Targeted Editing of the pp38 Gene in Marek’s Disease Virus-Transformed Cell Lines Using CRISPR/Cas9 System
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
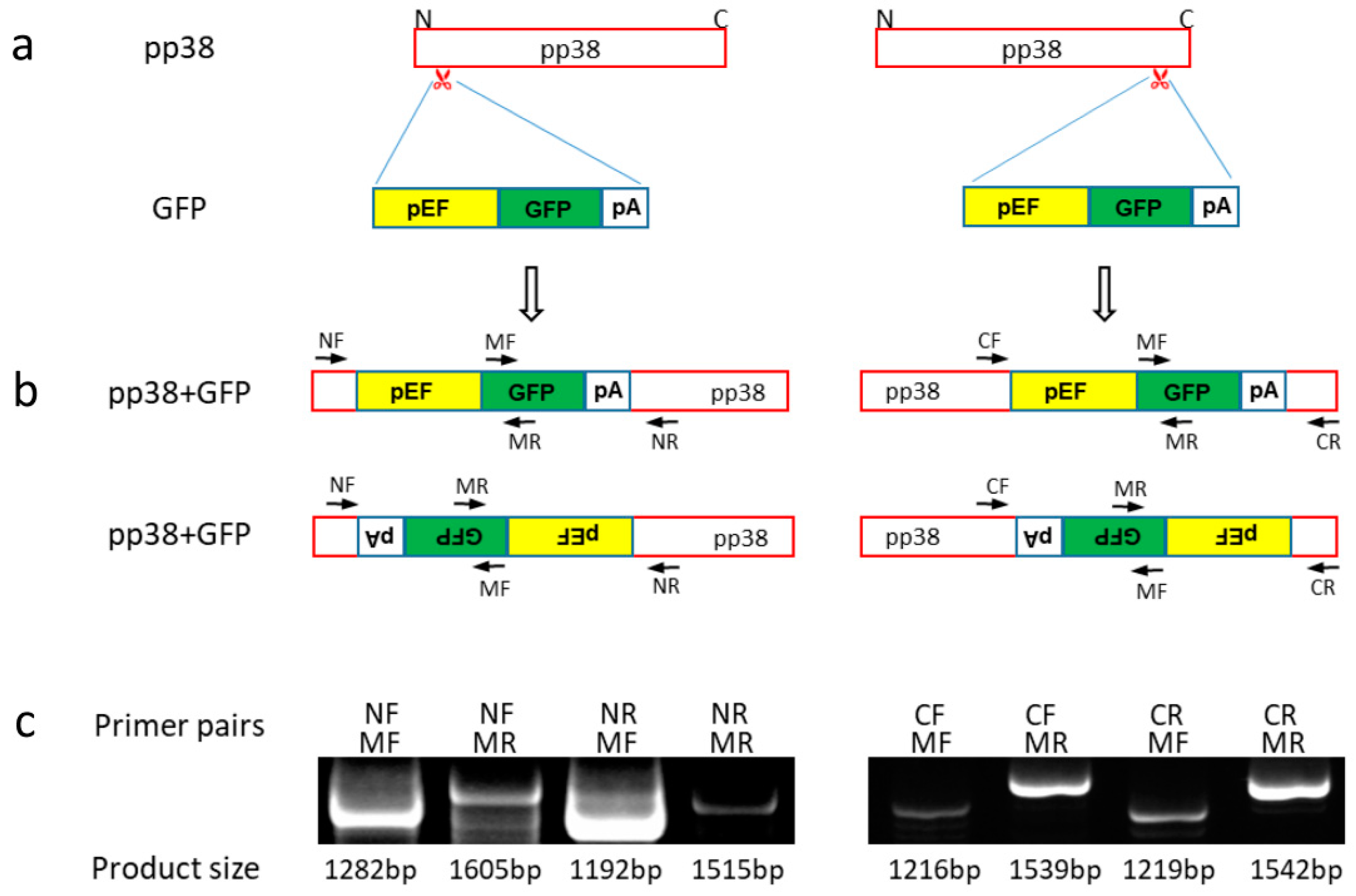
2.2. gRNAs and GFP Donor Template
2.3. Generation of Stable MSB-1 and HP8 Cell Lines Expressing Cas9
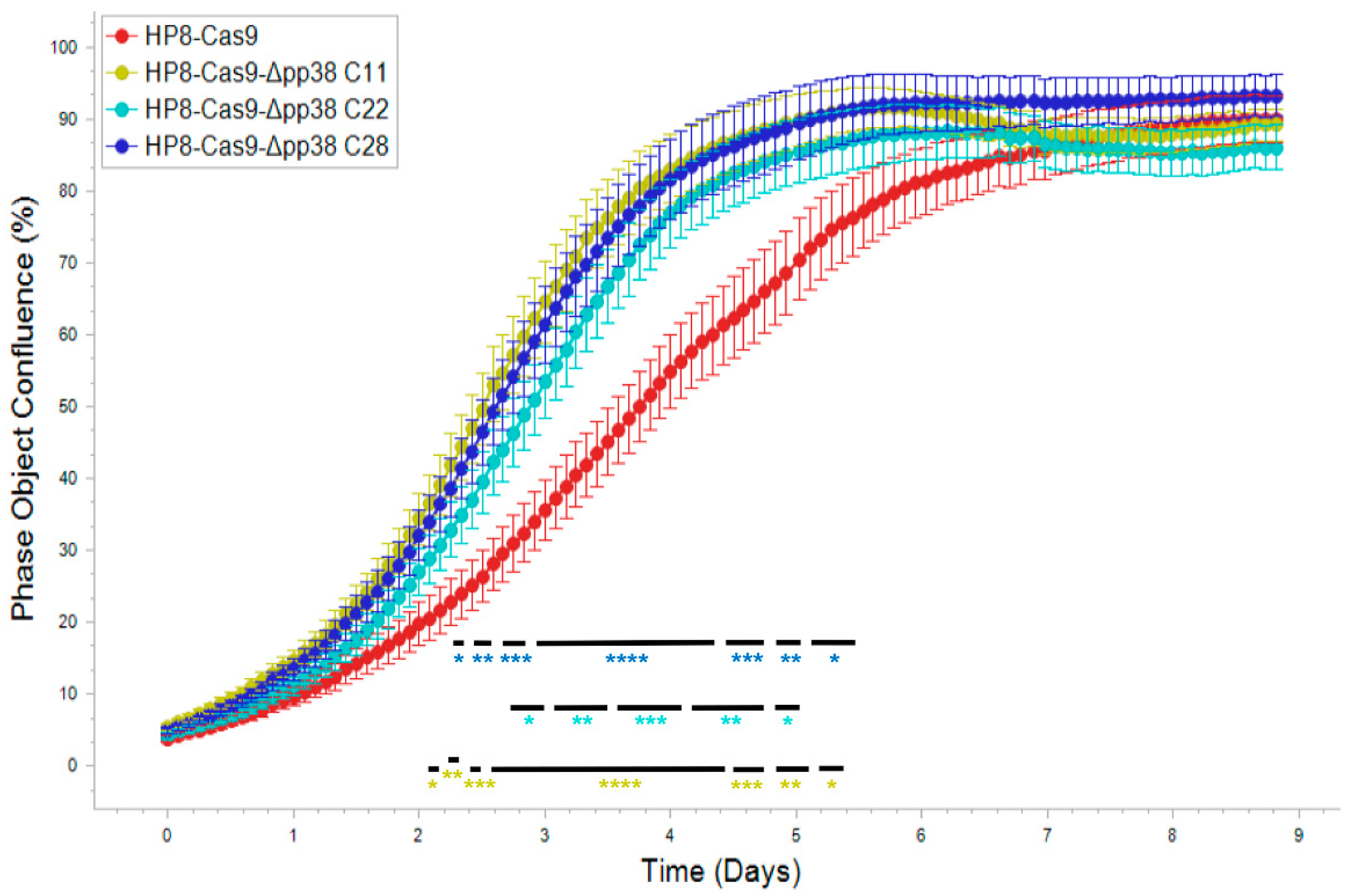
2.4. Generation and Characterization of pp38 Deletion Cell Line HP8-Δpp38
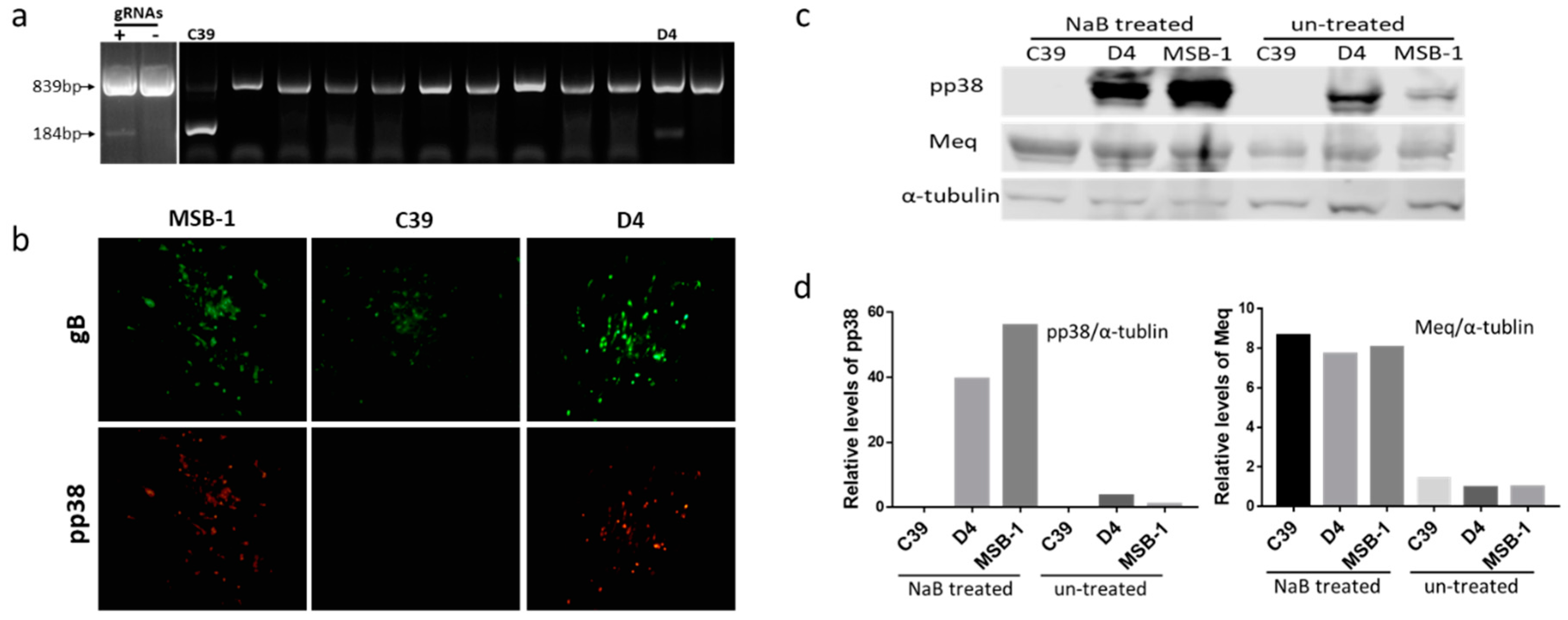
2.5. Reactivation of MDV from MSB-1-Δpp38
2.6. Western Blotting Analysis
2.7. Growth of HP8-Cas9-Δpp38 Cells
2.8. Single Cell Sorting and Flow Cytometry Analysis
2.9. q-PCR for GFP Copy Number
3. Results
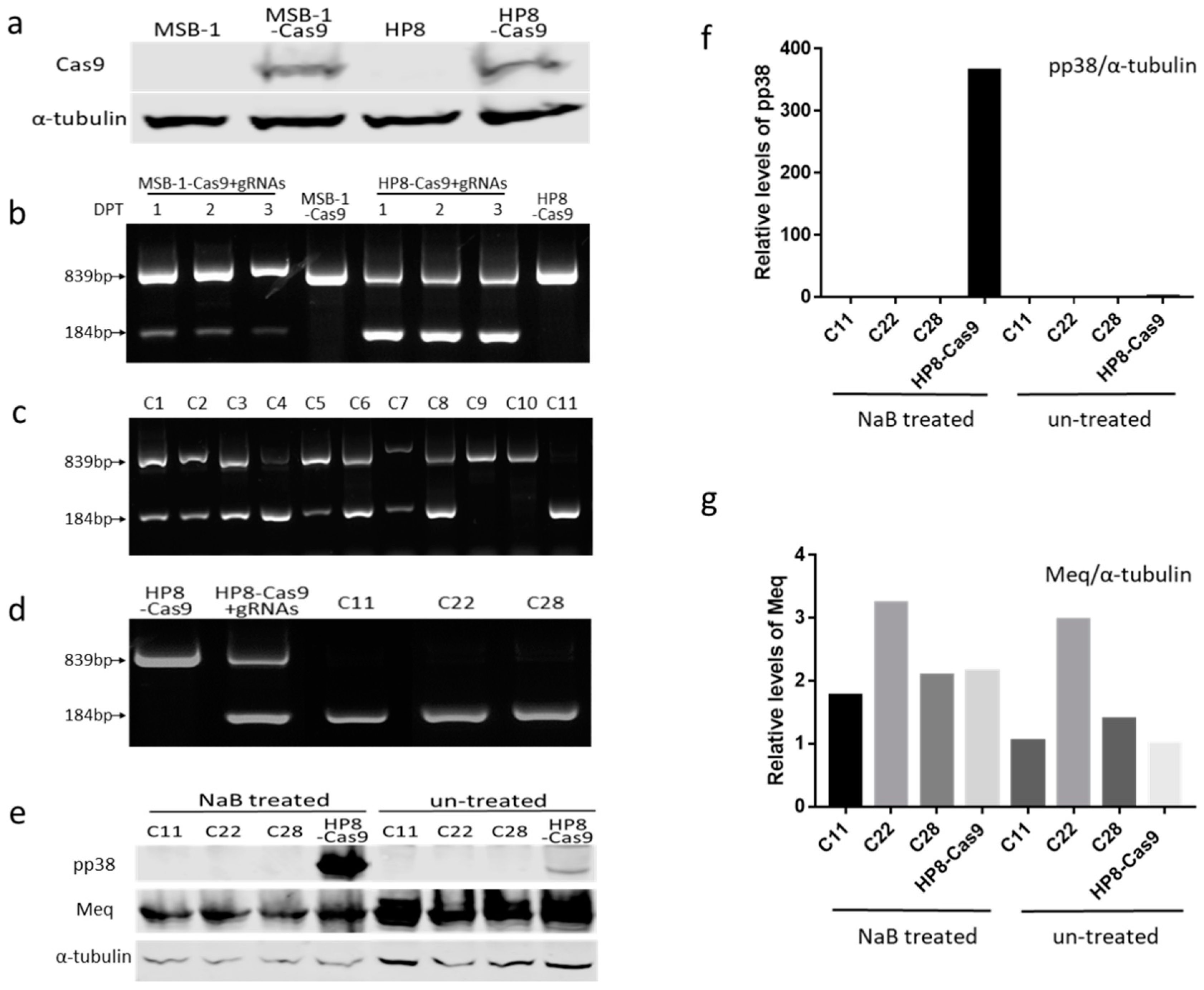
3.1. Editing of pp38 in MSB-1 Cells
3.2. Editing of pp38 in HP8 Cell Line
3.3. Insertion of Marker Gene into MDV Genome in HP8 Cells by NHEJ Pathway
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Delecluse, H.J.; Hammerschmidt, W. Status of marek’s disease virus in established lymphoma cell lines: Herpesvirus integration is common. J. Virol. 1993, 67, 82–92. [Google Scholar] [PubMed]
- Kaufer, B.B.; Jarosinski, K.W.; Osterrieder, N. Herpesvirus telomeric repeats facilitate genomic integration into host telomeres and mobilization of viral DNA during reactivation. J. Exp. Med. 2011, 208, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Delecluse, H.J.; Schuller, S.; Hammerschmidt, W. Latent marek’s disease virus can be activated from its chromosomally integrated state in herpesvirus-transformed lymphoma cells. EMBO J. 1993, 12, 3277–3286. [Google Scholar] [CrossRef] [PubMed]
- Greco, A.; Fester, N.; Engel, A.T.; Kaufer, B.B. Role of the short telomeric repeat region in marek’s disease virus replication, genomic integration, and lymphomagenesis. J. Virol. 2014, 88, 14138–14147. [Google Scholar] [CrossRef] [PubMed]
