Genomics Analyses of GIV and GVI Noroviruses Reveal the Distinct Clustering of Human and Animal Viruses
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. RNA Extraction and RT-PCR
2.3. Sequence and Phylogenetic Analyses
2.4. Structural Modeling
3. Results
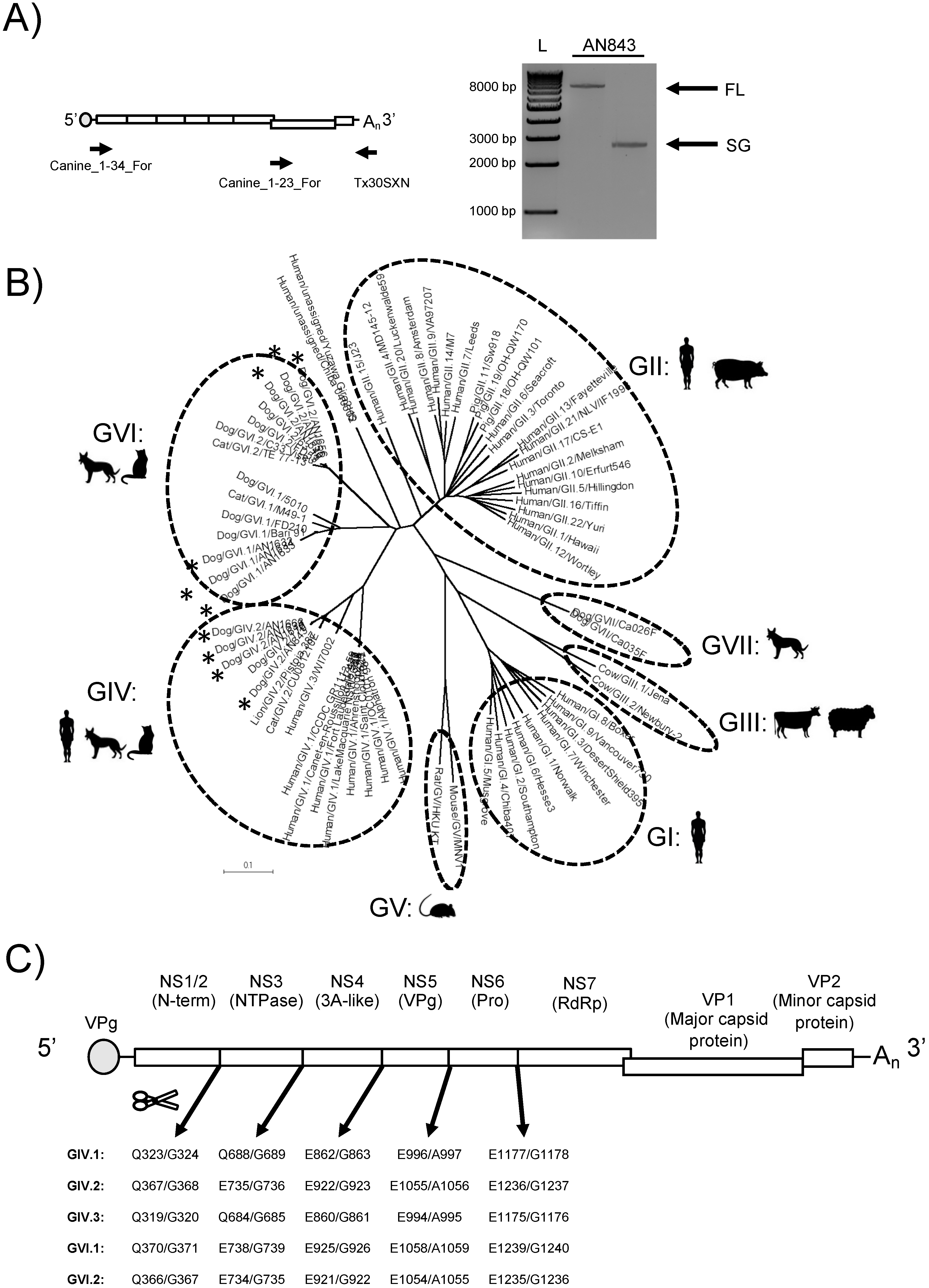
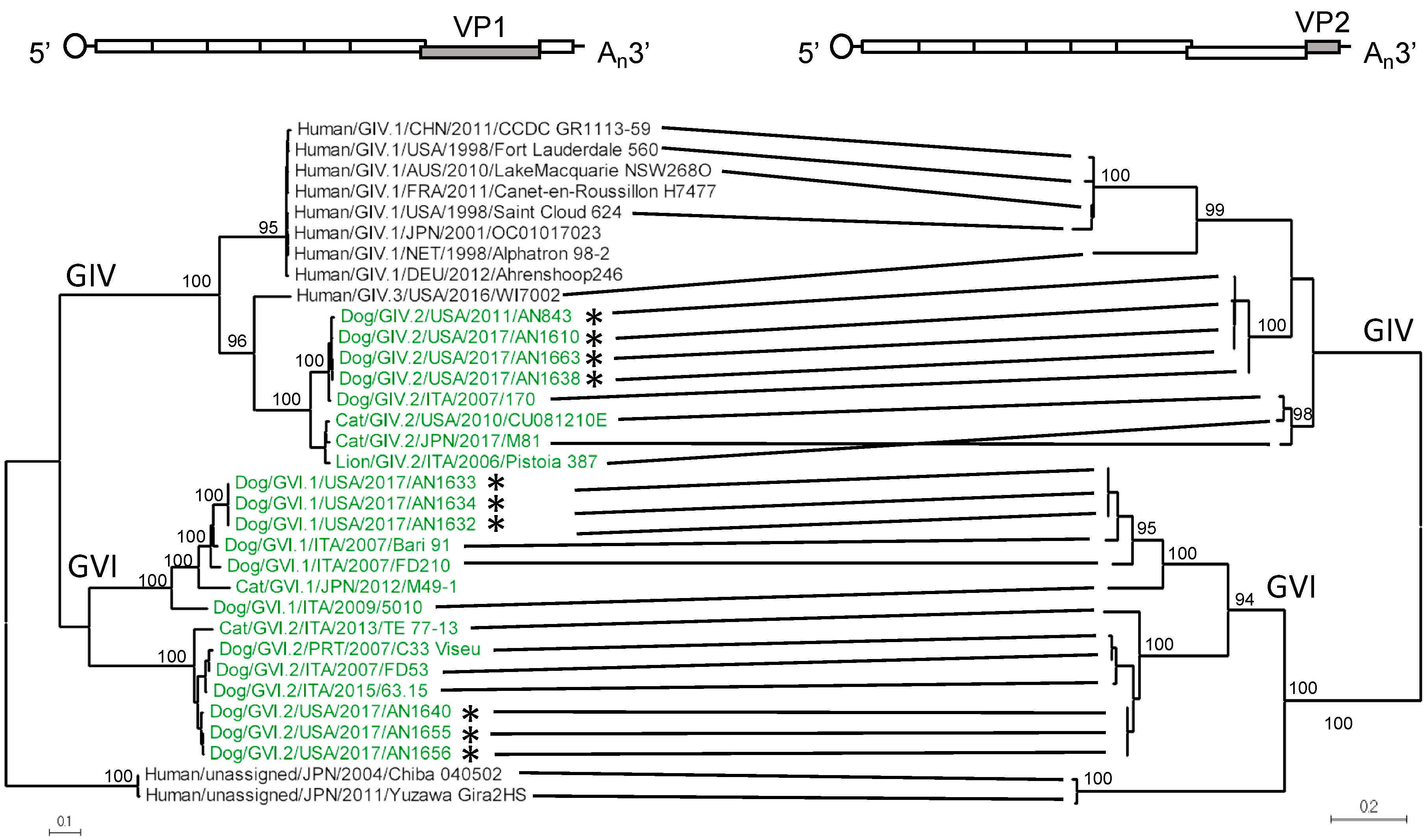
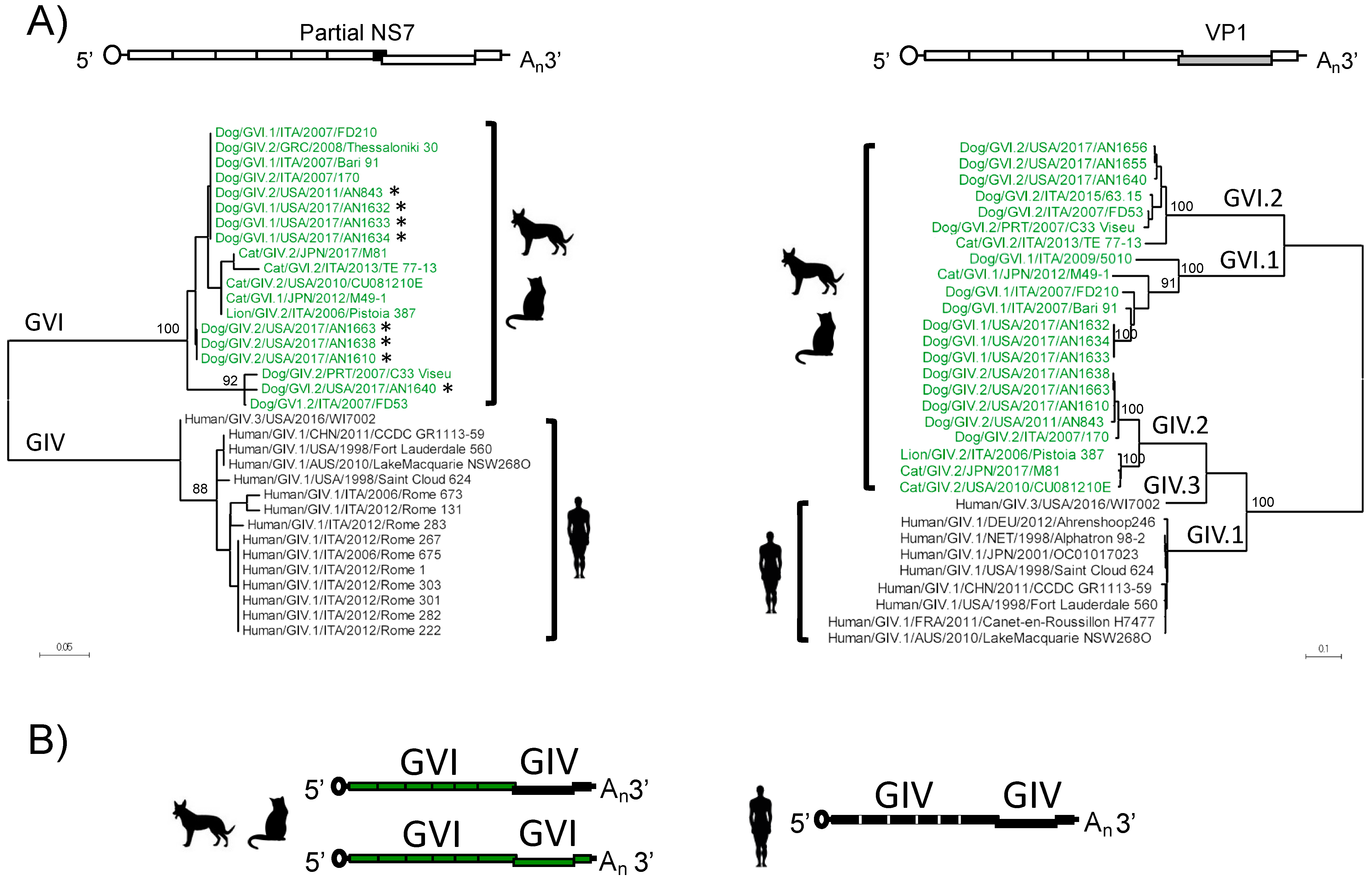
3.1. Amplification, Sequencing, and Characterization of Novel Canine Norovirus Strains
3.2. Structural Analyses of VP1 Reveal Species-Specific Differences between Human, Canine, and Feline Capsid Proteins
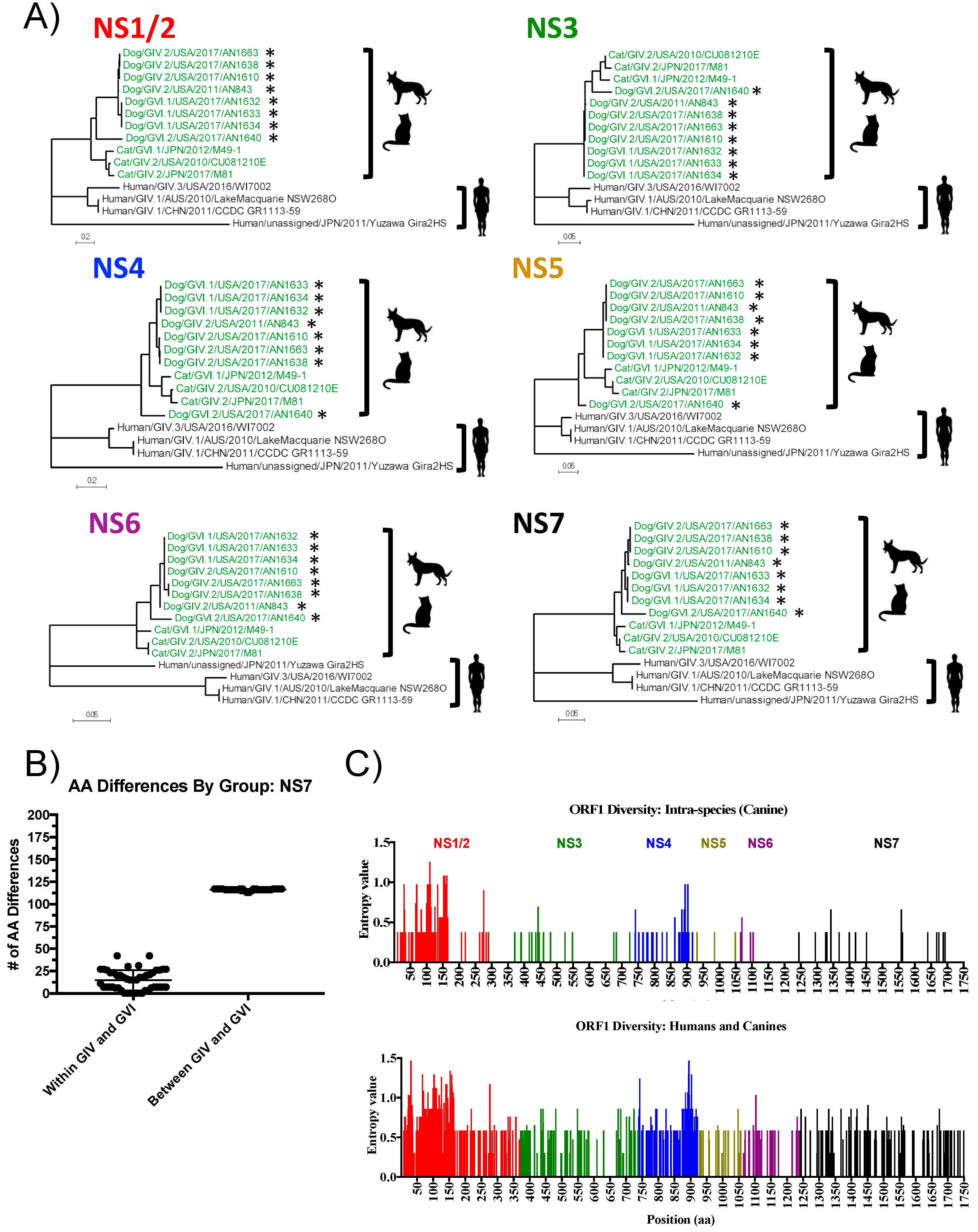
3.3. Analyses of Non-Structural Proteins Suggest Order-Specific Differences within the Replication Machinery
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Disclaimer
Conflicts of Interest
References
- Rockx, B.H.; Vennema, H.; Hoebe, C.J.; Duizer, E.; Koopmans, M.P. Association of histo-blood group antigens and susceptibility to norovirus infections. J. Infect. Dis. 2005, 191, 749–754. [Google Scholar] [CrossRef] [PubMed]
- Ettayebi, K.; Crawford, S.E.; Murakami, K.; Broughman, J.R.; Karandikar, U.; Tenge, V.R.; Neill, F.H.; Blutt, S.E.; Zeng, X.L.; et al. Replication of human noroviruses in stem cell-derived human enteroids. Science 2016, 353, 1387–1393. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.M.; Hutson, A.M.; Estes, M.K.; Prasad, B.V. Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus. Proc. Natl. Acad. Sci. USA 2008, 105, 9175–9180. [Google Scholar] [CrossRef] [PubMed]
- Prasad, B.V.; Hardy, M.E.; Dokland, T.; Bella, J.; Rossmann, M.G.; Estes, M.K. X-ray crystallographic structure of the Norwalk virus capsid. Science 1999, 286, 287–290. [Google Scholar] [CrossRef] [PubMed]
- Vongpunsawad, S.; Prasad, B.V.V.; Estes, M.K. Norwalk Virus Minor Capsid Protein VP2 Associates within the VP1 Shell Domain. J. Virol. 2013, 87, 4818–4825. [Google Scholar] [CrossRef] [PubMed]
- Glass, P.J.; White, L.J.; Ball, J.M.; Leparc-Goffart, I.; Hardy, M.E.; Estes, M.K. Norwalk virus open reading frame 3 encodes a minor structural protein. J. Virol. 2000, 74, 6581–6591. [Google Scholar] [CrossRef] [PubMed]
