Homburgvirus LP-018 Has a Unique Ability to Infect Phage-Resistant Listeria monocytogenes
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Bacteriophages
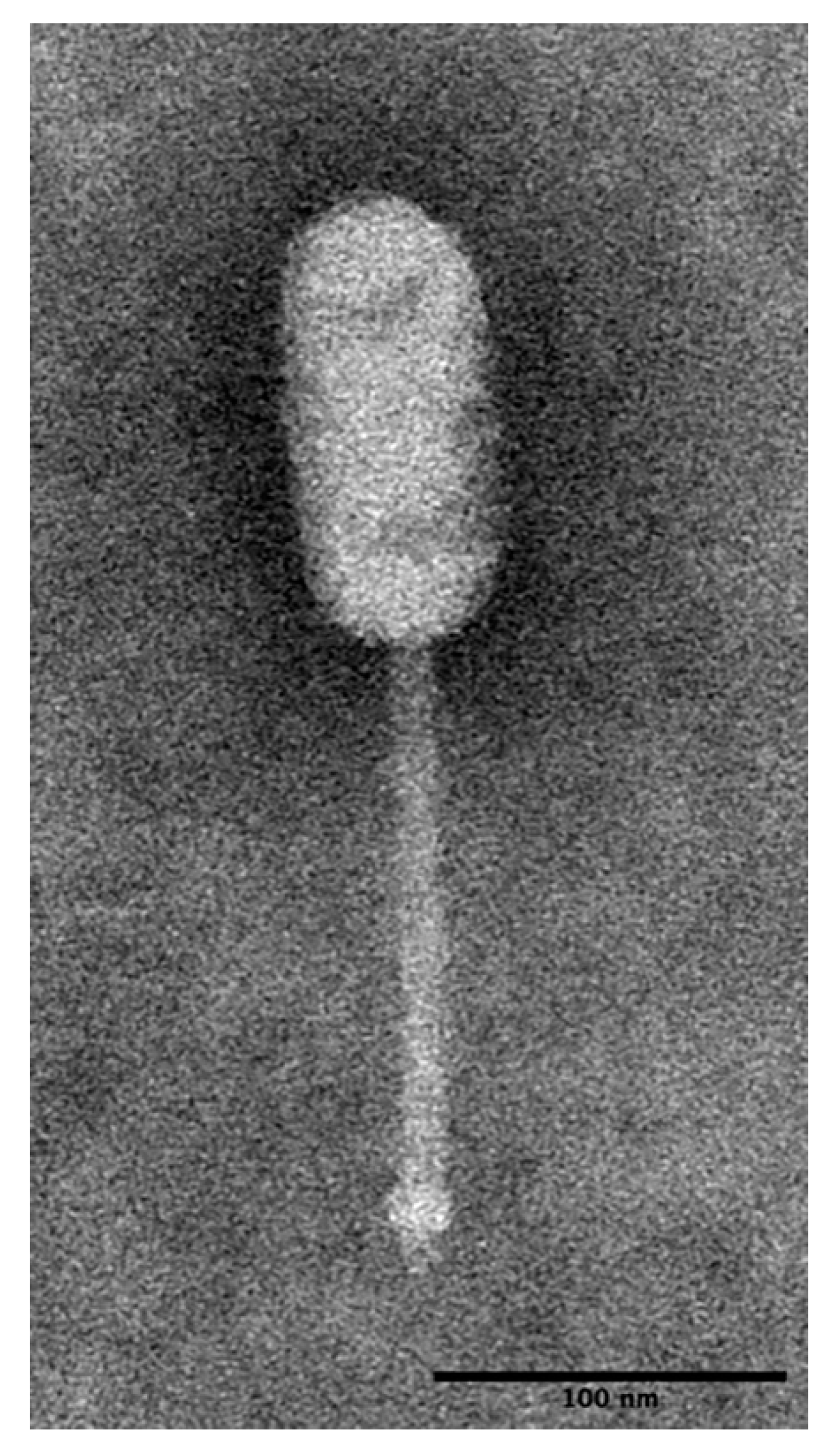
2.2. Morphological Observation of LP-018 by Transmission Electron Microscopy
2.3. Isolation of Phage-Resistant Mutants
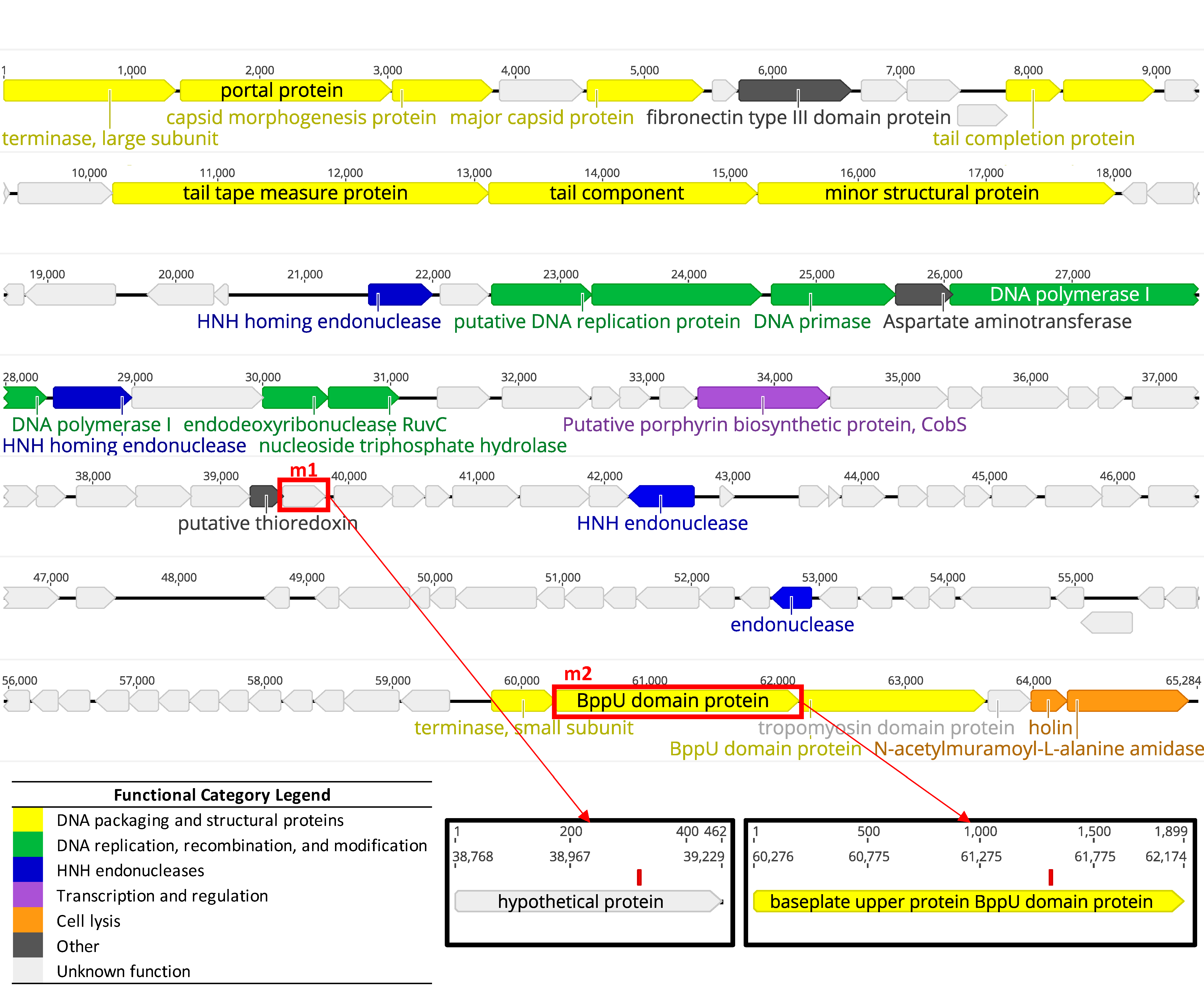
2.4. DNA Extraction and Genomic Analysis
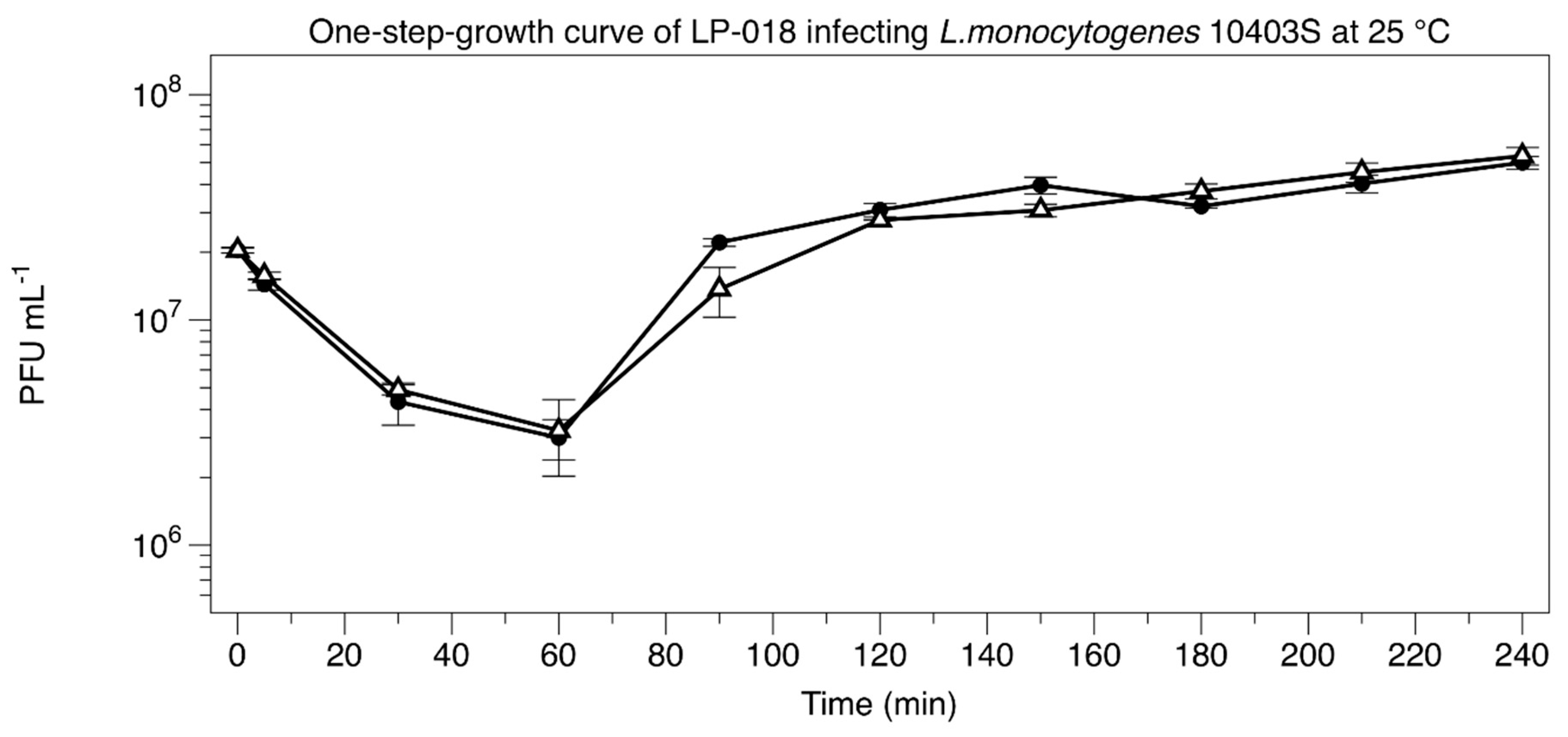
2.5. One-Step-Growth Experiment
2.6. Growth Inhibition Assay of Listeria monocytogenes by LP-018
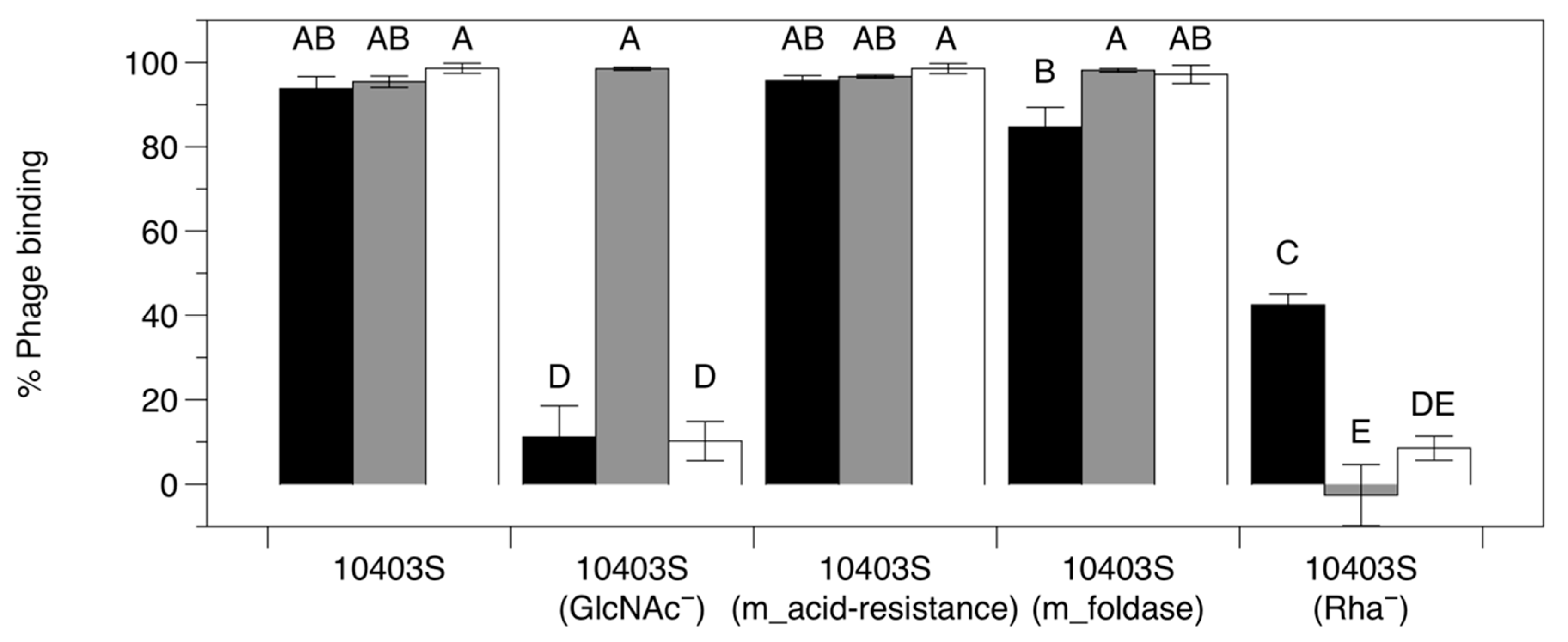
2.7. Phage Adsorption Assay
3. Results and Discussion
3.1. Characterization of LP-018
3.1.1. LP-018 Shares Morphology with Phages of the Genus Homburgvirus
3.1.2. Whole Genome Sequencing Confirms LP-018 is a Homburgvirus
3.2. Growth Characteristics of LP-018
3.2.1. One-Step-Growth Experiment
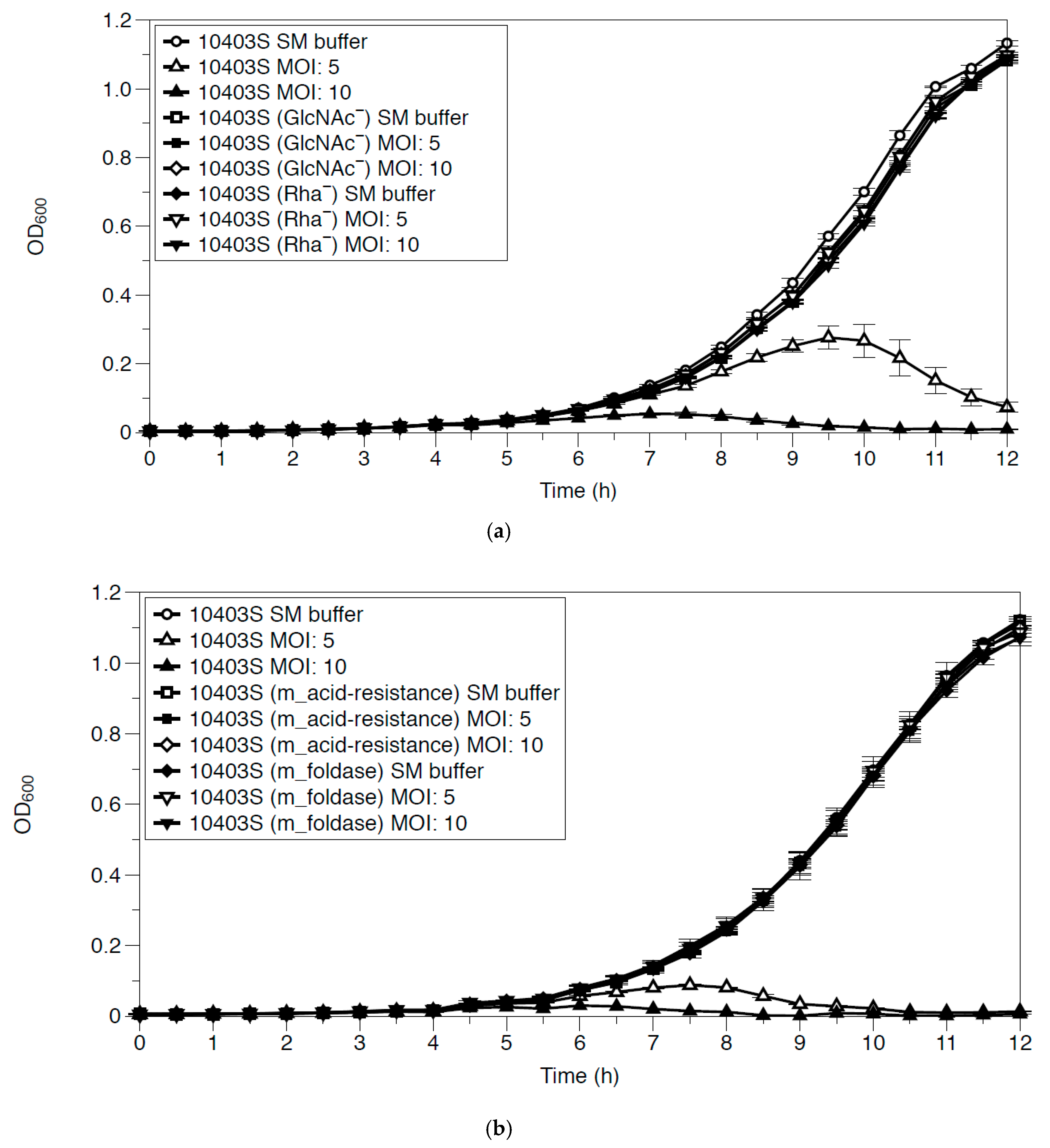
3.2.2. Growth Inhibition of Listeria monocytogenes by LP-018
3.3. Isolation and Characterization of LP-018-Resistant Mutants
3.3.1. Isolation of LP-018-Resistant Mutants
3.3.2. Genetic Characterization of LP-018-Resistant Mutants
3.3.3. Adsorption Assay with Phage-Resistant Mutants
3.4. Growth Inhibition of Phage-Resistant Mutants by LP-018