- Kaufer, B.B. Detection of integrated herpesvirus genomes by fluorescence in situ hybridization (fish). Methods Mol. Biol. 2013, 1064, 141–152. [Google Scholar]
- McPherson, M.C.; Cheng, H.H.; Delany, M.E. Marek’s disease herpesvirus vaccines integrate into chicken host chromosomes yet lack a virus-host phenotype associated with oncogenic transformation. Vaccine 2016, 34, 5554–5561. [Google Scholar] [CrossRef]
- Robinson, C.M.; Cheng, H.H.; Delany, M.E. Temporal kinetics of marek’s disease herpesvirus: Integration occurs early after infection in both b and t cells. Cytogenet. Genome Res. 2014, 144, 142–154. [Google Scholar] [CrossRef]
- Venugopal, K. Marek’s disease: An update on oncogenic mechanisms and control. Res. Vet. Sci. 2000, 69, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Biggs, P.M. The leeuwenhoek lecture, 1997. Marek’s disease herpesvirus: Oncogenesis and prevention. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1997, 352, 1951–1962. [Google Scholar] [CrossRef]
- Brown, A.C.; Baigent, S.J.; Smith, L.P.; Chattoo, J.P.; Petherbridge, L.J.; Hawes, P.; Allday, M.J.; Nair, V. Interaction of meq protein and c-terminal-binding protein is critical for induction of lymphomas by marek’s disease virus. Proc. Natl. Acad. Sci. USA 2006, 103, 1687–1692. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Lee, L.F.; Hunt, H.D.; Reed, W.M.; Lupiani, B.; Reddy, S.M. A marek’s disease virus vil-8 deletion mutant has attenuated virulence and confers protection against challenge with a very virulent plus strain. Avian Dis. 2005, 49, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Lee, L.F.; Reed, W.M.; Kung, H.J.; Reddy, S.M. Marek’s disease virus-encoded vil-8 gene is involved in early cytolytic infection but dispensable for establishment of latency. J. Virol. 2004, 78, 4753–4760. