Filovirus Virulence in Interferon α/β and γ Double Knockout Mice, and Treatment with Favipiravir
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Cells and Viruses
2.3. Animal Studies
2.4. Mouse Challenge and Observations
2.5. Susceptibility Study
2.6. Serial Dosing Study
2.7. Natural History Study
2.8. EBOV qRT-PCR
2.9. Hematology and Serum Chemistry
2.10. Histology and Immunohistochemistry
2.11. Favipiravir Treatment
3. Results
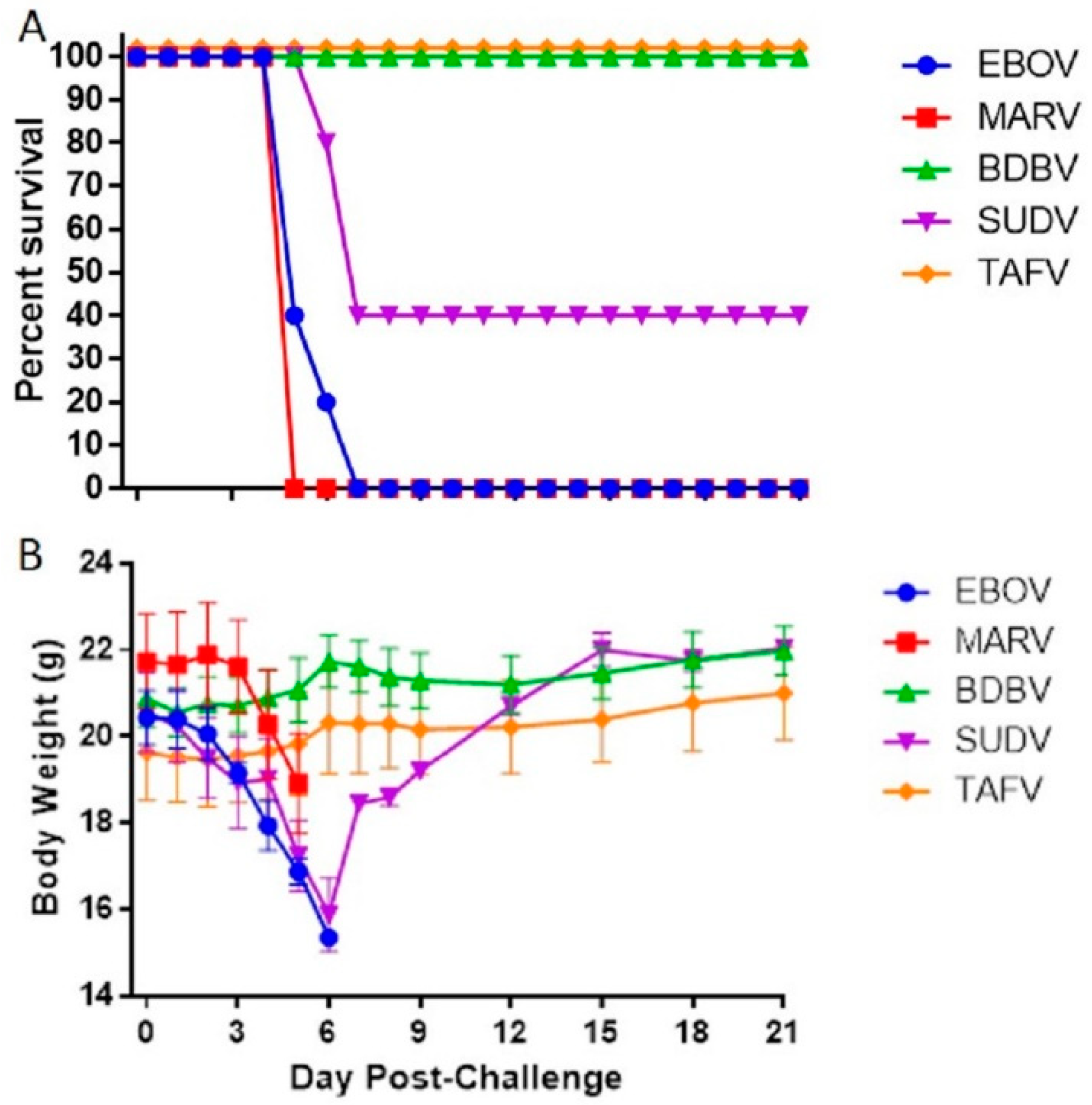
3.1. Susceptibility of IFNAGR KO Mice to Filoviruses
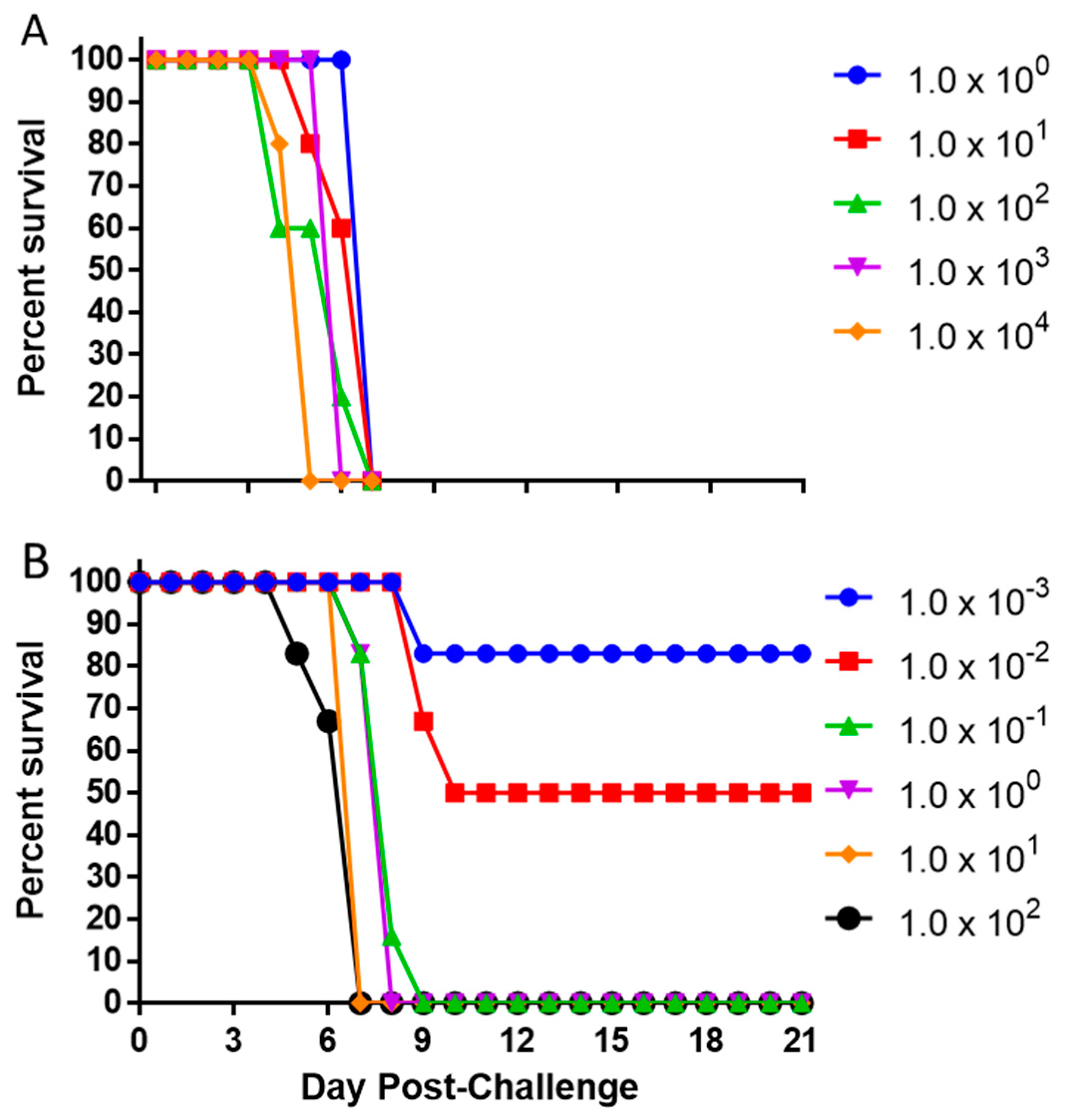
3.2. Determination of the Lethal Dose Range of EBOV in the IFNAGR KO Mouse Model
3.3. Progression of Ebola Virus Disease in the IFNAGR Mouse Model
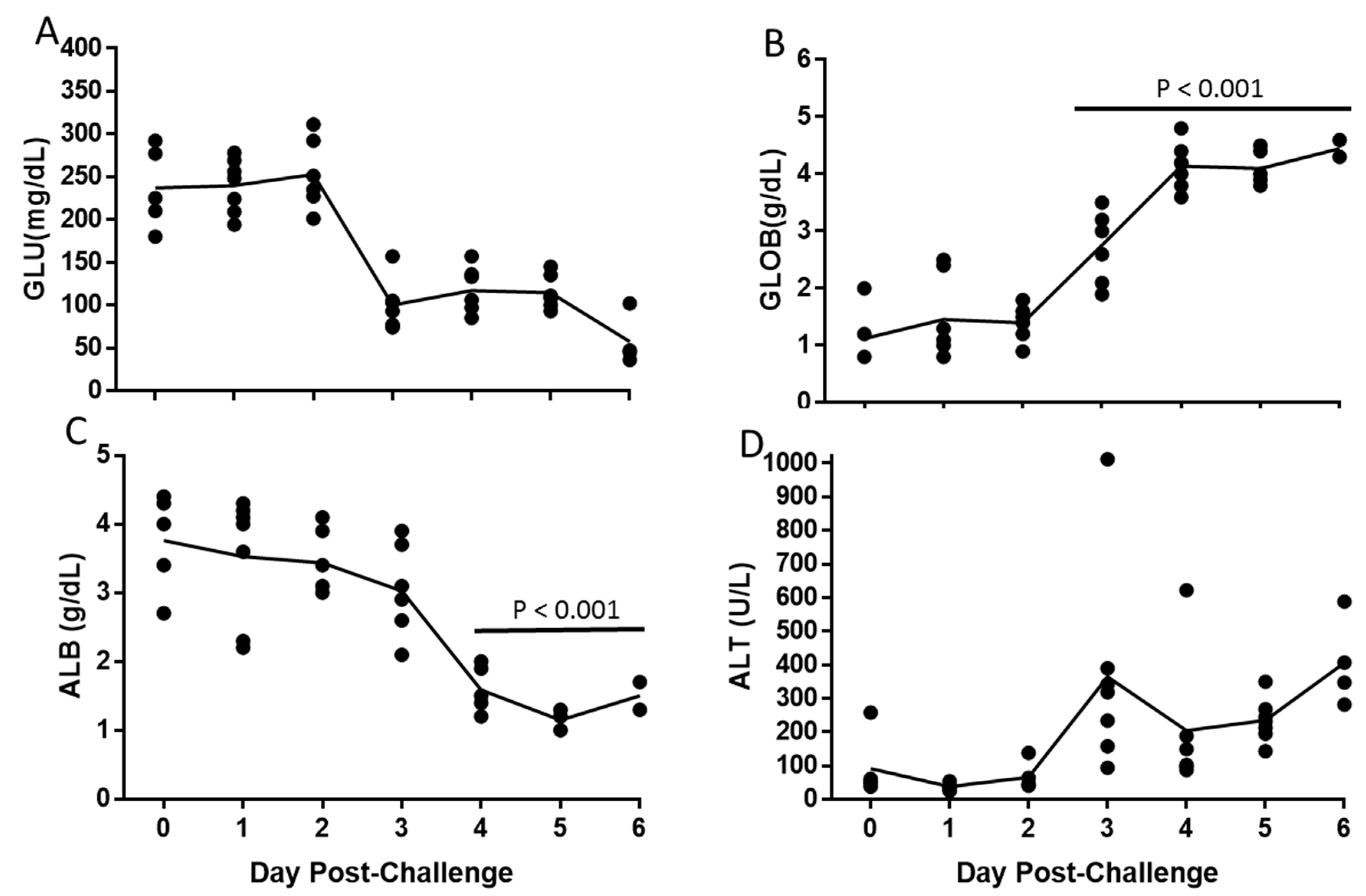
3.4. Alteration of Hematologic and Liver Parameters During the Course of EBOV Infection in IFANGR KO Mice