- Conley, M.J.; McElwee, M.; Azmi, L.; Gabrielsen, M.; Byron, O.; Goodfellow, I.G.; Bhella, D. Calicivirus VP2 forms a portal-like assembly following receptor engagement. Nature 2019, 565, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Green, K.Y. Caliciviridae: The Noroviruses. In Fields Virology; Knipe, P.M., Ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; pp. 582–608. [Google Scholar]
- Pires, S.M.; Fischer-Walker, C.L.; Lanata, C.F.; Devleesschauwer, B.; Hall, A.J.; Kirk, M.D.; Duarte, A.S.; Black, R.E.; Angulo, F.J. Aetiology-Specific Estimates of the Global and Regional Incidence and Mortality of Diarrhoeal Diseases Commonly Transmitted through Food. PLoS ONE 2015, 10, e0142927. [Google Scholar] [CrossRef] [PubMed]
- Martella, V.; Decaro, N.; Lorusso, E.; Radogna, A.; Moschidou, P.; Amorisco, F.; Lucente, M.S.; Desario, C.; Mari, V.; Elia, G.; et al. Genetic heterogeneity and recombination in canine noroviruses. J. Virol. 2009, 83, 11391–11396. [Google Scholar] [CrossRef] [PubMed]
- Bodnar, L.; Lorusso, E.; di Martino, B.; Catella, C.; Lanave, G.; Elia, G.; Banyai, K.; Buonavoglia, C.; Martella, V. Identification of a novel canine norovirus. Infect. Genet. Evol. 2017, 52, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Mesquita, J.R.; Barclay, L.; Nascimento, M.S.; Vinje, J. Novel norovirus in dogs with diarrhea. Emerg. Infect. Dis. 2010, 16, 980–982. [Google Scholar] [CrossRef] [PubMed]
- Pinto, P.; Wang, Q.; Chen, N.; Dubovi, E.J.; Daniels, J.B.; Millward, L.M.; Buonavoglia, C.; Martella, V.; Saif, L.J. Discovery and genomic characterization of noroviruses from a gastroenteritis outbreak in domestic cats in the US. PLoS ONE 2012, 7, e32739. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, B.; Marsilio, F.; di Profio, F.; Lorusso, E.; Friedrich, K.G.; Buonavoglia, C.; Martella, V. Detection of antibodies against norovirus genogroup GIV in carnivores. Clin. Vaccine Immunol. 2010, 17, 180–182. [Google Scholar] [CrossRef] [PubMed]
- Caddy, S.; Emmott, E.; El-Attar, L.; Mitchell, J.; de Rougemont, A.; Brownlie, J.; Goodfellow, I. Serological evidence for multiple strains of canine norovirus in the UK dog population. PLoS ONE 2013, 8, e81596. [Google Scholar] [CrossRef] [PubMed]