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.Z.; Lee, L.F.; Liu, J.L.; Kung, H.J. Structural analysis and transcriptional mapping of the marek’s disease virus gene encoding pp38, an antigen associated with transformed cells. J. Virol. 1991, 65, 6509–6515. [Google Scholar] [PubMed]
- Dorange, F.; Tischer, B.K.; Vautherot, J.F.; Osterrieder, N. Characterization of marek’s disease virus serotype 1 (mdv-1) deletion mutants that lack ul46 to ul49 genes: Mdv-1 ul49, encoding vp22, is indispensable for virus growth. J. Virol. 2002, 76, 1959–1970. [Google Scholar] [CrossRef] [PubMed]
- Fragnet, L.; Blasco, M.A.; Klapper, W.; Rasschaert, D. The rna subunit of telomerase is encoded by marek’s disease virus. J. Virol. 2003, 77, 5985–5996. [Google Scholar] [CrossRef]
- Jarosinski, K.W.; Osterrieder, N.; Nair, V.K.; Schat, K.A. Attenuation of marek’s disease virus by deletion of open reading frame rlorf4 but not rlorf5a. J. Virol. 2005, 79, 11647–11659. [Google Scholar] [CrossRef]
- Li, Y.; Sun, A.; Su, S.; Zhao, P.; Cui, Z.; Zhu, H. Deletion of the meq gene significantly decreases immunosuppression in chickens caused by pathogenic marek’s disease virus. Virol. J. 2011, 8, 2. [Google Scholar] [CrossRef]
- Lupiani, B.; Lee, L.F.; Cui, X.; Gimeno, I.; Anderson, A.; Morgan, R.W.; Silva, R.F.; Witter, R.L.; Kung, H.J.; Reddy, S.M. Marek’s disease virus-encoded meq gene is involved in transformation of lymphocytes but is dispensable for replication. Proc. Natl. Acad. Sci. USA 2004, 101, 11815–11820. [Google Scholar] [CrossRef]
- Parcells, M.S.; Lin, S.F.; Dienglewicz, R.L.; Majerciak, V.; Robinson, D.R.; Chen, H.C.; Wu, Z.; Dubyak, G.R.; Brunovskis, P.; Hunt, H.D.; et al. Marek’s disease virus (mdv) encodes an interleukin-8 homolog (vil-8): Characterization of the vil-8 protein and a vil-8 deletion mutant mdv. J. Virol. 2001, 75, 5159–5173. [Google Scholar] [CrossRef]