3.5. Isolation and Characterization of LP-018 Mutants with Different Host Ranges
3.6. Efficiency of Plaquing of Homburgviruses Against LP-018-Resistant Mutants
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gray, J.A.; Chandry, P.S.; Kaur, M.; Kocharunchitt, C.; Bowman, J.P.; Fox, E.M. Novel biocontrol methods for Listeria monocytogenes biofilms in food production facilities. Front. Microbiol. 2018, 9, 605. [Google Scholar] [CrossRef] [PubMed]
- Pizarro-Cerda, J.; Cossart, P. Listeria monocytogenes: Cell biology of invasion and intracellular growth. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Vivant, A.-L.; Garmyn, D.; Piveteau, P. Listeria monocytogenes, a down-to-earth pathogen. Front. Cell. Infect. Microbiol. 2013, 3, 87. [Google Scholar] [CrossRef] [PubMed]
- Sauders, B.D.; Overdevest, J.; Fortes, E.; Windham, K.; Schukken, Y.; Lembo, A.; Wiedmann, M. Diversity of Listeria species in urban and natural environments. Appl. Env. Microbiol. 2012, 78, 4420–4433. [Google Scholar] [CrossRef] [PubMed]
- Fenlon, D. Wild birds and silage as reservoirs of Listeria in the agricultural environment. J. Appl. Bacteriol. 1985, 59, 537–543. [Google Scholar] [CrossRef]
