Antiviral Role of IFITM Proteins in Classical Swine Fever Virus Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Virus
2.2. Real-Time Quantitative PCR (RT-qPCR)
2.3. Construction and Transfection of Plasmid
2.4. Acquisition and Titration of Lentivirus
2.5. Establishment and Detection of IFITM-Overexpression and -Knockdown Cell Lines
2.6. Western Blot
2.7. Cell Viability Assay
2.8. Virus Titration By Indirect Immunofluorescence Assay (IFA)
2.9. Confocal Immunofluorescence Microscopy
2.10. Statistical Analysis
3. Results
3.1. Overexpression of IFITMs Inhibits CSFV Replication in PAMs
3.2. Knockdown of IFITMs Mediated by shRNA Enhances CSFV Replication
3.3. Expression of IFITMs Is Induced by IFN-α Treatment
3.4. IFITM Expression is Downregulated By CSFV
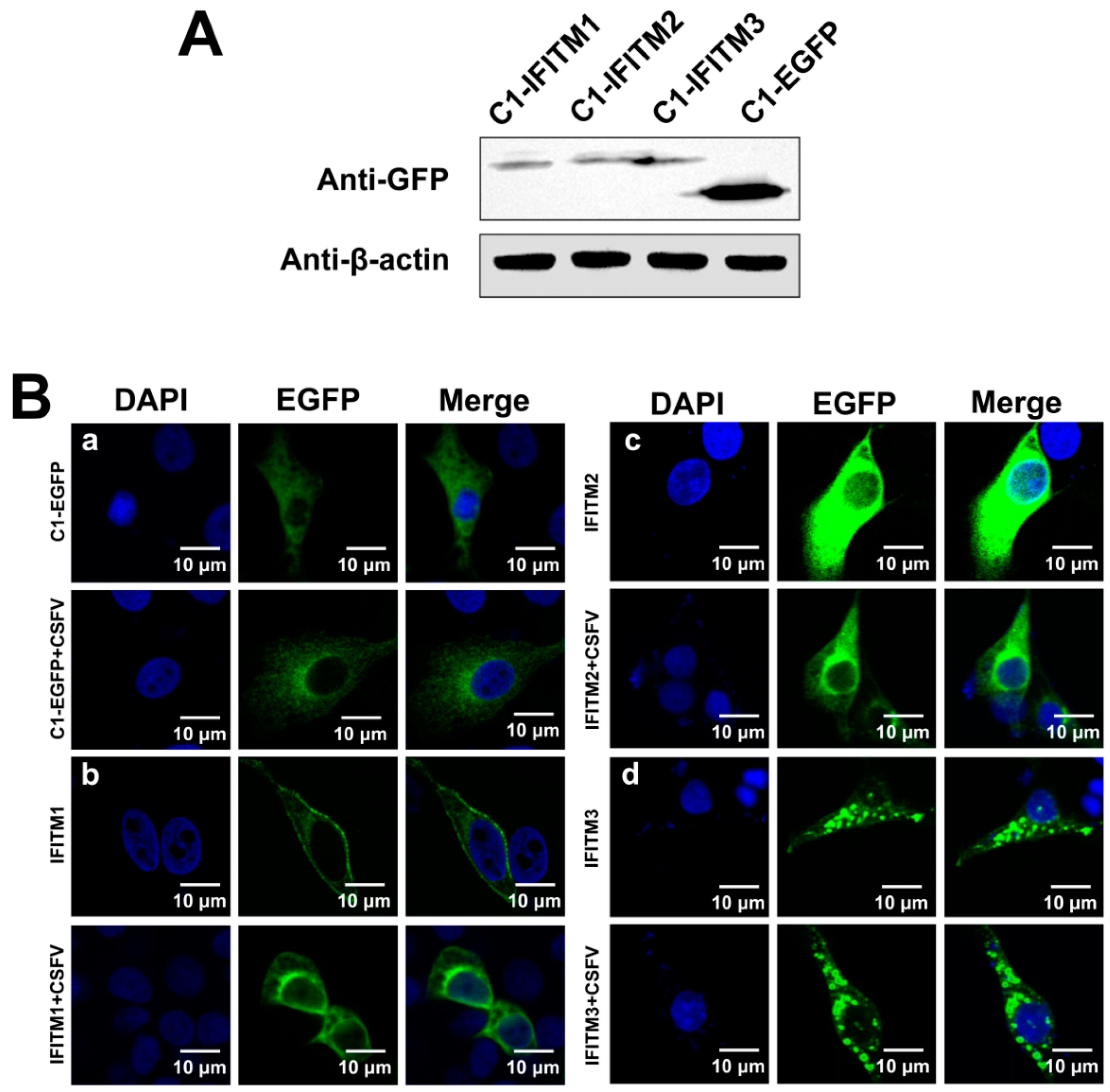
3.5. Distribution of IFITMs in CSFV-Infected PAMs
3.6. IFITMs Do Not Interfere with CSFV Binding But Restrict CSFV Entry
3.7. Colocalization of IFITMs with Rab5, Rab7, and Lamp1
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gladue, D.P.; Largo, E.; Holinka, L.G.; Ramirez-Medina, E.; Vuono, E.A.; Berggren, K.A.; Risatti, G.R.; Nieva, J.L.; Borca, M.V. Classical swine fever virus p7 protein interacts with host protein camlg and regulates calcium permeability at the endoplasmic reticulum. Viruses 2018, 10, 460. [Google Scholar] [CrossRef] [PubMed]
- Bohorquez, J.A.; Wang, M.; Perez-Simo, M.; Vidal, E.; Rosell, R.; Ganges, L. Low cd4/cd8 ratio in classical swine fever postnatal persistent infection generated at 3 weeks after birth. Transbound. Emerg. Dis. 2018. [Google Scholar] [CrossRef] [PubMed]
- Ji, W.; Guo, Z.; Ding, N.Z.; He, C.Q. Studying classical swine fever virus: Making the best of a bad virus. Virus Res. 2015, 197, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Ji, S.; Liu, Y.; Lei, J.L.; Xia, S.L.; Wang, Y.; Du, M.L.; Shao, L.; Meng, X.Y.; Zhou, M.; et al. Isolation and characterization of a moderately virulent classical swine fever virus emerging in china. Transbound. Emerg. Dis. 2017, 64, 1848–1857. [Google Scholar] [CrossRef]
- Yu, S.; Yin, C.; Song, K.; Li, S.; Zheng, G.L.; Li, L.F.; Wang, J.; Li, Y.; Luo, Y.; Sun, Y.; et al. Engagement of cellular cholesterol in the life cycle of classical swine fever virus: Its potential as an antiviral target. J. Gen. Virol. 2018. [Google Scholar] [CrossRef]
- Dai, J.; Pan, W.; Wang, P. Isg15 facilitates cellular antiviral response to dengue and west nile virus infection in vitro. Virol. J. 2011, 8, 468. [Google Scholar] [CrossRef]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef]