- Reddy, S.M.; Lupiani, B.; Gimeno, I.M.; Silva, R.F.; Lee, L.F.; Witter, R.L. Rescue of a pathogenic marek’s disease virus with overlapping cosmid dnas: Use of a pp38 mutant to validate the technology for the study of gene function. Proc. Natl. Acad. Sci. USA 2002, 99, 7054–7059. [Google Scholar] [CrossRef]
- Sun, A.; Lee, L.F.; Khan, O.A.; Heidari, M.; Zhang, H.; Lupiani, B.; Reddy, S.M. Deletion of marek’s disease virus large subunit of ribonucleotide reductase impairs virus growth in vitro and in vivo. Avian Dis. 2013, 57, 464–468. [Google Scholar] [CrossRef]
- Trapp, S.; Parcells, M.S.; Kamil, J.P.; Schumacher, D.; Tischer, B.K.; Kumar, P.M.; Nair, V.K.; Osterrieder, N. A virus-encoded telomerase rna promotes malignant t cell lymphomagenesis. J. Exp. Med. 2006, 203, 1307–1317. [Google Scholar] [CrossRef]
- Cortes, P.L.; Cardona, C.J. Pathogenesis of a marek’s disease virus mutant lacking vil-8 in resistant and susceptible chickens. Avian Dis. 2004, 48, 50–60. [Google Scholar] [CrossRef]
- Petherbridge, L.; Brown, A.C.; Baigent, S.J.; Howes, K.; Sacco, M.A.; Osterrieder, N.; Nair, V.K. Oncogenicity of virulent marek’s disease virus cloned as bacterial artificial chromosomes. J. Virol. 2004, 78, 13376–13380. [Google Scholar] [CrossRef]
- Nair, V.; Kung, H.J. Marek’s disease virus oncogenicity: Molecular mechanisms. In Marek’s Disease: An Evolving Problem, 1st ed.; Davison, F., Nair, V., Eds.; Elsevier Academic Press: London, UK, 2004; pp. 32–48. [Google Scholar]
- Yu, Z.H.; Teng, M.; Sun, A.J.; Yu, L.L.; Hu, B.; Qu, L.H.; Ding, K.; Cheng, X.C.; Liu, J.X.; Cui, Z.Z.; et al. Virus-encoded mir-155 ortholog is an important potential regulator but not essential for the development of lymphomas induced by very virulent marek’s disease virus. Virology 2014, 448, 55–64. [Google Scholar] [CrossRef]
