G.; Rambaut, A. Evolutionary analysis of the dynamics of viral infectious disease. Nat. Rev. Genet. 2009, 10, 540–550. [Google Scholar] [CrossRef]
- Pérez-Losada, M.; Arenas, M.; Galán, J.C.; Palero, F.; González-Candelas, F. Recombination in viruses: Mechanisms, methods of study, and evolutionary consequences. Infect. Genet. Evol. 2015, 30, 296–307. [Google Scholar] [CrossRef] [PubMed]
- Brandes, N.; Linial, M. Gene overlapping and size constraints in the viral world. Biol. Direct. 2016, 11, 26. [Google Scholar] [CrossRef] [PubMed]
- La Scola, B.; Audic, S.; Robert, C.; Jungang, L.; de Lamballerie, X.; Drancourt, M.; Birtles, R.; Claverie, J.M.; Raoult, D. A giant virus in amoebae. Science 2003, 299, 2033. [Google Scholar] [CrossRef]
- Raoult, D.; Audic, S.; Robert, C.; Abergel, C.; Renesto, P.; Ogata, H.; La Scola, B.; Suzan, M.; Claverie, J.M. The 1.2-megabase genome sequence of Mimivirus. Science 2004, 306, 1344–1350. [Google Scholar] [CrossRef]
- Xiao, C.; Chipman, P.R.; Battisti, A.J.; Bowman, V.D.; Renesto, P.; Raoult, D.; Rossmann, M.G. Cryo-electron microscopy of the giant Mimivirus. J. Mol. Biol. 2005, 353, 493–496. [Google Scholar] [CrossRef]
- Van Etten, J.L.; Meints, R.H. Giant viruses infecting algae. Annu. Rev. Microbiol. 1999, 53, 447–494. [Google Scholar] [CrossRef]
- Abergel, C.; Legendre, M.; Claverie, J.M. The rapidly expanding universe of giant viruses: Mimivirus, Pandoravirus, Pithovirus and Mollivirus. FEMS Microbiol. Rev. 2015, 39, 779–796. [Google Scholar] [CrossRef]
- Abrahão, J.S.; Dornas, F.P.; Silva, L.C.; Almeida, G.M.; Boratto, P.V.; Colson, P.; La Scola, B.; Kroon, E.G. Acanthamoeba polyphaga mimivirus and other giant viruses: An open field to outstanding discoveries. Virol. J. 2014, 11, 120. [Google Scholar] [CrossRef] [PubMed]
- Campos, R.K.; Boratto, P.V.; Assis, F.L.; Aguiar, E.R.; Silva, L.C.; Albarnaz, J.D.; Dornas, F.P.; Trindade, G.S.; Ferreira, P.P.; Marques, J.T. Samba virus: A novel mimivirus from a giant rain forest, the Brazilian Amazon. Virol. J. 2014, 11, 95. [Google Scholar] [CrossRef] [PubMed]
- Antwerpen, M.H.; Georgi, E.; Zoeller, L.; Woelfel, R.; Stoecker, K.; Scheid, P. Whole-genome sequencing of a pandoravirus isolated from keratitis-inducing acanthamoeba. Genome Announc. 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- Legendre, M.; Bartoli, J.; Shmakova, L.; Jeudy, S.; Labadie, K.; Adrait, A.; Lescot, M.; Poirot, O.; Bertaux, L.; Bruley, C.; et al. Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc. Natl. Acad. Sci. USA 2014, 111, 4274–4279. [Google Scholar] [CrossRef] [PubMed]
- Claverie, J.M.; Abergel, C. From extraordinary endocytobionts to pandoraviruses. Comment on Scheid et al.: Some secrets are revealed: Parasitic keratitis amoebae as vectors of the scarcely described pandoraviruses to humans. Parasitol. Res. 2015, 114, 1625–1627. [Google Scholar] [CrossRef] [PubMed]
- Andreani, J.; Verneau, J.; Raoult, D.; Levasseur, A.; La Scola, B. Deciphering viral presences: Two novel partial giant viruses detected in marine metagenome and in a mine drainage metagenome. Virol. J. 2018, 15, 66. [Google Scholar] [CrossRef]
- Aherfi, S.; Colson, P.; La Scola, B.; Raoult, D. Giant Viruses of Amoebas: An Update. Front. Microbiol. 2016, 7, 349. [Google Scholar] [CrossRef] [PubMed]
