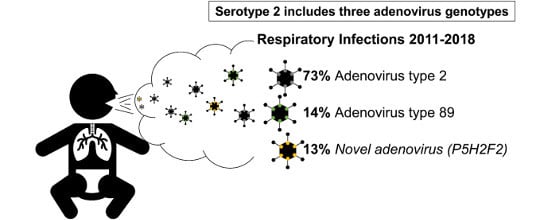
Pediatric Infections by Human mastadenovirus C Types 2, 89, and a Recombinant Type Detected in Japan between 2011 and 2018
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients, Clinical Data, and Adenovirus Sample Collection
2.2. Determination of Partial Sequences of Hexon Loop and Penton Base-Coding Regions
2.3. Multiplex PCR Using the Fiber Region
2.4. Virus Isolation and Neutralization
2.5. Whole Genome Sequence Analysis
2.6. Sequence Analysis
2.7. Ethical Considerations
3. Results
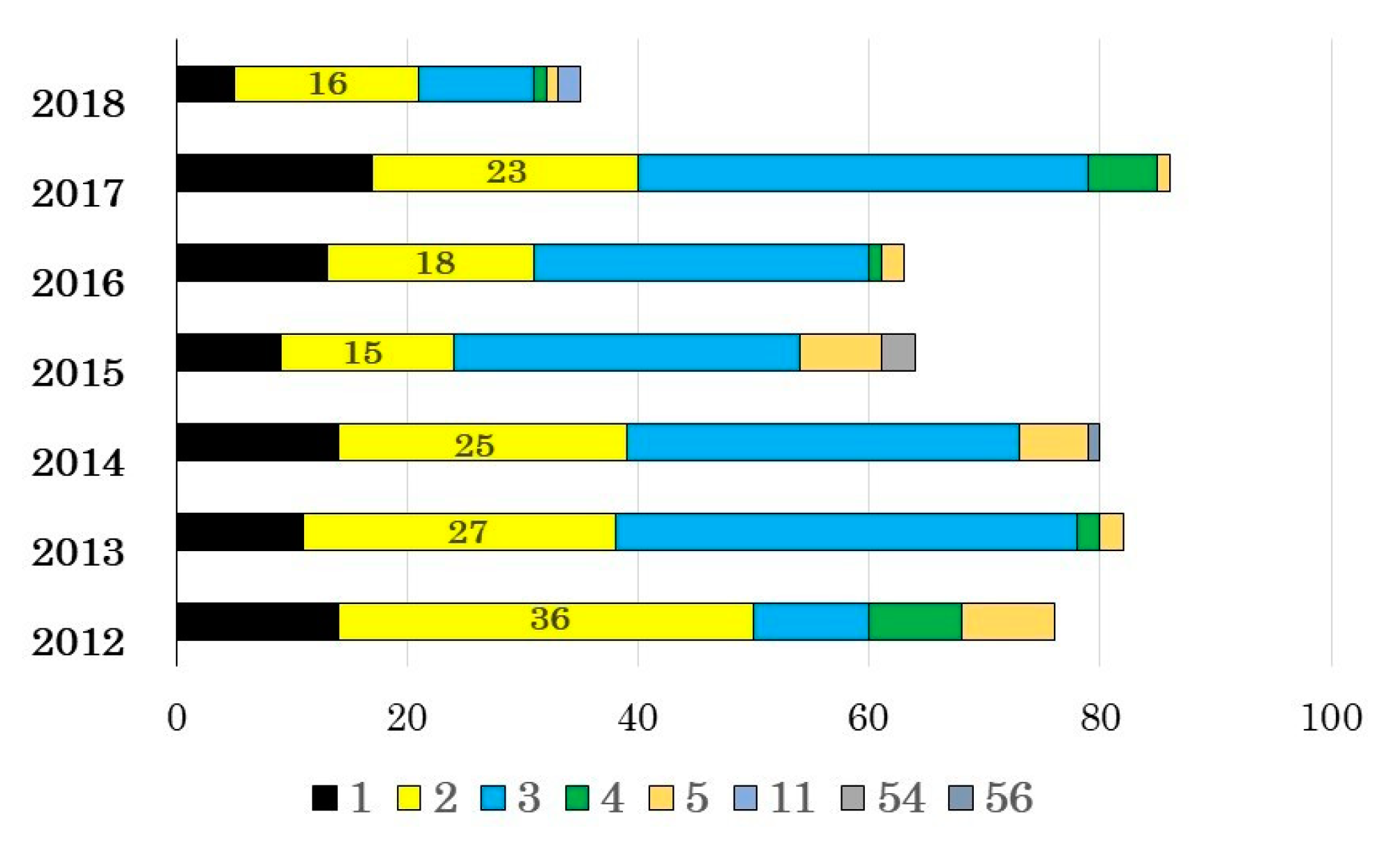
3.1. Adenovirus Detection and Typing by Hexon- and Fiber-Coding Regions
3.2. Nucleotide Sequencing of Penton Base- and Hexon-Coding Regions from Ad-2 Positive Samples