- Zhao, Y.; Xu, H.; Yao, Y.; Smith, L.P.; Kgosana, L.; Green, J.; Petherbridge, L.; Baigent, S.J.; Nair, V. Critical role of the virus-encoded microrna-155 ortholog in the induction of marek’s disease lymphomas. PLoS Pathog. 2011, 7, e1001305. [Google Scholar] [CrossRef]
- Kaufer, B.B.; Arndt, S.; Trapp, S.; Osterrieder, N.; Jarosinski, K.W. Herpesvirus telomerase rna (vtr) with a mutated template sequence abrogates herpesvirus-induced lymphomagenesis. PLoS Pathog. 2011, 7, e1002333. [Google Scholar] [CrossRef]
- Mwangi, W.N.; Smith, L.P.; Baigent, S.J.; Beal, R.K.; Nair, V.; Smith, A.L. Clonal structure of rapid-onset mdv-driven cd4+ lymphomas and responding cd8+ t cells. PLoS Pathog. 2011, 7, e1001337. [Google Scholar] [CrossRef]
- Brown, A.C.; Nair, V.; Allday, M.J. Epigenetic regulation of the latency-associated region of marek’s disease virus in tumor-derived t-cell lines and primary lymphoma. J. Virol. 2012, 86, 1683–1695. [Google Scholar] [CrossRef]
- Mwangi, W.N.; Vasoya, D.; Kgosana, L.B.; Watson, M.; Nair, V. Differentially expressed genes during spontaneous lytic switch of marek’s disease virus in lymphoblastoid cell lines determined by global gene expression profiling. J. Gen. Virol. 2017, 98, 779–790. [Google Scholar] [CrossRef]
- Greenfeld, H.; Takasaki, K.; Walsh, M.J.; Ersing, I.; Bernhardt, K.; Ma, Y.; Fu, B.; Ashbaugh, C.W.; Cabo, J.; Mollo, S.B.; et al. Traf1 coordinates polyubiquitin signaling to enhance epstein-barr virus lmp1-mediated growth and survival pathway activation. PLoS Pathog. 2015, 11, e1004890. [Google Scholar] [CrossRef]