- Mesquita, J.R.; Delgado, I.; Costantini, V.; Heenemann, K.; Vahlenkamp, T.W.; Vinje, J.; Nascimento, M.S. Seroprevalence of canine norovirus in 14 European countries. Clin. Vaccine Immunol. 2014, 21, 898–900. [Google Scholar] [CrossRef] [PubMed]
- Caddy, S.; Breiman, A.; le Pendu, J.; Goodfellow, I. Genogroup IV and VI canine noroviruses interact with histo-blood group antigens. J. Virol. 2014, 88, 10377–10391. [Google Scholar] [CrossRef] [PubMed]
- Caddy, S.L.; de Rougemont, A.; Emmott, E.; El-Attar, L.; Mitchell, J.A.; Hollinshead, M.; Belliot, G.; Brownlie, J.; le Pendu, J.; Goodfellow, I. Evidence for human norovirus infection of dogs in the United kingdom. J. Clin. Microbiol. 2015, 53, 1873–1883. [Google Scholar] [CrossRef] [PubMed]
- Summa, M.; von Bonsdorff, C.H.; Maunula, L. Pet dogs—A transmission route for human noroviruses? J. Clin. Virol. 2012, 53, 244–247. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, B.; di Profio, F.; Ceci, C.; di Felice, E.; Green, K.Y.; Bok, K.; de Grazia, S.; Giammanco, G.M.; Massirio, I.; Lorusso, E.; et al. Seroprevalence of norovirus genogroup IV antibodies among humans, Italy, 2010–2011. Emerg. Infect. Dis. 2014, 20, 1828–1832. [Google Scholar] [CrossRef] [PubMed]
- Katayama, K.; Shirato-Horikoshi, H.; Kojima, S.; Kageyama, T.; Oka, T.; Hoshino, F.; Fukushi, S.; Shinohara, M.; Uchida, K.; Suzuki, Y.; et al. Phylogenetic analysis of the complete genome of 18 Norwalk-like viruses. Virology 2002, 299, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Parra, G.I.; Squires, R.B.; Karangwa, C.K.; Johnson, J.A.; Lepore, C.J.; Sosnovtsev, S.V.; Green, K.Y. Static and Evolving Norovirus Genotypes: Implications for Epidemiology and Immunity. PLoS Pathog. 2017, 13, e1006136. [Google Scholar] [CrossRef] [PubMed]