- Andrade, A.; Arantes, T.S.; Rodrigues, R.A.L.; Machado, T.B.; Dornas, F.P.; Landell, M.F.; Furst, C.; Borges, L.G.A.; Dutra, L.A.L.; Almeida, G.; et al. Ubiquitous giants: A plethora of giant viruses found in Brazil and Antarctica. Virol. J. 2018, 15, 22. [Google Scholar] [CrossRef]
- Dornas, F.P.; Assis, F.L.; Aherfi, S.; Arantes, T.; Abrahao, J.S.; Colson, P.; La Scola, B. A Brazilian Marseillevirus Is the Founding Member of a Lineage in Family Marseilleviridae. Viruses 2016, 8, 76. [Google Scholar] [CrossRef] [PubMed]
- Schulz, F.; Alteio, L.; Goudeau, D.; Ryan, E.M.; Yu, F.B.; Malmstrom, R.R.; Blanchard, J.; Woyke, T. Hidden diversity of soil giant viruses. Nat. Commun. 2018, 9, 4881. [Google Scholar] [CrossRef]
- Backstrom, D.; Yutin, N.; Jorgensen, S.L.; Dharamshi, J.; Homa, F.; Zaremba-Niedwiedzka, K.; Spang, A.; Wolf, Y.I.; Koonin, E.V.; Ettema, T.J.G. Virus Genomes from Deep Sea Sediments Expand the Ocean Megavirome and Support Independent Origins of Viral Gigantism. MBio 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Colson, P.; La Scola, B.; Levasseur, A.; Caetano-Anolles, G.; Raoult, D. Mimivirus: Leading the way in the discovery of giant viruses of amoebae. Nat. Rev. Microbiol. 2017, 15, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Philippe, N.; Legendre, M.; Doutre, G.; Coute, Y.; Poirot, O.; Lescot, M.; Arslan, D.; Seltzer, V.; Bertaux, L.; Bruley, C.; et al. Pandoraviruses: Amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science 2013, 341, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Colson, P.; Pontarotti, P.; Raoult, D. Mimivirus inaugurated in the 21st century the beginning of a reclassification of viruses. Curr. Opin. Microbiol. 2016, 31, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Rolland, C.; Andreani, J.; Louazani, A.C.; Aherfi, S.; Francis, R.; Rodrigues, R.; Silva, L.S.; Sahmi, D.; Mougari, S.; Chelkha, N.; et al. Discovery and Further Studies on Giant Viruses at the IHU Mediterranee Infection That Modified the Perception of the Virosphere. Viruses 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Colson, P.; La Scola, B.; Raoult, D. Giant Viruses of Amoebae: A Journey Through Innovative Research and Paradigm Changes. Annu. Rev. Virol. 2017, 4, 61–85. [Google Scholar] [CrossRef]
- Claverie, J.M.; Abergel, C. Mimiviridae: An Expanding Family of Highly Diverse Large dsDNA Viruses Infecting a Wide Phylogenetic Range of Aquatic Eukaryotes. Viruses 2018, 10. [Google Scholar] [CrossRef]
- Colson, P.; Levasseur, A.; La Scola, B.; Sharma, V.; Nasir, A.; Pontarotti, P.; Caetano-Anolles, G.; Raoult, D. Ancestrality and Mosaicism of Giant Viruses Supporting the Definition of the Fourth TRUC of Microbes. Front. Microbiol. 2018, 9, 2668. [Google Scholar] [CrossRef] [PubMed]
- Legendre, M.; Santini, S.; Rico, A.; Abergel, C.; Claverie, J.M. Breaking the 1000-gene barrier for Mimivirus using ultra-deep genome and transcriptome sequencing. Virol. J. 2011, 8, 99. [Google Scholar] [CrossRef] [PubMed]
- Yutin, N.; Wolf, Y.I.; Raoult, D.; Koonin, E.V. Eukaryotic large nucleo-cytoplasmic DNA viruses: Clusters of orthologous genes and reconstruction of viral genome evolution. Virol. J. 2009, 6, 223. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Yutin, N. Evolution of the Large Nucleocytoplasmic DNA Viruses of Eukaryotes and Convergent Origins of Viral Gigantism. Adv. Virus Res. 2019, 103, 167–202. [Google Scholar] [CrossRef] [PubMed]