- Kramarenko, T.; Roasto, M.; Meremäe, K.; Kuningas, M.; Põltsama, P.; Elias, T. Listeria monocytogenes prevalence and serotype diversity in various foods. Food Control 2013, 30, 24–29. [Google Scholar] [CrossRef]
- Todd, E.; Notermans, S. Surveillance of listeriosis and its causative pathogen, Listeria monocytogenes. Food Control 2011, 22, 1484–1490. [Google Scholar] [CrossRef]
- Hoffman, S.; Maculloch, B.; Batz, M. Economic Burden of Major Foodborne Illnesses Acquired in the United States; United States Department of Agriculture: Wahington, DC, USA, 2015.
- Klontz, K.C.; Mccarthy, P.V.; Datta, A.R.; Lee, J.O.; Acheson, D.W.; Brackett, R.E. Role of the us food and drug administration in the regulatory management of human listeriosis in the united states. J. Food Prot. 2008, 71, 1277–1286. [Google Scholar] [CrossRef]
- De Noordhout, C.M.; Devleesschauwer, B.; Angulo, F.J.; Verbeke, G.; Haagsma, J.; Kirk, M.; Havelaar, A.; Speybroeck, N. The global burden of listeriosis: A systematic review and meta-analysis. Lancet Infect. Dis. 2014, 14, 1073–1082. [Google Scholar] [CrossRef]
- Olanya, O.M.; Hoshide, A.K.; Ijabadeniyi, O.A.; Ukuku, D.O.; Mukhopadhyay, S.; Niemira, B.A.; Ayeni, O. Cost estimation of listeriosis (Listeria monocytogenes) occurrence in south africa in 2017 and its food safety implications. Food Control 2019, 102, 231–239. [Google Scholar] [CrossRef]