- Li, L.F.; Yu, J.; Li, Y.; Wang, J.; Li, S.; Zhang, L.; Xia, S.L.; Yang, Q.; Wang, X.; Yu, S.; et al. Guanylate-binding protein 1, an interferon-induced gtpase, exerts an antiviral activity against classical swine fever virus depending on its gtpase activity. J. Virol. 2016, 90, 4412–4426. [Google Scholar] [CrossRef]
- Li, L.F.; Yu, J.; Zhang, Y.; Yang, Q.; Li, Y.; Zhang, L.; Wang, J.; Li, S.; Luo, Y.; Sun, Y.; et al. Interferon-inducible oligoadenylate synthetase-like protein acts as an antiviral effector against classical swine fever virus via the mda5-mediated type i interferon-signaling pathway. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Renson, P.; Blanchard, Y.; Le Dimna, M.; Felix, H.; Cariolet, R.; Jestin, A.; Le Potier, M.F. Acute induction of cell death-related ifn stimulated genes (isg) differentiates highly from moderately virulent csfv strains. Vet. Res. 2010, 41, 7. [Google Scholar] [CrossRef]
- Zhao, Y.; Pang, D.; Wang, T.; Yang, X.; Wu, R.; Ren, L.; Yuan, T.; Huang, Y.; Ouyang, H. Human mxa protein inhibits the replication of classical swine fever virus. Virus Res. 2011, 156, 151–155. [Google Scholar] [CrossRef] [PubMed]
- He, D.N.; Zhang, X.M.; Liu, K.; Pang, R.; Zhao, J.; Zhou, B.; Chen, P.Y. In vitro inhibition of the replication of classical swine fever virus by porcine mx1 protein. Antivir. Res. 2014, 104, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Narayana, S.K.; Helbig, K.J.; McCartney, E.M.; Eyre, N.S.; Bull, R.A.; Eltahla, A.; Lloyd, A.R.; Beard, M.R. The interferon-induced transmembrane proteins, ifitm1, ifitm2, and ifitm3 inhibit hepatitis c virus entry. J. Biol. Chem. 2015, 290, 25946–25959. [Google Scholar] [CrossRef] [PubMed]
- Desai, T.M.; Marin, M.; Chin, C.R.; Savidis, G.; Brass, A.L.; Melikyan, G.B. Ifitm3 restricts influenza a virus entry by blocking the formation of fusion pores following virus-endosome hemifusion. PLoS Pathog. 2014, 10, e1004048. [Google Scholar] [CrossRef] [PubMed]
- Wrensch, F.; Karsten, C.B.; Gnirss, K.; Hoffmann, M.; Lu, K.; Takada, A.; Winkler, M.; Simmons, G.; Pohlmann, S. Interferon-induced transmembrane protein-mediated inhibition of host cell entry of ebolaviruses. J. Infect. Dis. 2015, 212 (Suppl. 2), S210–S218. [Google Scholar] [CrossRef]
- Chan, Y.K.; Huang, I.C.; Farzan, M. Ifitm proteins restrict antibody-dependent enhancement of dengue virus infection. PLoS ONE 2012, 7, e34508. [Google Scholar] [CrossRef]
- Brass, A.L.; Huang, I.C.; Benita, Y.; John, S.P.; Krishnan, M.N.; Feeley, E.M.; Ryan, B.J.; Weyer, J.L.; van der Weyden, L.; Fikrig, E.; et al. The ifitm proteins mediate cellular resistance to influenza a h1n1 virus, west nile virus, and dengue virus. Cell 2009, 139, 1243–1254. [Google Scholar] [CrossRef]