- Andreani, J.; Aherfi, S.; Bou Khalil, J.Y.; Di Pinto, F.; Bitam, I.; Raoult, D.; Colson, P.; La Scola, B. Cedratvirus, a Double-Cork Structured Giant Virus, is a Distant Relative of Pithoviruses. Viruses 2016, 8. [Google Scholar] [CrossRef]
- Wilhelm, S.W.; Coy, S.R.; Gann, E.R.; Moniruzzaman, M.; Stough, J.M. Standing on the Shoulders of Giant Viruses: Five Lessons Learned about Large Viruses Infecting Small Eukaryotes and the Opportunities They Create. PLoS Pathog. 2016, 12, e1005752. [Google Scholar] [CrossRef] [PubMed]
- Iyer, L.M.; Aravind, L.; Koonin, E.V. Common origin of four diverse families of large eukaryotic DNA viruses. J. Virol. 2001, 75, 11720–11734. [Google Scholar] [CrossRef] [PubMed]
- Nasir, A.; Kim, K.M.; Caetano-Anolles, G. Giant viruses coexisted with the cellular ancestors and represent a distinct supergroup along with superkingdoms Archaea, Bacteria and Eukarya. BMC Evol. Biol. 2012, 12, 156. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.G.; Allen, M.J.; Wilson, W.H.; Suttle, C.A. Giant virus with a remarkable complement of genes infects marine zooplankton. Proc. Natl. Acad. Sci. USA 2010, 107, 19508–19513. [Google Scholar] [CrossRef]
- Yutin, N.; Wolf, Y.I.; Koonin, E.V. Origin of giant viruses from smaller DNA viruses not from a fourth domain of cellular life. Virology 2014, 466–467, 38–52. [Google Scholar] [CrossRef]
- Koonin, E.V.; Yutin, N. Multiple evolutionary origins of giant viruses. F1000Research 2018, 7. [Google Scholar] [CrossRef]
- Arslan, D.; Legendre, M.; Seltzer, V.; Abergel, C.; Claverie, J.M. Distant Mimivirus relative with a larger genome highlights the fundamental features of Megaviridae. Proc. Natl. Acad. Sci. USA 2011, 108, 17486–17491. [Google Scholar] [CrossRef]
- Iyer, L.M.; Balaji, S.; Koonin, E.V.; Aravind, L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006, 117, 156–184. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, F.; Ueki, S. Evolution and Phylogeny of Large DNA Viruses, Mimiviridae and Phycodnaviridae Including Newly Characterized Heterosigma akashiwo Virus. Front. Microbiol. 2016, 7, 1942. [Google Scholar] [CrossRef]
- Filee, J.; Chandler, M. Gene exchange and the origin of giant viruses. Intervirology 2010, 53, 354–361. [Google Scholar] [CrossRef]
- Boyer, M.; Yutin, N.; Pagnier, I.; Barrassi, L.; Fournous, G.; Espinosa, L.; Robert, C.; Azza, S.; Sun, S.; Rossmann, M.G.; et al. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc. Natl. Acad. Sci. USA 2009, 106, 21848–21853. [Google Scholar] [CrossRef]
- Moreira, D.; Brochier-Armanet, C. Giant viruses, giant chimeras: The multiple evolutionary histories of Mimivirus genes. BMC Evol. Biol. 2008, 8, 12. [Google Scholar] [CrossRef]
- Gallot-Lavallee, L.; Blanc, G.; Claverie, J.M. Comparative Genomics of Chrysochromulina Ericina Virus and Other Microalga-Infecting Large DNA Viruses Highlights Their Intricate Evolutionary Relationship with the Established Mimiviridae Family. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Yoosuf, N.; Pagnier, I.; Fournous, G.; Robert, C.; La Scola, B.; Raoult, D.; Colson, P. Complete genome sequence of Courdo11 virus, a member of the family Mimiviridae. Virus Genes. 2014, 48, 218–223. [Google Scholar] [CrossRef]
- Rodrigues, R.A.L.; Abrahao, J.S.; Drumond, B.P.; Kroon, E.G. Giants among larges: How gigantism impacts giant virus entry into amoebae. Curr. Opin. Microbiol. 2016, 31, 88–93. [Google Scholar] [CrossRef]