- Moye, Z.; Woolston, J.; Sulakvelidze, A. Bacteriophage applications for food production and processing. Viruses 2018, 10, 205. [Google Scholar] [CrossRef] [PubMed]
- Chernomordik, A. Bacteriophages and their therapeutic-prophylactic use. Meditsinskaia Sestra 1989, 48, 44–47. [Google Scholar] [PubMed]
- Carlton, R.M. Phage therapy: Past history and future prospects. Arch. Immunol. Ther. Exp. 1999, 47, 267–274. [Google Scholar]
- Denes, T.; den Bakker, H.C.; Tokman, J.I.; Guldimann, C.; Wiedmann, M. Selection and characterization of phage-resistant mutant strains of Listeria monocytogenes reveal host genes linked to phage adsorption. Appl. Environ. Microbiol. 2015, 81, 4295–4305. [Google Scholar] [CrossRef]
- Fister, S.; Fuchs, S.; Stessl, B.; Schoder, D.; Wagner, M.; Rossmanith, P. Screening and characterisation of bacteriophage p100 insensitive Listeria monocytogenes isolates in austrian dairy plants. Food Control 2016, 59, 108–117. [Google Scholar] [CrossRef]
- Eugster, M.R.; Morax, L.S.; Hüls, V.J.; Huwiler, S.G.; Leclercq, A.; Lecuit, M.; Loessner, M.J. Bacteriophage predation promotes serovar diversification in Listeria monocytogenes. Mol. Microbiol. 2015, 97, 33–46. [Google Scholar] [CrossRef]