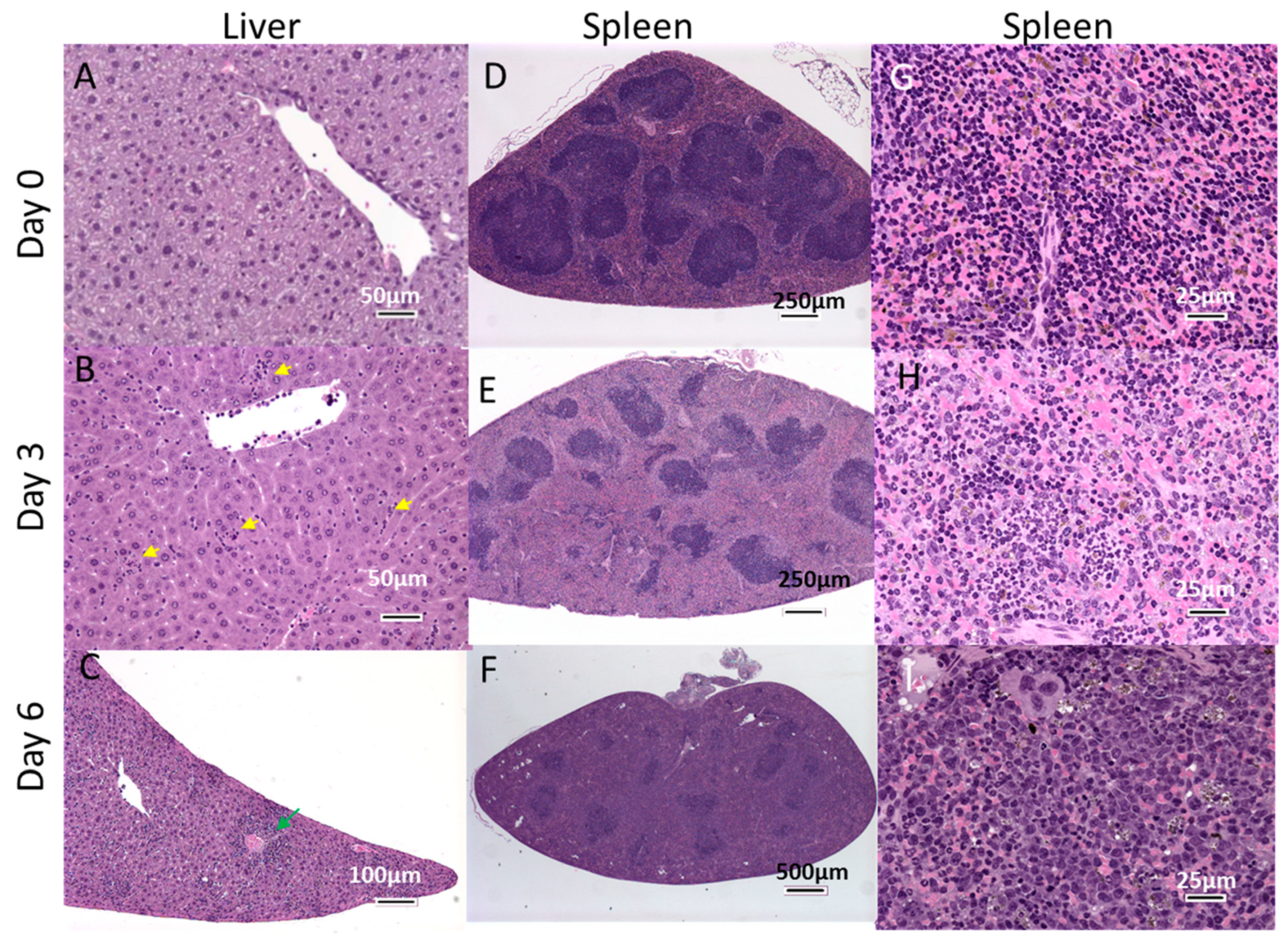
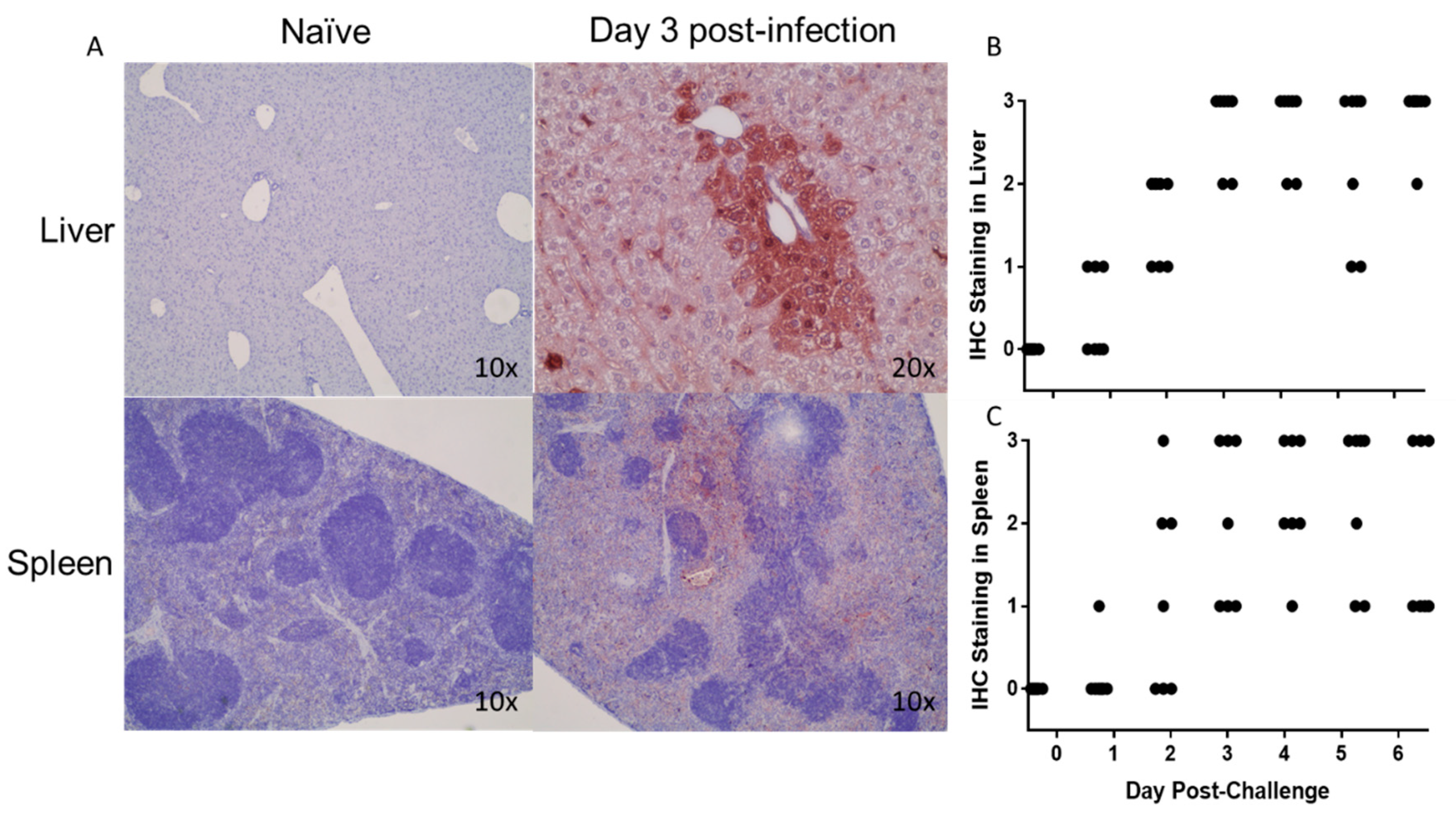
3.5. Histopathological Changes in EBOV-Infected IFNAGR KO Mice
3.6. EBOV-Challenged IFNAGR KO Mice as a Screen Model for Antivirals
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kuhn, J.H.; Bao, Y.; Bavari, S.; Becker, S.; Bradfute, S.; Brauburger, K.; Rodney Brister, J.; Bukreyev, A.A.; Cai, Y.; Chandran, K.; et al. Virus nomenclature below the species level: A standardized nomenclature for filovirus strains and variants rescued from cdna. Arch. Virol. 2014, 159, 1229–1237. [Google Scholar] [CrossRef] [PubMed]
- CDC. 2014 Ebola Outbreak in West Africa—Case Counts. Available online: https://www.cdc.gov/vhf/ebola/history/2014-2016-outbreak/case-counts.html (accessed on 27 June 2018).