- Van Diemen, F.R.; Kruse, E.M.; Hooykaas, M.J.; Bruggeling, C.E.; Schurch, A.C.; van Ham, P.M.; Imhof, S.M.; Nijhuis, M.; Wiertz, E.J.; Lebbink, R.J. Crispr/cas9-mediated genome editing of herpesviruses limits productive and latent infections. PLoS Pathog. 2016, 12, e1005701. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Walsh, M.J.; Bernhardt, K.; Ashbaugh, C.W.; Trudeau, S.J.; Ashbaugh, I.Y.; Jiang, S.; Jiang, C.; Zhao, B.; Root, D.E.; et al. Crispr/cas9 screens reveal epstein-barr virus-transformed b cell host dependency factors. Cell Host Microbe 2017, 21, 580–591.e7. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Zhou, H.; Liang, J.; Gerdt, C.; Wang, C.; Ke, L.; Schmidt, S.C.S.; Narita, Y.; Ma, Y.; Wang, S.; et al. The epstein-barr virus regulome in lymphoblastoid cells. Cell Host Microbe 2017, 22, 561–573.e4. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Bassett, A.; Nair, V. Targeted editing of avian herpesvirus vaccine vector using crispr/cas9 nucleases. J. Vaccine Technol. 2016, 1, 1–7. [Google Scholar]
- Tang, N.; Zhang, Y.; Pedrera, M.; Chang, P.; Baigent, S.; Moffat, K.; Shen, Z.; Nair, V.; Yao, Y. A simple and rapid approach to develop recombinant avian herpesvirus vectored vaccines using crispr/cas9 system. Vaccine 2018, 36, 716–722. [Google Scholar] [CrossRef] [PubMed]
- Gimeno, I.M.; Witter, R.L.; Hunt, H.D.; Reddy, S.M.; Lee, L.F.; Silva, R.F. The pp38 gene of marek’s disease virus (mdv) is necessary for cytolytic infection of b cells and maintenance of the transformed state but not for cytolytic infection of the feather follicle epithelium and horizontal spread of mdv. J. Virol. 2005, 79, 4545–4549. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tang, N.; Sadigh, Y.; Baigent, S.; Shen, Z.; Nair, V.; Yao, Y. Application of crispr/cas9 gene editing system on mdv-1 genome for the study of gene function. Viruses 2018, 10, 279. [Google Scholar] [CrossRef] [PubMed]
- Nazerian, K.; Witter, R.L. Properties of a chicken lymphoblastoid cell line from marek’s disease tumor. J. Natl. Cancer Inst. 1975, 54, 453–458. [Google Scholar]