- Trudelle, D.M.; Bryan, D.W.; Hudson, L.K.; Denes, T.G. Cross-resistance to phage infection in Listeria monocytogenes serotype 1/2a mutants. Food Microbiol. 2019, 84. [Google Scholar] [CrossRef]
- Sumrall, E.T.; Shen, Y.; Keller, A.P.; Rismondo, J.; Pavlou, M.; Eugster, M.R.; Boulos, S.; Disson, O.; Thouvenot, P.; Kilcher, S. Phage resistance at the cost of virulence: Listeria monocytogenes serovar 4b requires galactosylated teichoic acids for inlb-mediated invasion. PLoS Pathog. 2019, 15, e1008032. [Google Scholar] [CrossRef]
- Hodgson, D.A. Generalized transduction of serotype 1/2 and serotype 4b strains of Listeria monocytogenes. Mol. Microbiol. 2000, 35, 312–323. [Google Scholar] [CrossRef]
- Bishop, D.; Hinrichs, D. Adoptive transfer of immunity to Listeria monocytogenes. The influence of in vitro stimulation on lymphocyte subset requirements. J. Immunol. 1987, 139, 2005–2009. [Google Scholar]
- Vongkamjan, K.; Switt, A.M.; den Bakker, H.C.; Fortes, E.D.; Wiedmann, M. Silage collected from dairy farms harbors an abundance of listeriaphages with considerable host range and genome size diversity. Appl. Environ. Microbiol. 2012, 78, 8666–8675. [Google Scholar] [CrossRef] [PubMed]
- Denes, T.; Vongkamjan, K.; Ackermann, H.W.; Moreno Switt, A.I.; Wiedmann, M.; den Bakker, H.C. Comparative genomic and morphological analyses of Listeria phages isolated from farm environments. Appl. Environ. Microbiol. 2014, 80, 4616–4625. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Extraction of bacteriophage λ DNA from large-scale cultures using proteinase k and sds. Csh Protoc. 2006, 2006, pdb-prot3972. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Andrews, S. Fastqc. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 11 March 2019).
- Turner, I.; Garimella, K.V.; Iqbal, Z.; McVean, G. Integrating long-range connectivity information into de bruijn graphs. Bioinformatics 2018, 34, 2556–2565. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, snpeff: Snps in the genome of drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Completing bacterial genome assemblies with multiplex minion sequencing. Microb. Genom. 2017, 3, e000132. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. Spades: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Bushnell, B. Bbtools: A Suite of Fast, Multithreaded Bioinformatics Tools Designed for Analysis of DNA and rna Sequence Data; Joint Genome Institute: Berkeley, CA, USA, 2018. Available online: https://jgi. doe.gov/data-and-tools/bbtools (accessed on 11 March 2019).