- Simonyan, V.; Chumakov, K.; Dingerdissen, H.; Faison, W.; Goldweber, S.; Golikov, A.; Gulzar, N.; Karagiannis, K.; Lam, P.V.N.; Maudru, T.; et al. High-performance integrated virtual environment (HIVE): A robust infrastructure for next-generation sequence data analysis. Database (Oxford) 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Simonyan, V.; Mazumder, R. High-Performance Integrated Virtual Environment (HIVE) Tools and Applications for Big Data Analysis. Genes (Basel) 2014, 5, 957–981. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Huang, P.W.; Zhong, W.M.; Farkas, T.; Cubitt, D.W.; Matson, D.O. Design and evaluation of a primer pair that detects both Norwalk- and Sapporo-like caliciviruses by RT-PCR. J. Virol. Methods 1999, 83, 145–154. [Google Scholar] [CrossRef]
- Bull, R.A.; Hansman, G.S.; Clancy, L.E.; Tanaka, M.M.; Rawlinson, W.D.; White, P.A. Norovirus recombination in ORF1/ORF2 overlap. Emerg. Infect. Dis. 2005, 11, 1079–1085. [Google Scholar] [CrossRef] [PubMed]
- Thackray, L.B.; Wobus, C.E.; Chachu, K.A.; Liu, B.; Alegre, E.R.; Henderson, K.S.; Kelley, S.T.; Virgin, H.W.T. Murine noroviruses comprising a single genogroup exhibit biological diversity despite limited sequence divergence. J. Virol. 2007, 81, 10460–10473. [Google Scholar] [CrossRef] [PubMed]
- Eden, J.S.; Tanaka, M.M.; Boni, M.F.; Rawlinson, W.D.; White, P.A. Recombination within the pandemic norovirus GII.4 lineage. J. Virol. 2013, 87, 6270–6282. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.K.; Glatt, S.; Ferrer, J.L.; Koromyslova, A.D.; Leuthold, M.M.; Dunder, J.; Hansman, G.S. Structural analysis of a feline norovirus protruding domain. Virology 2015, 474, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Cheetham, S.; Souza, M.; Meulia, T.; Grimes, S.; Han, M.G.; Saif, L.J. Pathogenesis of a genogroup II human norovirus in gnotobiotic pigs. J. Virol. 2006, 80, 10372–10381. [Google Scholar] [CrossRef] [PubMed]
- Bok, K.; Parra, G.I.; Mitra, T.; Abente, E.; Shaver, C.K.; Boon, D.; Engle, R.; Yu, C.; Kapikian, A.Z.; Sosnovtsev, S.V.; et al. Chimpanzees as an animal model for human norovirus infection and vaccine development. Proc. Natl. Acad. Sci. USA 2011, 108, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Rockx, B.H.; Bogers, W.M.; Heeney, J.L.; van Amerongen, G.; Koopmans, M.P. Experimental norovirus infections in non-human primates. J. Med. Virol. 2005, 75, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Bui, T.; Kocher, J.; Li, Y.; Wen, K.; Li, G.; Liu, F.; Yang, X.; LeRoith, T.; Tan, M.; Xia, M.; et al. Median infectious dose of human norovirus GII.4 in gnotobiotic pigs is decreased by simvastatin treatment and increased by age. J. Gen. Virol. 2013, 94, 2005–2016. [Google Scholar] [CrossRef] [PubMed]