- Akiyama, Y.; Kato, S. Two cell lines from lymphomas of marek’s disease. Biken J. 1974, 17, 105–116. [Google Scholar]
- Tang, N.; Zhang, Y.; Pedrera, M.; Chang, P.; Baigent, S.; Moffat, K.; Shen, Z.; Nair, V.; Yao, Y. Generating recombinant avian herpesvirus vectors with crispr/cas9 gene editing. J. Vis. Exp. 2019, 143, e58193. [Google Scholar] [CrossRef] [PubMed]
- Baigent, S.J.; Petherbridge, L.J.; Howes, K.; Smith, L.P.; Currie, R.J.; Nair, V.K. Absolute quantitation of marek’s disease virus genome copy number in chicken feather and lymphocyte samples using real-time pcr. J. Virol. Methods 2005, 123, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.T.N.; Nair, V.; Yao, Y.; The Pirbright Institute, Pirbright, UK. pp38 sequence edited by CRISPR/Cas9 system. Unpublished work. 2018. [Google Scholar]
- Zhang, Y.T.N.; Nair, V.; Yao, Y.; The Pirbright Institute, Pirbright, UK. Editing of pp38 in mdv cell line msb-1 by crispr/cas9 system. Unpublished work. 2018. [Google Scholar]
- Xie, Q.; Anderson, A.S.; Morgan, R.W. Marek’s disease virus (mdv) icp4, pp38, and meq genes are involved in the maintenance of transformation of mdcc-msb1 mdv-transformed lymphoblastoid cells. J. Virol. 1996, 70, 1125–1131. [Google Scholar] [PubMed]
- Cohrs, R.J.; Gilden, D.H. Human herpesvirus latency. Brain Pathol. 2001, 11, 465–474. [Google Scholar] [CrossRef]
- Van Diemen, F.R.; Lebbink, R.J. Crispr/cas9, a powerful tool to target human herpesviruses. Cell. Microbiol. 2017, 19. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.N.V.; The Pirbright Institute, Pirbright, UK. Mdv integration determined by fish. Unpublished work. 2013. [Google Scholar]