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and samtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. Quast: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D. Rasttk: A modular and extensible implementation of the rast algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G. Interproscan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R.; Oliver Glöckner, F.; Peplies, J. Jspeciesws: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2015, 32, 929–931. [Google Scholar] [CrossRef] [PubMed]
- Schmuki, M.M.; Erne, D.; Loessner, M.J.; Klumpp, J. Bacteriophage p70: Unique morphology and unrelatedness to other listeria bacteriophages. J. Virol. 2012, 86, 13099–13102. [Google Scholar] [CrossRef] [PubMed]
- Hudson, L.K.; Peters, T.L.; Song, Y.; Denes, T.G. Complete genome sequences and transmission electron micrographs of Listeria phages of the genus Homburgvirus. Microbiol. Resour. Announc. 2019, 8, e00825-19. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, H.-W.; Caprioli, T.; Kasatiya, S.S. A large new Streptococcus bacteriophage. Can. J. Microbiol. 1975, 21, 571–574. [Google Scholar] [CrossRef]
- Adriaenssens, E.; Brister, J.R. How to name and classify your phage: An informal guide. Viruses 2017, 9, 70. [Google Scholar] [CrossRef]
- Bishop-Lilly, K.A.; Plaut, R.D.; Chen, P.E.; Akmal, A.; Willner, K.M.; Butani, A.; Dorsey, S.; Mokashi, V.; Mateczun, A.J.; Chapman, C. Whole genome sequencing of phage resistant Bacillus anthracis mutants reveals an essential role for cell surface anchoring protein CsaB in phage ap50c adsorption. Virol. J. 2012, 9, 246. [Google Scholar] [CrossRef]
- Tiensuu, T.; Andersson, C.; Rydén, P.; Johansson, J. Cycles of light and dark co-ordinate reversible colony differentiation in Listeria monocytogenes. Mol. Microbiol. 2013, 87, 909–924. [Google Scholar] [CrossRef]
- Liu, Y.; Orsi, R.H.; Gaballa, A.; Wiedmann, M.; Boor, K.J.; Guariglia-Oropeza, V. Systematic review of the Listeria monocytogenes σb regulon supports a role in stress response, virulence and metabolism. Future Microbiol. 2019, 14, 801–828. [Google Scholar] [CrossRef]
- Denes, T.; Wiedmann, M. Environmental responses and phage susceptibility in foodborne pathogens: Implications for improving applications in food safety. Curr. Opin. Biotechnol. 2014, 26, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Alonzo, F.; Port, G.C.; Cao, M.; Freitag, N.E. The posttranslocation chaperone prsa2 contributes to multiple facets of Listeria monocytogenes pathogenesis. Infect. Immun. 2009, 77, 2612–2623. [Google Scholar] [CrossRef] [PubMed]
- Zemansky, J.; Kline, B.C.; Woodward, J.J.; Leber, J.H.; Marquis, H.; Portnoy, D.A. Development of a mariner-based transposon and identification of Listeria monocytogenes determinants, including the peptidyl-prolyl isomerase prsa2, that contribute to its hemolytic phenotype. J. Bacteriol. 2009, 191, 3950–3964. [Google Scholar] [CrossRef] [PubMed]
- Alonzo, F.; Freitag, N.E. Listeria monocytogenes prsa2 is required for virulence factor secretion and bacterial viability within the host cell cytosol. Infect. Immun. 2010, 78, 4944–4957. [Google Scholar] [CrossRef]
- Beveridge, T.J.; Matias, V.R. Ultrastructure of gram-positive cell walls. In Gram-Positive Pathogens, 2nd ed.; American Society of Microbiology: Washington, DC, USA, 2006; pp. 3–11. [Google Scholar]
- Fernandes, S.; São-José, C. Enzymes and mechanisms employed by tailed bacteriophages to breach the bacterial cell barriers. Viruses 2018, 10, 396. [Google Scholar] [CrossRef]
- Veesler, D.; Spinelli, S.; Mahony, J.; Lichière, J.; Blangy, S.; Bricogne, G.; Legrand, P.; Ortiz-Lombardia, M.; Campanacci, V.; van Sinderen, D. Structure of the phage tp901-1 1.8 mda baseplate suggests an alternative host adhesion mechanism. Proc. Natl. Acad. Sci. USA 2012, 109, 8954–8958. [Google Scholar] [CrossRef]