- Smith, S.; Weston, S.; Kellam, P.; Marsh, M. Ifitm proteins-cellular inhibitors of viral entry. Curr. Opin. Virol. 2014, 4, 71–77. [Google Scholar] [CrossRef]
- Diamond, M.S.; Farzan, M. The broad-spectrum antiviral functions of ifit and ifitm proteins. Nat. Rev. Immunol. 2013, 13, 46–57. [Google Scholar] [CrossRef]
- Yount, J.S.; Karssemeijer, R.A.; Hang, H.C. S-palmitoylation and ubiquitination differentially regulate interferon-induced transmembrane protein 3 (ifitm3)-mediated resistance to influenza virus. J. Biol. Chem. 2012, 287, 19631–19641. [Google Scholar] [CrossRef]
- Miller, L.C.; Jiang, Z.; Sang, Y.; Harhay, G.P.; Lager, K.M. Evolutionary characterization of pig interferon-inducible transmembrane gene family and member expression dynamics in tracheobronchial lymph nodes of pigs infected with swine respiratory disease viruses. Vet. Immunol. Immunopathol. 2014, 159, 180–191. [Google Scholar] [CrossRef] [PubMed]
- Lanz, C.; Yanguez, E.; Andenmatten, D.; Stertz, S. Swine interferon-inducible transmembrane proteins potently inhibit influenza a virus replication. J. Virol. 2015, 89, 863–869. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Markosyan, R.M.; Zheng, Y.M.; Golfetto, O.; Bungart, B.; Li, M.; Ding, S.; He, Y.; Liang, C.; Lee, J.C.; et al. Ifitm proteins restrict viral membrane hemifusion. PLoS Pathog. 2013, 9, e1003124. [Google Scholar] [CrossRef] [PubMed]
- Mudhasani, R.; Tran, J.P.; Retterer, C.; Radoshitzky, S.R.; Kota, K.P.; Altamura, L.A.; Smith, J.M.; Packard, B.Z.; Kuhn, J.H.; Costantino, J.; et al. Ifitm-2 and ifitm-3 but not ifitm-1 restrict rift valley fever virus. J. Virol. 2013, 87, 8451–8464. [Google Scholar] [CrossRef] [PubMed]
- Perreira, J.M.; Chin, C.R.; Feeley, E.M.; Brass, A.L. Ifitms restrict the replication of multiple pathogenic viruses. J. Mol. Biol. 2013, 425, 4937–4955. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, C.; Zhou, L.; Zhang, N.; Wang, X.; Ge, X.; Guo, X.; Yang, H. Porcine reproductive and respiratory syndrome virus counteracts the porcine intrinsic virus restriction factors-ifitm1 and tetherin in marc-145 cells. Virus Res. 2014, 191, 92–100. [Google Scholar] [CrossRef] [PubMed]
- John, S.P.; Chin, C.R.; Perreira, J.M.; Feeley, E.M.; Aker, A.M.; Savidis, G.; Smith, S.E.; Elia, A.E.; Everitt, A.R.; Vora, M.; et al. The cd225 domain of ifitm3 is required for both ifitm protein association and inhibition of influenza a virus and dengue virus replication. J. Virol. 2013, 87, 7837–7852. [Google Scholar] [CrossRef] [PubMed]