- Kuhn, J.H. Filoviruses. A compendium of 40 years of epidemiological, clinical, and laboratory studies. Arch. Virol. Suppl. 2008, 20, 13–360. [Google Scholar] [PubMed]
- Towner, J.S.; Khristova, M.L.; Sealy, T.K.; Vincent, M.J.; Erickson, B.R.; Bawiec, D.A.; Hartman, A.L.; Comer, J.A.; Zaki, S.R.; Stroher, U.; et al. Marburgvirus genomics and association with a large hemorrhagic fever outbreak in angola. J. Virol. 2006, 80, 6497–6516. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control (CDC). Update: Filovirus infection in animal handlers. MMWR Morb. Mortal. Wkly. Rep. 1990, 39, 221. [Google Scholar]
- Miranda, M.E.; White, M.E.; Dayrit, M.M.; Hayes, C.G.; Ksiazek, T.G.; Burans, J.P. Seroepidemiological study of filovirus related to ebola in the philippines. Lancet 1991, 337, 425–426. [Google Scholar] [CrossRef]
- Lawrence, P.; Danet, N.; Reynard, O.; Volchkova, V.; Volchkov, V. Human transmission of ebola virus. Curr. Opin. Virol. 2017, 22, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Baseler, L.; Chertow, D.S.; Johnson, K.M.; Feldmann, H.; Morens, D.M. The pathogenesis of ebola virus disease. Annu. Rev. Pathol. Mech. Dis. 2017, 12, 387–418. [Google Scholar] [CrossRef]
- Matassov, D.; Mire, C.E.; Latham, T.; Geisbert, J.B.; Xu, R.; Ota-Setlik, A.; Agans, K.N.; Kobs, D.J.; Wendling, M.Q.S.; Burnaugh, A.; et al. Single dose trivalent vesiculovax vaccine protects macaques from lethal ebolavirus and marburgvirus challenge. J. Virol. 2017. [Google Scholar] [CrossRef]
- Nakayama, E.; Saijo, M. Animal models for ebola and marburg virus infections. Front. Microbiol. 2013, 4, 267. [Google Scholar] [CrossRef] [PubMed]
- Cross, R.W.; Mire, C.E.; Borisevich, V.; Geisbert, J.B.; Fenton, K.A.; Geisbert, T.W. The domestic ferret (mustela putorius furo) as a lethal infection model for 3 species of ebolavirus. J. Infect. Dis. 2016, 214, 565–569. [Google Scholar] [CrossRef]
- Kozak, R.; He, S.; Kroeker, A.; de La Vega, M.A.; Audet, J.; Wong, G.; Urfano, C.; Antonation, K.; Embury-Hyatt, C.; Kobinger, G.P.; et al. Ferrets infected with bundibugyo virus or ebola virus recapitulate important aspects of human filovirus disease. J. Virol. 2016, 90, 9209–9223. [Google Scholar] [CrossRef] [PubMed]
- Kroeker, A.; He, S.; de La Vega, M.A.; Wong, G.; Embury-Hyatt, C.; Qiu, X. Characterization of sudan ebolavirus infection in ferrets. Oncotarget 2017, 8, 46262–46272. [Google Scholar] [CrossRef] [PubMed]
- Cross, R.W.; Fenton, K.A.; Geisbert, T.W. Small animal models of filovirus disease: Recent advances and future directions. Expert Opin. Drug Discov. 2018, 13, 1027–1040. [Google Scholar] [CrossRef] [PubMed]
- Ebihara, H.; Zivcec, M.; Gardner, D.; Falzarano, D.; LaCasse, R.; Rosenke, R.; Long, D.; Haddock, E.; Fischer, E.; Kawaoka, Y.; et al. A syrian golden hamster model recapitulating ebola hemorrhagic fever. J. Infect. Dis. 2013, 207, 306–318. [Google Scholar] [CrossRef] [PubMed]
- Marzi, A.; Banadyga, L.; Haddock, E.; Thomas, T.; Shen, K.; Horne, E.J.; Scott, D.P.; Feldmann, H.; Ebihara, H. A hamster model for marburg virus infection accurately recapitulates marburg hemorrhagic fever. Sci. Rep. 2016, 6, 39214. [Google Scholar] [CrossRef] [PubMed]
- De Wit, E.; Munster, V.J.; Metwally, S.A.; Feldmann, H. Assessment of rodents as animal models for reston ebolavirus. J. Infect. Dis. 2011, 204 (Suppl. 3), S968–S972. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, Y.; Safronetz, D.; Brown, K.; LaCasse, R.; Marzi, A.; Ebihara, H.; Feldmann, H. Protective efficacy of a bivalent recombinant vesicular stomatitis virus vaccine in the syrian hamster model of lethal ebola virus infection. J. Infect. Dis. 2011, 204 (Suppl. 3), S1090–S1097. [Google Scholar] [CrossRef]
- Cheresiz, S.V.; Semenova, E.A.; Chepurnov, A.A. Adapted lethality: What we can learn from guinea pig-adapted ebola virus infection model. Adv. Virol. 2016, 2016, 8059607. [Google Scholar] [CrossRef]
- Connolly, B.M.; Steele, K.E.; Davis, K.J.; Geisbert, T.W.; Kell, W.M.; Jaax, N.K.; Jahrling, P.B. Pathogenesis of experimental ebola virus infection in guinea pigs. J. Infect. Dis. 1999, 179, S203–S217. [Google Scholar] [CrossRef]