- Li, X.; Koç, C.; Kühner, P.; Stierhof, Y.-D.; Krismer, B.; Enright, M.C.; Penadés, J.R.; Wolz, C.; Stehle, T.; Cambillau, C. An essential role for the baseplate protein gp45 in phage adsorption to Staphylococcus aureus. Sci. Rep. 2016, 6, 26455. [Google Scholar] [CrossRef]
- Tokman, J.I.; Kent, D.J.; Wiedmann, M.; Denes, T. Temperature significantly affects the plaquing and adsorption efficiencies of Listeria phages. Front. Microbiol. 2016, 7, 631. [Google Scholar] [CrossRef]


| Strain or Phage | Description | Reference or Original |
|---|---|---|
| Listeria monocytogenes Strain | ||
| MACK | Lineage II, Serotype 1/2a | Hodgson, 2000 [20] |
| 10403S | Lineage II, Serotype 1/2a | Bishop and Hinrichs, 1987 [21] |
| FSL D4-0014 | 10403S mutant; referred to here as 10403S (GlcNAc−); nonsense mutation in LMRG_00541; deficient of N-acetyl glucosamine in cell wall teichoic acids | Denes et al., 2015 [15] |
| FSL D4-0119 | 10403S mutant; referred to here as 10403S (Rha−); nonsense mutation in LMRG_00542; deficient of rhamnose in cell wall teichoic acids | Denes et al., 2015 [15] |
| UTK S1-0004 | 10403S mutant; referred to here as 10403S (m_acid-resistance); nonsense mutation in LMRG_00278 (encodes acid-resistance family protein HdeD) | This study |
| UTK S1-0010 | 10403S mutant; referred to here as 10403S (m_foldase); nonsense mutation in LMRG_01613 (encodes foldase PrsA precursor) | This study |
| Listeria Phage | ||
| LP-018 | Homburgvirus | Vongkamjan et al., 2012 [22] |
| LP-026 | Homburgvirus | Vongkamjan et al., 2012 [22] |
| LP-037 | Homburgvirus | Vongkamjan et al., 2012 [22] |
| LP-110 | Homburgvirus | Vongkamjan et al., 2012 [22] |
| LP-114 | Homburgvirus | Vongkamjan et al., 2012 [22] |
| LP-048 | Pecentumvirus | Vongkamjan et al., 2012; Denes et al., 2014 [22,23] |
| LP-125 | Pecentumvirus | Vongkamjan et al., 2012; Denes et al., 2014 [22,23] |
| LP-026 | LP-037 | LP-110 | LP-114 | P70 a | LP-018 | |
|---|---|---|---|---|---|---|
| LP-026 | * | 97.60 (93.40) | 96.63 (93.26) | 96.38 (93.62) | 96.29 (91.95) | 97.59 (93.72) |
| LP-037 | 97.60 (96.74) | * | 96.83 (96.56) | 97.49 (95.52) | 96.29 (95.73) | 97.71 (97.50) |
| LP-110 | 96.63 (95.96) | 96.83 (95.92) | * | 97.34 (93.17) | 96.68 (95.42) | 97.45 (95.62) |
| LP-114 | 96.38 (94.17) | 97.49 (92.89) | 97.34 (91.08) | * | 96.31 (93.88) | 97.41 (92.41) |
| P70 a | 96.29 (92.05) | 96.29 (92.46) | 96.68 (92.75) | 96.31 (93.11) | * | 96.31 (91.47) |
| LP-018 | 97.59 (96.21) | 97.71 (96.74) | 97.45 (95.37) | 97.41 (94.31) | 96.33 (93.90) | * |
| Phage | Listeria monocytogenes Strain | ||||
|---|---|---|---|---|---|
| 10403S | FSL D4-0014 | FSL D4-0119 | UTK-S1-0004 | UTK-S1-0010 | |
| 10403S (GlcNAc−) | 10403S (Rha−) | 10403S (m_acid-resistance) | 10403S (m_foldase) | ||
| LP-018 | 1.3 (0.36) | 0.0016 (0.0013) | 0.061 (0.018) | 0 | 0 |
| LP-018_m1 | 1.5 (0.64) | 0 | 0.13 (0.064) | 0 | 0 |
| LP-018_m2 | 0.91 (0.19) | 0.49 (0.13) | 0 | 0 | 0 |
| LP-048 | 0.70 (0.11) | 0.55 (0.21) | 0 | 0.35 (0.091) | 0.35 (0.24) |
| LP-125 | 0.80 (0.34) | 0 | 0 | 0.76 (0.52) | 0.78 (0.45) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, Y.; Peters, T.L.; Bryan, D.W.; Hudson, L.K.; Denes, T.G. Homburgvirus LP-018 Has a Unique Ability to Infect Phage-Resistant Listeria monocytogenes. Viruses 2019, 11, 1166. https://doi.org/10.3390/v11121166
Song Y, Peters TL, Bryan DW, Hudson LK, Denes TG. Homburgvirus LP-018 Has a Unique Ability to Infect Phage-Resistant Listeria monocytogenes. Viruses. 2019; 11(12):1166. https://doi.org/10.3390/v11121166
Chicago/Turabian StyleSong, Yaxiong, Tracey L. Peters, Daniel W. Bryan, Lauren K. Hudson, and Thomas G. Denes. 2019. "Homburgvirus LP-018 Has a Unique Ability to Infect Phage-Resistant Listeria monocytogenes" Viruses 11, no. 12: 1166. https://doi.org/10.3390/v11121166
APA StyleSong, Y., Peters, T. L., Bryan, D. W., Hudson, L. K., & Denes, T. G. (2019). Homburgvirus LP-018 Has a Unique Ability to Infect Phage-Resistant Listeria monocytogenes. Viruses, 11(12), 1166. https://doi.org/10.3390/v11121166