- Taube, S.; Kolawole, A.O.; Hohne, M.; Wilkinson, J.E.; Handley, S.A.; Perry, J.W.; Thackray, L.B.; Akkina, R.; Wobus, C.E. A mouse model for human norovirus. MBio 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Liu, B.; Tao, Y.; Li, C.; Xia, M.; Zhong, W.; Jiang, X.; Liu, H.; Tan, M. Norovirus GII.17 Natural Infections in Rhesus Monkeys, China. Emerg. Infect. Dis. 2017, 23, 316–319. [Google Scholar] [CrossRef] [PubMed]
- Han, M.G.; Cheetham, S.; Azevedo, M.; Thomas, C.; Saif, L.J. Immune responses to bovine norovirus-like particles with various adjuvants and analysis of protection in gnotobiotic calves. Vaccine 2006, 24, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Takano, T.; Hiramatsu, K.; Matsuyama, M.; Mutoh, K.; Matsumoto, Y.; Fukushima, T.; Doki, T.; Kusuhara, H.; Hohdatsu, T. Viral shedding and clinical status of feline-norovirus-infected cats after reinfection with the same strain. Arch. Virol. 2018, 163, 1503–1510. [Google Scholar] [CrossRef] [PubMed]
- Matthijnssens, J.; van Ranst, M. Genotype constellation and evolution of group A rotaviruses infecting humans. Curr. Opin. Virol. 2012, 2, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Fujii, Y.; Shimoike, T.; Takagi, H.; Murakami, K.; Todaka-Takai, R.; Park, Y.; Katayama, K. Amplification of all 11 RNA segments of group A rotaviruses based on reverse transcription polymerase chain reaction. Microbiol. Immunol. 2012, 56, 630–638. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Donnelly, M.E.; Scholes, D.T.; George, K.S.; Hatta, M.; Kawaoka, Y.; Wentworth, D.E. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and Swine origin human influenza a viruses. J. Virol. 2009, 83, 10309–10313. [Google Scholar] [CrossRef] [PubMed]