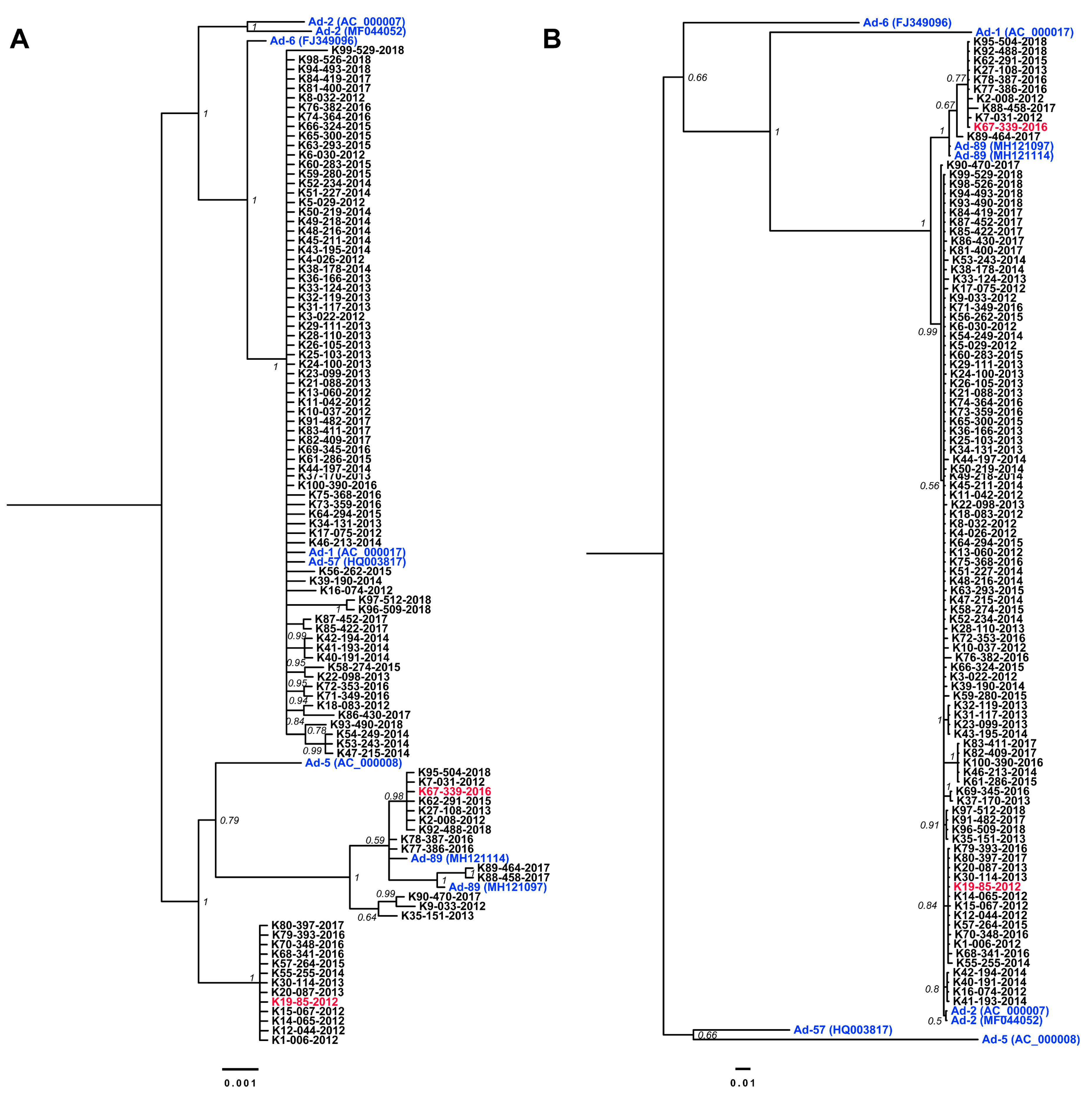
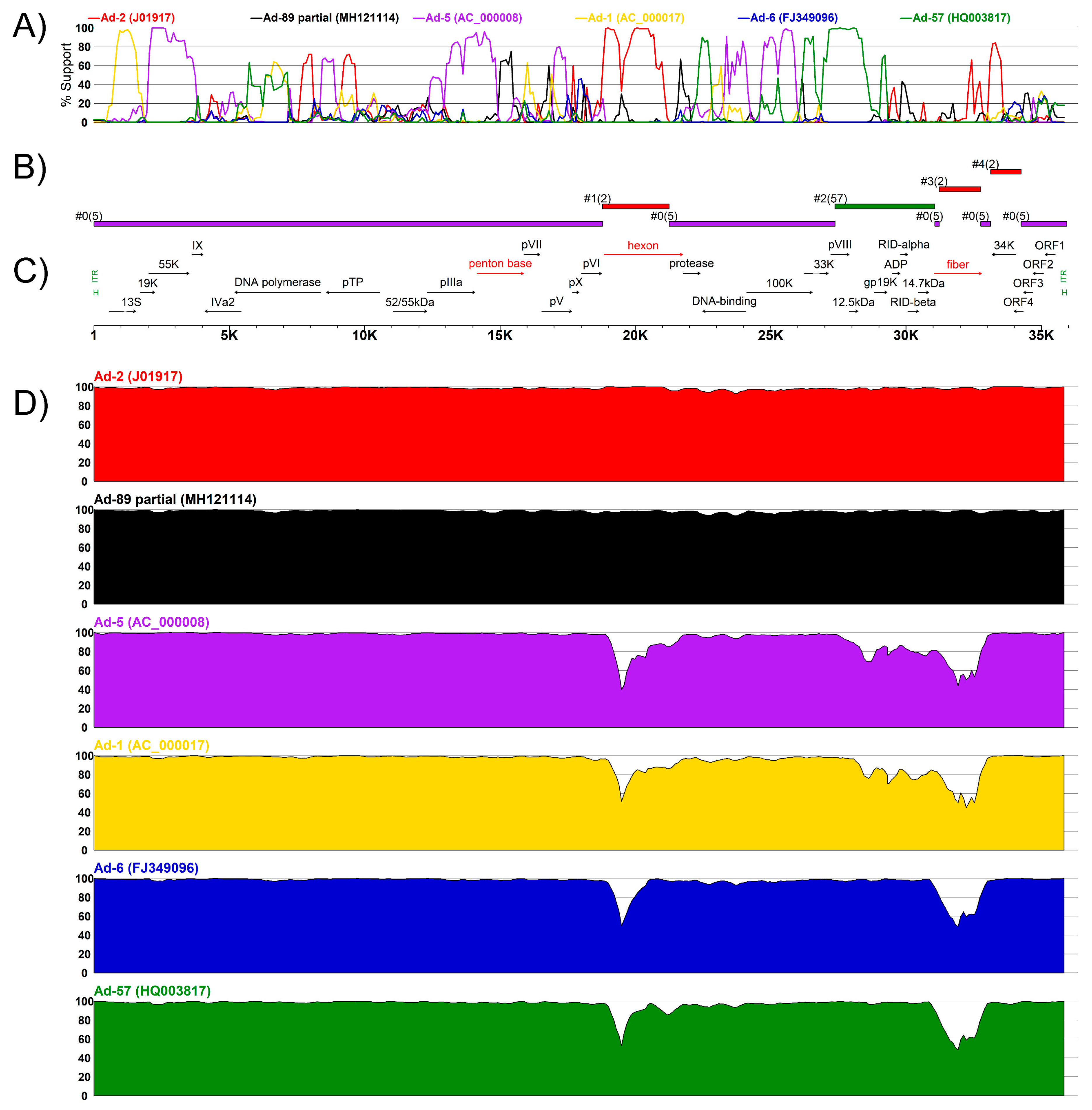
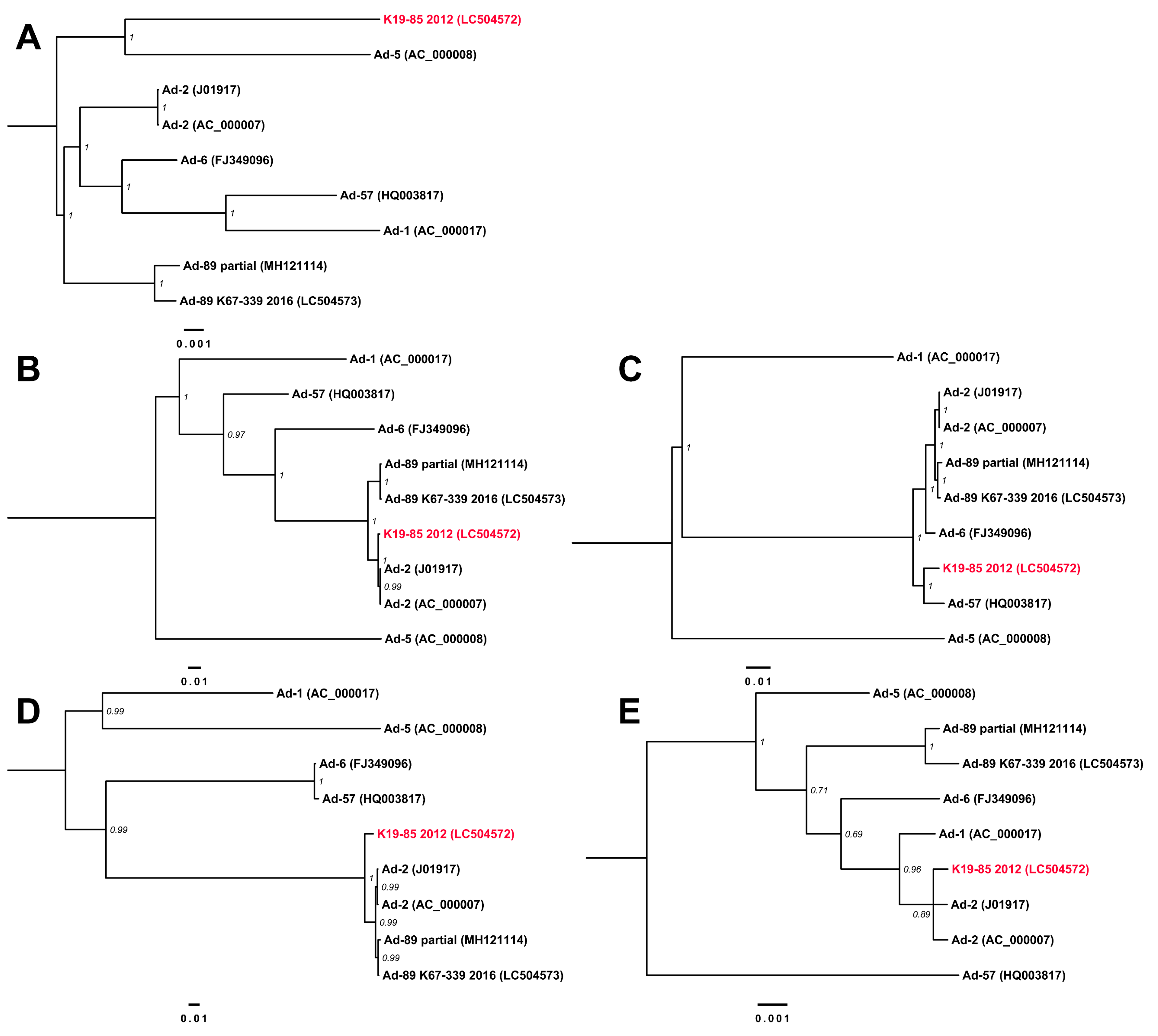
3.3. Complete Genome Sequences for Ad-89 and a Putatively Recombinant Ad Type
3.4. Clinical Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Harrach, B.; Benkö, M.; Both, G.W.; Brown, M.; Davison, A.J.; Echavarría, M.; Hess, M.; Jones, M.S.; Kajon, A.; Lehmkuhl, H.D.; et al. Family Adenoviridae. In Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Lefkowitz, E.J., Carstens, E.B., Eds.; Elsevier Academic Press: San Diego, CA, USA, 2011; pp. 125–141. [Google Scholar]
- Wold, W.S.; Ison, M.G. Adenovirus. In Fields Virology, 6th ed.; Lippincott, Williams & Wilkins: Philadelphia, PA, USA, 2013; pp. 1732–1767. [Google Scholar]