- Feeley, E.M.; Sims, J.S.; John, S.P.; Chin, C.R.; Pertel, T.; Chen, L.M.; Gaiha, G.D.; Ryan, B.J.; Donis, R.O.; Elledge, S.J.; et al. Ifitm3 inhibits influenza a virus infection by preventing cytosolic entry. PLoS Pathog. 2011, 7, e1002337. [Google Scholar] [CrossRef]
- Amini-Bavil-Olyaee, S.; Choi, Y.J.; Lee, J.H.; Shi, M.; Huang, I.C.; Farzan, M.; Jung, J.U. The antiviral effector ifitm3 disrupts intracellular cholesterol homeostasis to block viral entry. Cell Host Microbe 2013, 13, 452–464. [Google Scholar] [CrossRef]
- Wilkins, C.; Woodward, J.; Lau, D.T.; Barnes, A.; Joyce, M.; McFarlane, N.; McKeating, J.A.; Tyrrell, D.L.; Gale, M., Jr. Ifitm1 is a tight junction protein that inhibits hepatitis c virus entry. Hepatology 2013, 57, 461–469. [Google Scholar] [CrossRef]
- Shi, B.J.; Liu, C.C.; Zhou, J.; Wang, S.Q.; Gao, Z.C.; Zhang, X.M.; Zhou, B.; Chen, P.Y. Entry of classical swine fever virus into pk-15 cells via a ph-, dynamin-, and cholesterol-dependent, clathrin-mediated endocytic pathway that requires rab5 and rab7. J. Virol. 2016, 90, 9194–9208. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.N.; Liu, Y.Y.; Xiao, F.C.; Liu, C.C.; Liang, X.D.; Chen, J.; Zhou, J.; Baloch, A.S.; Kan, L.; Zhou, B.; et al. Rab5, rab7, and rab11 are required for caveola-dependent endocytosis of classical swine fever virus in porcine alveolar macrophages. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Randall, R.E.; Goodbourn, S. Interferons and viruses: An interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 2008, 89, 1–47. [Google Scholar] [CrossRef] [PubMed]
- Pervolaraki, K.; Rastgou Talemi, S.; Mendoza, J.L.; McLauchlan, J.; Hofer, T.; Stanifer, M.L.; Boulant, S.; Garcia, K.C.; Bormann, F.; Bamford, C.; et al. Differential induction of interferon stimulated genes between type i and type iii interferons is independent of interferon receptor abundance. PLoS Pathog. 2018, 14, e1007420. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Qian, P.; Wu, Q.; Liu, S.; Fan, W.; Zhang, K.; Wang, R.; Zhang, H.; Chen, H.; Li, X. Swine interferon-induced transmembrane protein, sifitm3, inhibits foot-and-mouth disease virus infection in vitro and in vivo. Antivir. Res. 2014, 109, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Weidner, J.M.; Jiang, D.; Pan, X.B.; Chang, J.; Block, T.M.; Guo, J.T. Interferon-induced cell membrane proteins, ifitm3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. J. Virol. 2010, 84, 12646–12657. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; He, W.R.; Shen, L.; Dong, H.; Yu, J.; Wang, X.; Yu, S.; Li, Y.; Li, S.; Luo, Y.; et al. The laminin receptor is a cellular attachment receptor for classical swine fever virus. J. Virol. 2015, 89, 4894–4906. [Google Scholar] [CrossRef] [PubMed]