- Cross, R.W.; Fenton, K.A.; Geisbert, J.B.; Mire, C.E.; Geisbert, T.W. Modeling the disease course of zaire ebolavirus infection in the outbred guinea pig. J. Infect. Dis. 2015, 212 (Suppl. 2), S305–S315. [Google Scholar] [CrossRef]
- Dowall, S.D.; Matthews, D.A.; Garcia-Dorival, I.; Taylor, I.; Kenny, J.; Hertz-Fowler, C.; Hall, N.; Corbin-Lickfett, K.; Empig, C.; Schlunegger, K.; et al. Elucidating variations in the nucleotide sequence of ebola virus associated with increasing pathogenicity. Genome Biol. 2014, 15, 540. [Google Scholar] [CrossRef] [PubMed]
- Mateo, M.; Carbonnelle, C.; Reynard, O.; Kolesnikova, L.; Nemirov, K.; Page, A.; Volchkova, V.A.; Volchkov, V.E. Vp24 is a molecular determinant of ebola virus virulence in guinea pigs. J. Infect. Dis. 2011, 204 (Suppl. 3), S1011–S1020. [Google Scholar] [CrossRef] [PubMed]
- Mire, C.E.; Geisbert, J.B.; Versteeg, K.M.; Mamaeva, N.; Agans, K.N.; Geisbert, T.W.; Connor, J.H. A single-vector, single-injection trivalent filovirus vaccine: Proof of concept study in outbred guinea pigs. J. Infect. Dis. 2015, 212 (Suppl. 2), S384–S388. [Google Scholar] [CrossRef] [PubMed]
- Twenhafel, N.A.; Shaia, C.I.; Bunton, T.E.; Shamblin, J.D.; Wollen, S.E.; Pitt, L.M.; Sizemore, D.R.; Ogg, M.M.; Johnston, S.C. Experimental aerosolized guinea pig-adapted zaire ebolavirus (variant: Mayinga) causes lethal pneumonia in guinea pigs. Vet. Pathol. 2015, 52, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Volchkova, V.A.; Dolnik, O.; Martinez, M.J.; Reynard, O.; Volchkov, V.E. Genomic rna editing and its impact on ebola virus adaptation during serial passages in cell culture and infection of guinea pigs. J. Infect. Dis. 2011, 204 (Suppl. 3), S941–S946. [Google Scholar] [CrossRef]
- Wong, G.; He, S.; Wei, H.; Kroeker, A.; Audet, J.; Leung, A.; Cutts, T.; Graham, J.; Kobasa, D.; Embury-Hyatt, C.; et al. Development and characterization of a guinea pig-adapted sudan virus. J. Virol. 2016, 90, 392–399. [Google Scholar] [CrossRef]
- Bradfute, S.B.; Warfield, K.L.; Bray, M. Mouse models for filovirus infections. Viruses 2012, 4, 1477–1508. [Google Scholar] [CrossRef]
- Bray, M. The role of the type i interferon response in the resistance of mice to filovirus infection. J. Gen. Virol. 2001, 82, 1365–1373. [Google Scholar] [CrossRef]
- Bray, M.; Davis, K.; Geisbert, T.; Schmaljohn, C.; Huggins, J. A mouse model for evaluation of prophylaxis and therapy of ebola hemorrhagic fever. J. Infect. Dis. 1999, 179 (Suppl. 1), S248–S258. [Google Scholar] [CrossRef]
- Banadyga, L.; Dolan, M.A.; Ebihara, H. Rodent-adapted filoviruses and the molecular basis of pathogenesis. J. Mol. Biol. 2016, 428, 3449–3466. [Google Scholar] [CrossRef]
- Zumbrun, E.E.; Abdeltawab, N.F.; Bloomfield, H.A.; Chance, T.B.; Nichols, D.K.; Harrison, P.E.; Kotb, M.; Nalca, A. Development of a murine model for aerosolized ebolavirus infection using a panel of recombinant inbred mice. Viruses 2012, 4, 3468–3493. [Google Scholar] [CrossRef] [PubMed]
- Haddock, E.; Feldmann, H.; Marzi, A. Ebola virus infection in commonly used laboratory mouse strains. J. Infect. Dis. 2018, 218, S453–S457. [Google Scholar] [CrossRef] [PubMed]
- St Claire, M.C.; Ragland, D.R.; Bollinger, L.; Jahrling, P.B. Animal models of ebolavirus infection. Comp. Med. 2017, 67, 253–262. [Google Scholar] [PubMed]
- Groseth, A.; Eickmann, M.; Ebihara, H.; Becker, S.; Hoenen, T. Filoviruses: Ebola, marburg and disease. In Encyclopedia of Life Sciences; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2001. [Google Scholar]
- Wong, G.; Qiu, X.G. Type i interferon receptor knockout mice as models for infection of highly pathogenic viruses with outbreak potential. Zool. Res. 2018, 39, 3–14. [Google Scholar] [PubMed]
- Raymond, J.; Bradfute, S.; Bray, M. Filovirus infection of stat-1 knockout mice. J. Infect. Dis. 2011, 204 (Suppl. 3), S986–S990. [Google Scholar] [CrossRef] [PubMed]