- Haga, K.; Fujimoto, A.; Takai-Todaka, R.; Miki, M.; Doan, Y.H.; Murakami, K.; Yokoyama, M.; Murata, K.; Nakanishi, A.; Katayama, K. Functional receptor molecules CD300lf and CD300ld within the CD300 family enable murine noroviruses to infect cells. Proc. Natl. Acad. Sci. USA 2016, 113, E6248–E6255. [Google Scholar] [CrossRef] [PubMed]
- Orchard, R.C.; Wilen, C.B.; Doench, J.G.; Baldridge, M.T.; McCune, B.T.; Lee, Y.C.; Lee, S.; Pruett-Miller, S.M.; Nelson, C.A.; Fremont, D.H.; et al. Discovery of a proteinaceous cellular receptor for a norovirus. Science 2016, 353, 933–936. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Lou, Z.; Tan, M.; Chen, Y.; Liu, Y.; Zhang, Z.; Zhang, X.C.; Jiang, X.; Li, X.; Rao, Z. Structural basis for the recognition of blood group trisaccharides by norovirus. J. Virol. 2007, 81, 5949–5957. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Fengling, L.; Lianzhu, W.; Yuxiu, Z.; Yanhua, J. Function of VP2 protein in the stability of the secondary structure of virus-like particles of genogroup II norovirus at different pH levels: Function of VP2 protein in the stability of NoV VLPs. J. Microbiol. 2014, 52, 970–975. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Regev, D.; Watanabe, M.; Hickman, D.; Moussatche, N.; Jesus, D.M.; Kahan, S.M.; Napthine, S.; Brierley, I.; Hunter, R.N., 3rd; et al. Identification of immune and viral correlates of norovirus protective immunity through comparative study of intra-cluster norovirus strains. PLoS Pathog. 2013, 9, e1003592. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, B.; Marsilio, F. Feline calicivirus VP2 is involved in the self-assembly of the capsid protein into virus-like particles. Res. Vet. Sci. 2010, 89, 279–281. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.C.; Lee, N.; Ho, W.S.; Law, C.O.; Lau, T.C.; Tsui, S.K.; Sung, J.J. Covariation of major and minor viral capsid proteins in norovirus genogroup II genotype 4 strains. J. Virol. 2012, 86, 1227–1232. [Google Scholar] [CrossRef] [PubMed]
- Sosnovtsev, S.V.; Green, K.Y. Identification and genomic mapping of the ORF3 and VPg proteins in feline calicivirus virions. Virology 2000, 277, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Ludwig-Begall, L.F.; Mauroy, A.; Thiry, E. Norovirus recombinants: Recurrent in the field, recalcitrant in the lab—A scoping review of recombination and recombinant types of noroviruses. J. Gen. Virol. 2018, 99, 970–988. [Google Scholar] [CrossRef] [PubMed]