- Hulst, M.M.; van Gennip, H.G.; Moormann, R.J. Passage of classical swine fever virus in cultured swine kidney cells selects virus variants that bind to heparan sulfate due to a single amino acid change in envelope protein e(rns). J. Virol. 2000, 74, 9553–9561. [Google Scholar] [CrossRef]
- Rink, J.; Ghigo, E.; Kalaidzidis, Y.; Zerial, M. Rab conversion as a mechanism of progression from early to late endosomes. Cell 2005, 122, 735–749. [Google Scholar] [CrossRef]
- Alfajaro, M.M.; Cho, E.H.; Kim, D.S.; Kim, J.Y.; Park, J.G.; Soliman, M.; Baek, Y.B.; Park, C.H.; Kang, M.I.; Park, S.I.; et al. Early infection of porcine sapovirus disrupts tight junction and uses occludin as a co-receptor. J. Virol. 2018. [Google Scholar] [CrossRef]
- Vale-Costa, S.; Amorim, M.J. Recycling endosomes and viral infection. Viruses 2016, 8, 64. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, W.; Li, Y.; Guo, Y.; Wang, X.; Duan, M.; Guan, Z.; Liu, Z.; Zhang, M. Rabies virus co-localizes with early (rab5) and late (rab7) endosomal proteins in neuronal and sh-sy5y cells. Virol. Sin. 2017, 32, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Wang, C.; Zhang, L.; Wang, T.; Zhang, J.; Liang, W.; Li, C.; Qian, G.; Ouyang, Y.; Guo, K.; et al. Rab5 enhances classical swine fever virus proliferation and interacts with viral ns4b protein to facilitate formation of ns4b related complex. Front. Microbiol. 2017, 8, 1468. [Google Scholar] [CrossRef] [PubMed]
- Bailey, C.C.; Zhong, G.; Huang, I.C.; Farzan, M. Ifitm-family proteins: The cell’s first line of antiviral defense. Annu. Rev. Virol. 2014, 1, 261–283. [Google Scholar] [CrossRef] [PubMed]
- Savidis, G.; Perreira, J.M.; Portmann, J.M.; Meraner, P.; Guo, Z.; Green, S.; Brass, A.L. The ifitms inhibit zika virus replication. Cell Rep. 2016, 15, 2323–2330. [Google Scholar] [CrossRef] [PubMed]
- Raychaudhuri, S.; Prinz, W.A. The diverse functions of oxysterol-binding proteins. Annu. Rev. Cell Dev. Biol. 2010, 26, 157–177. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Moreno, R.; Cuesta-Geijo, M.A.; Martinez-Romero, C.; Barrado-Gil, L.; Galindo, I.; Garcia-Sastre, A.; Alonso, C. Antiviral role of ifitm proteins in african swine fever virus infection. PLoS ONE 2016, 11, e0154366. [Google Scholar] [CrossRef] [PubMed]
| Primers | Sequence (5′–3′) | Purpose |