- Brannan, J.M.; Froude, J.W.; Prugar, L.I.; Bakken, R.R.; Zak, S.E.; Daye, S.P.; Wilhelmsen, C.E.; Dye, J.M. Interferon alpha/beta receptor-deficient mice as a model for ebola virus disease. J. Infect. Dis. 2015, 212 (Suppl. 2), S282–S294. [Google Scholar] [CrossRef] [PubMed]
- Bradfute, S.B.; Braun, D.R.; Shamblin, J.D.; Geisbert, J.B.; Paragas, J.; Garrison, A.; Hensley, L.E.; Geisbert, T.W. Lymphocyte death in a mouse model of ebola virus infection. J. Infect. Dis. 2007, 196 (Suppl. 2), S296–S304. [Google Scholar] [CrossRef] [PubMed]
- Wong, G.; Leung, A.; He, S.; Cao, W.; De La Vega, M.A.; Griffin, B.D.; Soule, G.; Kobinger, G.P.; Kobasa, D.; Qiu, X. The makona variant of ebola virus is highly lethal to immunocompromised mice and immunocompetent ferrets. J. Infect. Dis. 2018, 218, S466–S470. [Google Scholar] [CrossRef] [PubMed]
- Marzi, A.; Kercher, L.; Marceau, J.; York, A.; Callsion, J.; Gardner, D.J.; Geisbert, T.W.; Feldmann, H. Stat1-deficient mice are not an appropriate model for efficacy testing of recombinant vesicular stomatitis virus-based filovirus vaccines. J. Infect. Dis. 2015, 212 (Suppl. 2), S404–S409. [Google Scholar] [CrossRef] [PubMed]
- Ekins, S. Efficacy of tilorone dihydrochloride against ebola virus infection. Antimicrob Agents Chemother. 2018, 62, e01711-17. [Google Scholar] [CrossRef] [PubMed]
- Martines, R.B.; Ng, D.L.; Greer, P.W.; Rollin, P.E.; Zaki, S.R. Tissue and cellular tropism, pathology and pathogenesis of ebola and marburg viruses. J. Pathol. 2015, 235, 153–174. [Google Scholar] [CrossRef] [PubMed]
- Geisbert, T.W.; Hensley, L.E.; Larsen, T.; Young, H.A.; Reed, D.S.; Geisbert, J.B.; Scott, D.P.; Kagan, E.; Jahrling, P.B.; Davis, K.J. Pathogenesis of ebola hemorrhagic fever in cynomolgus macaques: Evidence that dendritic cells are early and sustained targets of infection. Am. J. Pathol. 2003, 163, 2347–2370. [Google Scholar] [CrossRef]
- Madelain, V.; Oestereich, L.; Graw, F.; Nguyen, T.H.; de Lamballerie, X.; Mentre, F.; Gunther, S.; Guedj, J. Ebola virus dynamics in mice treated with favipiravir. Antivir. Res. 2015, 123, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Smither, S.J.; Eastaugh, L.S.; Steward, J.A.; Nelson, M.; Lenk, R.P.; Lever, M.S. Post-exposure efficacy of oral t-705 (favipiravir) against inhalational ebola virus infection in a mouse model. Antivir. Res. 2014, 104, 153–155. [Google Scholar] [CrossRef] [PubMed]
- Oestereich, L.; Ludtke, A.; Wurr, S.; Rieger, T.; Munoz-Fontela, C.; Gunther, S. Successful treatment of advanced ebola virus infection with t-705 (favipiravir) in a small animal model. Antivir. Res. 2014, 105, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Bixler, S.L.; Bocan, T.M.; Wells, J.; Wetzel, K.S.; Van Tongeren, S.A.; Garza, N.L.; Donnelly, G.; Cazares, L.H.; Soloveva, V.; Welch, L.; et al. Intracellular conversion and in vivo dose response of favipiravir (t-705) in rodents infected with ebola virus. Antivir. Res. 2018, 151, 50–54. [Google Scholar] [CrossRef]
- Zhu, W.; Zhang, Z.; He, S.; Wong, G.; Banadyga, L.; Qiu, X. Successful treatment of marburg virus with orally administrated t-705 (favipiravir) in a mouse model. Antivir. Res. 2018, 151, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Rahim, M.N.; Zhang, Z.; He, S.; Zhu, W.; Banadyga, L.; Safronetz, D.; Qiu, X. Postexposure protective efficacy of t-705 (favipiravir) against sudan virus infection in guinea pigs. J. Infect. Dis. 2018, 218, S649–S657. [Google Scholar] [CrossRef]
- Guedj, J.; Piorkowski, G.; Jacquot, F.; Madelain, V.; Nguyen, T.H.T.; Rodallec, A.; Gunther, S.; Carbonnelle, C.; Mentre, F.; Raoul, H.; et al. Antiviral efficacy of favipiravir against ebola virus: A translational study in cynomolgus macaques. PLoS Med. 2018, 15, e1002535. [Google Scholar] [CrossRef]
- Bixler, S.L.; Bocan, T.M.; Wells, J.; Wetzel, K.S.; Van Tongeren, S.A.; Dong, L.; Garza, N.L.; Donnelly, G.; Cazares, L.H.; Nuss, J.; et al. Efficacy of favipiravir (t-705) in nonhuman primates infected with ebola virus or marburg virus. Antivir. Res. 2018, 151, 97–104. [Google Scholar] [CrossRef]