- Bull, R.A.; Tanaka, M.M.; White, P.A. Norovirus recombination. J. Gen. Virol. 2007, 88, 3347–3359. [Google Scholar] [CrossRef] [PubMed]
- Bossert, B.; Conzelmann, K.K. Respiratory syncytial virus (RSV) nonstructural (NS) proteins as host range determinants: A chimeric bovine RSV with NS genes from human RSV is attenuated in interferon-competent bovine cells. J. Virol. 2002, 76, 4287–4293. [Google Scholar] [CrossRef] [PubMed]
- Yin, F.H.; Lomax, N.B. Host range mutants of human rhinovirus in which nonstructural proteins are altered. J. Virol. 1983, 48, 410–418. [Google Scholar] [PubMed]
- Naffakh, N.; Massin, P.; Escriou, N.; Crescenzo-Chaigne, B.; van der Werf, S. Genetic analysis of the compatibility between polymerase proteins from human and avian strains of influenza A viruses. J. Gen. Virol. 2000, 81, 1283–1291. [Google Scholar] [CrossRef] [PubMed]
- Long, J.S.; Giotis, E.S.; Moncorge, O.; Frise, R.; Mistry, B.; James, J.; Morisson, M.; Iqbal, M.; Vignal, A.; Skinner, M.A.; et al. Species difference in ANP32A underlies influenza A virus polymerase host restriction. Nature 2016, 529, 101–104. [Google Scholar] [CrossRef] [PubMed]
- Hyde, J.L.; Mackenzie, J.M. Subcellular localization of the MNV-1 ORF1 proteins and their potential roles in the formation of the MNV-1 replication complex. Virology 2010, 406, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Doerflinger, S.Y.; Cortese, M.; Romero-Brey, I.; Menne, Z.; Tubiana, T.; Schenk, C.; White, P.A.; Bartenschlager, R.; Bressanelli, S.; Hansman, G.S.; et al. Membrane alterations induced by nonstructural proteins of human norovirus. PLoS Pathog. 2017, 13, e1006705. [Google Scholar] [CrossRef] [PubMed]