|---|---|---|
| CMV-IFITM1-F | CGGAATTCTATGATCAAGAGCCAGCACGA | Amplification of IFITM1 |
| CMV-IFITM1-R | CGGGATCCGTAGCCTCTGTTACTCTTTGCG | |
| CMV-IFITM2-F | CGGAATTCTATGAACTGCGCTTCCCAGC | Amplification of IFITM2 |
| CMV-IFITM2-R | CGGGATCCGTAGCCTCTGTTACTCTTTGCGC | |
| CMV-IFITM3-F | CGGAATTCTATGAATTGCGCTTCCCAGC | Amplification of IFITM3 |
| CMV-IFITM3-R | CGGGATCCGTAGCCTCTGTAATCCTTTATGAGCT | |
| CSFV-F | GAGAAGGACAGCAGAACTAAGC | RT-qPCR for detection of CSFV |
| CSFV-R | TTACCGCCCATGCCAATAGG | |
| β-actin-F | CAAGGACCTCTACGCCAACAC | RT-qPCR for detection of β-actin |
| β-actin-R | TGGAGGCGCGATGATCTT | |
| shN-F | GATCCGCTTAAACGCATAGTAGGACTCAAGAGAGTCCTACTATGCGTTTAAGCTTTTTG | Negative control of knockdown |
| shN-R | AATTCAAAAAGCTTAAACGCATAGTAGGACTCTCTTGAGTCCTACTATGCGTTTAAGCG | |
| shIFTIM-F | GATCCGCAAAGAGTAACAGAGGCTACCAAGAGGTAGCCTCTGTTACTCTTTGCTTTTTG | Knockdown of IFITMs |
| shIFITM-R | AATTCAAAAAGCAAAGAGTAACAGAGGCTACCTCTTGGTAGCCTCTGTTACTCTTTGCG | |
| IFITM1-F | TGGCTTTCGCCTACTCCG | RT-qPCR for detection of IFITM1 |
| IFITM1-R | ACAGTGGCTCCGATGGTCAG | |
| IFITM2/3-F | TCAACATCCGAAGCGAGACC | RT-qPCR for detection of IFITM2 and IFITM3 |
| IFITM2/3-R | GAGTAGGCGAAAGCCACGAA | |
| C1-IFITM1-F | CGGAATTCATGATCAAGAGCCAGCACGA | Amplification of IFITM1 |
| C1-IFITM1-R | CGGGATCCCTAGTAGCCTCTGTTACTCTTTGCG | |
| C1-IFITM2-F | CGGAATTCATGAACTGCGCTTCCCAGC | Amplification of IFITM2 |
| C1-IFITM2-R | CGGGATCCCTAGTAGCCTCTGTTACTCTTTGCGC | |
| C1-IFITM3-F | CGGAATTCATGAATTGCGCTTCCCAGC | Amplification of IFITM3 |
| C1-IFITM3-R | CGGGATCCCTAGTAGCCTCTGTAATCCTTTATGAGCT | |
| Mx1-F | TCTGTAAGCAGGAGACCATCAACT | RT-qPCR for detection |
| Mx1-R | TTTCTCGCCACGTCCACTATC | of Mx1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Zheng, H.; Wang, Y.; Dong, W.; Liu, Y.; Zhang, L.; Zhang, Y. Antiviral Role of IFITM Proteins in Classical Swine Fever Virus Infection. Viruses 2019, 11, 126. https://doi.org/10.3390/v11020126
Li C, Zheng H, Wang Y, Dong W, Liu Y, Zhang L, Zhang Y. Antiviral Role of IFITM Proteins in Classical Swine Fever Virus Infection. Viruses. 2019; 11(2):126. https://doi.org/10.3390/v11020126
Chicago/Turabian StyleLi, Cheng, Hongqing Zheng, Yifan Wang, Wang Dong, Yaru Liu, Liang Zhang, and Yanming Zhang. 2019. "Antiviral Role of IFITM Proteins in Classical Swine Fever Virus Infection" Viruses 11, no. 2: 126. https://doi.org/10.3390/v11020126
APA StyleLi, C., Zheng, H., Wang, Y., Dong, W., Liu, Y., Zhang, L., & Zhang, Y. (2019). Antiviral Role of IFITM Proteins in Classical Swine Fever Virus Infection. Viruses, 11(2), 126. https://doi.org/10.3390/v11020126