- Silva, L.; Andrade, A.; Dornas, F.P.; Rodrigues, R.A.L.; Arantes, T.; Kroon, E.G.; Bonjardim, C.A.; Abrahao, J.S. Cedratvirus getuliensis replication cycle: An in-depth morphological analysis. Sci. Rep. 2018, 8, 4000. [Google Scholar] [CrossRef]
- Suzan-Monti, M.; La Scola, B.; Barrassi, L.; Espinosa, L.; Raoult, D. Ultrastructural characterization of the giant volcano-like virus factory of Acanthamoeba polyphaga Mimivirus. PLoS ONE 2007, 2, e328. [Google Scholar] [CrossRef]
- Diesend, J.; Kruse, J.; Hagedorn, M.; Hammann, C. Amoebae, Giant Viruses, and Virophages Make Up a Complex, Multilayered Threesome. Front. Cell. Infect. Microbiol. 2017, 7, 527. [Google Scholar] [CrossRef]
- Schulz, F.; Yutin, N.; Ivanova, N.N.; Ortega, D.R.; Lee, T.K.; Vierheilig, J.; Daims, H.; Horn, M.; Wagner, M.; Jensen, G.J.; et al. Giant viruses with an expanded complement of translation system components. Science 2017, 356, 82–85. [Google Scholar] [CrossRef] [PubMed]
- Abrahao, J.S.; Araujo, R.; Colson, P.; La Scola, B. The analysis of translation-related gene set boosts debates around origin and evolution of mimiviruses. PLoS Genet. 2017, 13, e1006532. [Google Scholar] [CrossRef] [PubMed]
- Raoult, D.; Forterre, P. Redefining viruses: Lessons from Mimivirus. Nat. Rev. Microbiol. 2008, 6, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Abrahao, J.; Silva, L.; Silva, L.S.; Khalil, J.Y.B.; Rodrigues, R.; Arantes, T.; Assis, F.; Boratto, P.; Andrade, M.; Kroon, E.G.; et al. Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere. Nat. Commun. 2018, 9, 749. [Google Scholar] [CrossRef] [PubMed]
- Bahir, I.; Fromer, M.; Prat, Y.; Linial, M. Viral adaptation to host: A proteome-based analysis of codon usage and amino acid preferences. Mol. Syst. Biol. 2009, 5, 311. [Google Scholar] [CrossRef] [PubMed]
- Barik, S. A Family of Novel Cyclophilins, Conserved in the Mimivirus Genus of the Giant DNA Viruses. Comput. Struct. Biotechnol. J. 2018, 16, 231–236. [Google Scholar] [CrossRef]
- Yutin, N.; Colson, P.; Raoult, D.; Koonin, E.V. Mimiviridae: Clusters of orthologous genes, reconstruction of gene repertoire evolution and proposed expansion of the giant virus family. Virol. J. 2013, 10, 106. [Google Scholar] [CrossRef]
- Van Etten, J.L. Unusual life style of giant chlorella viruses. Annu. Rev. Genet. 2003, 37, 153–195. [Google Scholar] [CrossRef] [PubMed]
- Deeg, C.M.; Chow, C.T.; Suttle, C.A. The kinetoplastid-infecting Bodo saltans virus (BsV), a window into the most abundant giant viruses in the sea. Elife 2018, 7. [Google Scholar] [CrossRef]
- Shukla, A.; Chatterjee, A.; Kondabagil, K. The number of genes encoding repeat domain-containing proteins positively correlates with genome size in amoebal giant viruses. Virus Evol. 2018, 4, vex039. [Google Scholar] [CrossRef]
- Bekliz, M.; Colson, P.; La Scola, B. The Expanding Family of Virophages. Viruses 2016, 8. [Google Scholar] [CrossRef]
- Desnues, C.; La Scola, B.; Yutin, N.; Fournous, G.; Robert, C.; Azza, S.; Jardot, P.; Monteil, S.; Campocasso, A.; Koonin, E.V.; et al. Provirophages and transpovirons as the diverse mobilome of giant viruses. Proc. Natl. Acad. Sci. USA 2012, 109, 18078–18083. [Google Scholar] [CrossRef]
- Fischer, M.G. Sputnik and Mavirus: More than just satellite viruses. Nat. Rev. Microbiol. 2012, 10, 78. [Google Scholar] [CrossRef]