- Chen, X.B.; Sondermeijer, P.J.; Velicer, L.F. Identification of a unique marek’s disease virus gene which encodes a 38-kilodalton phosphoprotein and is expressed in both lytically infected cells and latently infected lymphoblastoid tumor cells. J. Virol. 1992, 66, 85–94. [Google Scholar] [PubMed]
- Sadign, Y.T.N.; Nair, V.; Yao, Y.; The Pirbright Institute, Pirbright, UK. Off-target effect of HVT genome editing by CRISPR/Cas9 system. Unpublished work. 2019. [Google Scholar]
| Primer | Sequence (5′-3′) |
|---|---|
| NF | TTGGAATAGCCCCCTTCCCC |
| NR | TTCGAAGCAGAACACGAAGGG |
| CF | GATTCCACCTCCCCAGAATCC |
| CR | CAGAGAATGCAACAATGCGT |
| MF | ATGGTGAGCAAGGGCGA |
| MR | CCGGTGGTGCAGATGAAC |
| ovoF | CACTGCCACTGGGCTCTGT |
| ovoR | GCAATGGCAATAAACCTCCAA |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Luo, J.; Tang, N.; Teng, M.; Reddy, V.R.A.P.; Moffat, K.; Shen, Z.; Nair, V.; Yao, Y. Targeted Editing of the pp38 Gene in Marek’s Disease Virus-Transformed Cell Lines Using CRISPR/Cas9 System. Viruses 2019, 11, 391. https://doi.org/10.3390/v11050391
Zhang Y, Luo J, Tang N, Teng M, Reddy VRAP, Moffat K, Shen Z, Nair V, Yao Y. Targeted Editing of the pp38 Gene in Marek’s Disease Virus-Transformed Cell Lines Using CRISPR/Cas9 System. Viruses. 2019; 11(5):391. https://doi.org/10.3390/v11050391
Chicago/Turabian StyleZhang, Yaoyao, Jun Luo, Na Tang, Man Teng, Vishwanatha R.A.P. Reddy, Katy Moffat, Zhiqiang Shen, Venugopal Nair, and Yongxiu Yao. 2019. "Targeted Editing of the pp38 Gene in Marek’s Disease Virus-Transformed Cell Lines Using CRISPR/Cas9 System" Viruses 11, no. 5: 391. https://doi.org/10.3390/v11050391
APA StyleZhang, Y., Luo, J., Tang, N., Teng, M., Reddy, V. R. A. P., Moffat, K., Shen, Z., Nair, V., & Yao, Y. (2019). Targeted Editing of the pp38 Gene in Marek’s Disease Virus-Transformed Cell Lines Using CRISPR/Cas9 System. Viruses, 11(5), 391. https://doi.org/10.3390/v11050391