- Warfield, K.L.; Bradfute, S.B.; Wells, J.; Lofts, L.; Cooper, M.T.; Alves, D.A.; Reed, D.K.; VanTongeren, S.A.; Mech, C.A.; Bavari, S. Development and characterization of a mouse model for marburg hemorrhagic fever. J. Virol. 2009, 83, 6404–6415. [Google Scholar] [CrossRef] [PubMed]
- Ebihara, H.; Takada, A.; Kobasa, D.; Jones, S.; Neumann, G.; Theriault, S.; Bray, M.; Feldmann, H.; Kawaoka, Y. Molecular determinants of ebola virus virulence in mice. PLoS Pathog. 2006, 2, e73. [Google Scholar] [CrossRef] [PubMed]
- Valmas, C.; Basler, C.F. Marburg virus vp40 antagonizes interferon signaling in a species-specific manner. J. Virol. 2011, 85, 4309–4317. [Google Scholar] [CrossRef] [PubMed]
- Atkins, C.; Miao, J.; Kalveram, B.; Juelich, T.; Smith, J.K.; Perez, D.; Zhang, L.; Westover, J.L.B.; Van Wettere, A.J.; Gowen, B.B.; et al. Natural history and pathogenesis of wild-type marburg virus infection in stat2 knockout hamsters. J. Infect. Dis. 2018, 218, S438–S447. [Google Scholar] [CrossRef] [PubMed]
- Lever, M.S.; Piercy, T.J.; Steward, J.A.; Eastaugh, L.; Smither, S.J.; Taylor, C.; Salguero, F.J.; Phillpotts, R.J. Lethality and pathogenesis of airborne infection with filoviruses in a129 alpha/beta -/- interferon receptor-deficient mice. J. Med. Microbiol. 2012, 61, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Rhein, B.A.; Powers, L.S.; Rogers, K.; Anantpadma, M.; Singh, B.K.; Sakurai, Y.; Bair, T.; Miller-Hunt, C.; Sinn, P.; Davey, R.A.; et al. Interferon-gamma inhibits ebola virus infection. PLoS Pathog. 2015, 11, e1005263. [Google Scholar] [CrossRef]
- Alfson, K.J.; Avena, L.E.; Beadles, M.W.; Worwa, G.; Amen, M.; Patterson, J.L.; Carrion, R., Jr.; Griffiths, A. Intramuscular exposure of macaca fascicularis to low doses of low passage- or cell culture-adapted sudan virus or ebola virus. Viruses 2018, 10, 642. [Google Scholar] [CrossRef]
- Fisher-Hoch, S.P.; Platt, G.S.; Neild, G.H.; Southee, T.; Baskerville, A.; Raymond, R.T.; Lloyd, G.; Simpson, D.I. Pathophysiology of shock and hemorrhage in a fulminating viral infection (ebola). J. Infect. Dis. 1985, 152, 887–894. [Google Scholar] [CrossRef]
- Ebihara, H.; Rockx, B.; Marzi, A.; Feldmann, F.; Haddock, E.; Brining, D.; LaCasse, R.A.; Gardner, D.; Feldmann, H. Host response dynamics following lethal infection of rhesus macaques with zaire ebolavirus. J. Infect. Dis. 2011, 204 (Suppl. 3), S991–S999. [Google Scholar] [CrossRef]
- Bray, M.; Hatfill, S.; Hensley, L.; Huggins, J.W. Haematological, biochemical and coagulation changes in mice, guinea-pigs and monkeys infected with a mouse-adapted variant of ebola zaire virus. J. Comp. Pathol. 2001, 125, 243–253. [Google Scholar] [CrossRef]
- Furuta, Y.; Gowen, B.B.; Takahashi, K.; Shiraki, K.; Smee, D.F.; Barnard, D.L. Favipiravir (t-705), a novel viral rna polymerase inhibitor. Antivir. Res. 2013, 100, 446–454. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Comer, J.E.; Escaffre, O.; Neef, N.; Brasel, T.; Juelich, T.L.; Smith, J.K.; Smith, J.; Kalveram, B.; Perez, D.D.; Massey, S.; et al. Filovirus Virulence in Interferon α/β and γ Double Knockout Mice, and Treatment with Favipiravir. Viruses 2019, 11, 137. https://doi.org/10.3390/v11020137
Comer JE, Escaffre O, Neef N, Brasel T, Juelich TL, Smith JK, Smith J, Kalveram B, Perez DD, Massey S, et al. Filovirus Virulence in Interferon α/β and γ Double Knockout Mice, and Treatment with Favipiravir. Viruses. 2019; 11(2):137. https://doi.org/10.3390/v11020137
Chicago/Turabian StyleComer, Jason E., Olivier Escaffre, Natasha Neef, Trevor Brasel, Terry L. Juelich, Jennifer K. Smith, Jeanon Smith, Birte Kalveram, David D. Perez, Shane Massey, and et al. 2019. "Filovirus Virulence in Interferon α/β and γ Double Knockout Mice, and Treatment with Favipiravir" Viruses 11, no. 2: 137. https://doi.org/10.3390/v11020137
APA StyleComer, J. E., Escaffre, O., Neef, N., Brasel, T., Juelich, T. L., Smith, J. K., Smith, J., Kalveram, B., Perez, D. D., Massey, S., Zhang, L., & Freiberg, A. N. (2019). Filovirus Virulence in Interferon α/β and γ Double Knockout Mice, and Treatment with Favipiravir. Viruses, 11(2), 137. https://doi.org/10.3390/v11020137