- Fischer, M.G.; Hackl, T. Host genome integration and giant virus-induced reactivation of the virophage mavirus. Nature 2016, 540, 288–291. [Google Scholar] [CrossRef] [PubMed]
- Levasseur, A.; Bekliz, M.; Chabriere, E.; Pontarotti, P.; La Scola, B.; Raoult, D. MIMIVIRE is a defence system in mimivirus that confers resistance to virophage. Nature 2016, 531, 249–252. [Google Scholar] [CrossRef] [PubMed]
- Dou, C.; Yu, M.; Gu, Y.; Wang, J.; Yin, K.; Nie, C.; Zhu, X.; Qi, S.; Wei, Y.; Cheng, W. Structural and Mechanistic Analyses Reveal a Unique Cas4-like Protein in the Mimivirus Virophage Resistance Element System. iScience 2018, 3, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Krupovic, M. Polintons, virophages and transpovirons: A tangled web linking viruses, transposons and immunity. Curr. Opin. Virol. 2017, 25, 7–15. [Google Scholar] [CrossRef]
- Campbell, S.; Aswad, A.; Katzourakis, A. Disentangling the origins of virophages and polintons. Curr. Opin. Virol. 2017, 25, 59–65. [Google Scholar] [CrossRef] [PubMed]
- La Scola, B.; Desnues, C.; Pagnier, I.; Robert, C.; Barrassi, L.; Fournous, G.; Merchat, M.; Suzan-Monti, M.; Forterre, P.; Koonin, E.; et al. The virophage as a unique parasite of the giant mimivirus. Nature 2008, 455, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Suttle, C.A. Marine viruses--major players in the global ecosystem. Nat. Rev. Microbiol. 2007, 5, 801–812. [Google Scholar] [CrossRef]
- Chatterjee, A.; Sicheritz-Ponten, T.; Yadav, R.; Kondabagil, K. Genomic and metagenomic signatures of giant viruses are ubiquitous in water samples from sewage, inland lake, waste water treatment plant, and municipal water supply in Mumbai, India. Sci. Rep. 2019, 9, 3690. [Google Scholar] [CrossRef]
- Yoshikawa, G.; Blanc-Mathieu, R.; Song, C.; Kayama, Y.; Mochizuki, T.; Murata, K.; Ogata, H.; Takemura, M. Medusavirus, a novel large DNA virus discovered from hot spring water. J. Virol. 2019. [Google Scholar] [CrossRef]
- Wilhelm, S.W.; Bird, J.T.; Bonifer, K.S.; Calfee, B.C.; Chen, T.; Coy, S.R.; Gainer, P.J.; Gann, E.R.; Heatherly, H.T.; Lee, J.; et al. A Student’s Guide to Giant Viruses Infecting Small Eukaryotes: From Acanthamoeba to Zooxanthellae. Viruses 2017, 9. [Google Scholar] [CrossRef]
- Watanabe, T.; Yamazaki, S.; Maita, C.; Matushita, M.; Matsuo, J.; Okubo, T.; Yamaguchi, H. Lateral Gene Transfer Between Protozoa-Related Giant Viruses of Family Mimiviridae and Chlamydiae. Evol. Bioinform. Online 2018, 14. [Google Scholar] [CrossRef]
- Silva, L.C.; Almeida, G.M.; Oliveira, D.B.; Dornas, F.P.; Campos, R.K.; La Scola, B.; Ferreira, P.C.; Kroon, E.G.; Abrahao, J.S. A resourceful giant: APMV is able to interfere with the human type I interferon system. Microbes Infect. 2014, 16, 187–195. [Google Scholar] [CrossRef]
- Scarpellini, E.; Ianiro, G.; Attili, F.; Bassanelli, C.; De Santis, A.; Gasbarrini, A. The human gut microbiota and virome: Potential therapeutic implications. Dig. Liver Dis. 2015, 47, 1007–1012. [Google Scholar] [CrossRef]
- Popgeorgiev, N.; Temmam, S.; Raoult, D.; Desnues, C. Describing the silent human virome with an emphasis on giant viruses. Intervirology 2013, 56, 395–412. [Google Scholar] [CrossRef]
- Colson, P.; Fancello, L.; Gimenez, G.; Armougom, F.; Desnues, C.; Fournous, G.; Yoosuf, N.; Million, M.; La Scola, B.; Raoult, D. Evidence of the megavirome in humans. J. Clin. Virol. 2013, 57, 191–200. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).