- Cotton, B.T.; Hyde, J.L.; Sarvestani, S.T.; Sosnovtsev, S.V.; Green, K.Y.; White, P.A.; Mackenzie, J.M. The Norovirus NS3 Protein Is a Dynamic Lipid- and Microtubule-Associated Protein Involved in Viral RNA Replication. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Emmott, E.; Sorgeloos, F.; Caddy, S.L.; Vashist, S.; Sosnovtsev, S.; Lloyd, R.; Heesom, K.; Locker, N.; Goodfellow, I. Norovirus-Mediated Modification of the Translational Landscape via Virus and Host-Induced Cleavage of Translation Initiation Factors. Mol. Cell Proteom. 2017, 16, S215–S229. [Google Scholar] [CrossRef] [PubMed]
- Yen, J.B.; Wei, L.H.; Chen, L.W.; Chen, L.Y.; Hung, C.H.; Wang, S.S.; Chang, P.J. Subcellular Localization and Functional Characterization of GII.4 Norovirus-Encoded NTPase. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Lateef, Z.; Gimenez, G.; Baker, E.S.; Ward, V.K. Transcriptomic analysis of human norovirus NS1-2 protein highlights a multifunctional role in murine monocytes. BMC Genom. 2017, 18, 39. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Vega, V.; Sosnovtsev, S.V.; Belliot, G.; King, A.D.; Mitra, T.; Gorbalenya, A.; Green, K.Y. Norwalk virus N-terminal nonstructural protein is associated with disassembly of the Golgi complex in transfected cells. J. Virol. 2004, 78, 4827–4837. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ford-Siltz, L.A.; Mullis, L.; Sanad, Y.M.; Tohma, K.; Lepore, C.J.; Azevedo, M.; Parra, G.I. Genomics Analyses of GIV and GVI Noroviruses Reveal the Distinct Clustering of Human and Animal Viruses. Viruses 2019, 11, 204. https://doi.org/10.3390/v11030204
Ford-Siltz LA, Mullis L, Sanad YM, Tohma K, Lepore CJ, Azevedo M, Parra GI. Genomics Analyses of GIV and GVI Noroviruses Reveal the Distinct Clustering of Human and Animal Viruses. Viruses. 2019; 11(3):204. https://doi.org/10.3390/v11030204
Chicago/Turabian StyleFord-Siltz, Lauren A., Lisa Mullis, Yasser M. Sanad, Kentaro Tohma, Cara J. Lepore, Marli Azevedo, and Gabriel I. Parra. 2019. "Genomics Analyses of GIV and GVI Noroviruses Reveal the Distinct Clustering of Human and Animal Viruses" Viruses 11, no. 3: 204. https://doi.org/10.3390/v11030204
APA StyleFord-Siltz, L. A., Mullis, L., Sanad, Y. M., Tohma, K., Lepore, C. J., Azevedo, M., & Parra, G. I. (2019). Genomics Analyses of GIV and GVI Noroviruses Reveal the Distinct Clustering of Human and Animal Viruses. Viruses, 11(3), 204. https://doi.org/10.3390/v11030204