- Fox, J.P.; Brandt, C.D.; Wassermann, F.E.; Hall, C.E.; Spigland, I.; Kogon, A.; Elveback, L.R. The Virus Watch Program: A Continuing Surveillance of Viral Infections in Metropolitan New York Families: Observations of Adenovirus Infections: Virus Excretion Patterns, Antibody Response, Efficiency of Surveillance, Patterns of Infection, and Relation to Illness. Am. J. Epidemiol. 1969, 89, 25–50. [Google Scholar]
- Avila, M.M.; Carballal, G.; Rovaletti, H.; Ebekian, B.; Cusminsky, M.; Weissenbacher, M. Viral etiology in acute lower respiratory infections in children from a closed community. Am. Rev. Respir. Dis. 1989, 140, 634–637. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.P.; Hall, C.E.; Cooney, M.K. The Seattle virus watch: VII. Observations of adenovirus infections. Am. J. Epidemiol. 1977, 105, 362–386. [Google Scholar] [CrossRef] [PubMed]
- Tunkel, A.R.; Baron, E.L.; Buch, K.A.; Marty, F.M.; Martinez-Lage, M. Case 31-2019: A 45-Year-Old Woman with Headache and Somnolence. N. Engl. J. Med. 2019, 381, 1459–1470. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Fujisawa, T.; Suga, S.; Taniguchi, K.; Nagao, M.; Ito, M.; Ochiai, H.; Konagaya, M.; Hanaoka, N.; Fujimoto, T. Species differences in circulation and inflammatory responses in children with common respiratory adenovirus infections. J. Med. Virol. 2018, 90, 873–880. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Adenoviruses | Surveillance Data for National Adenovirus Type Reporting System (NATRS). Available online: https://www.cdc.gov/adenovirus/reporting-surveillance/natrs/surveillance-data.html (accessed on 31 October 2019).
- National Institute of Infectious Disease and Tuberculosis and Infectious Diseases Control Division, Ministry of Health, Labour and Welfare. Adenovirus infections, 2008 to June 2017, Japan; Infectious Agents Surveillance Report.2017; National Institute of Infectious Disease: Tokyo, Japan, 2017; pp. 133–135. Available online: https://www.niid.go.jp/niid/en/iasr-vol38-e/865-iasr/7390-449te.html (accessed on 5 December 2019).
- Jones, M.S., 2nd.; Harrach, B.; Ganac, R.D.; Gozum, M.M.; Dela Cruz, W.P.; Riedel, B.; Pan, C.; Delwart, E.L.; Schnurr, D.P. New adenovirus species found in a patient presenting with gastroenteritis. J. Virol. 2007, 81, 5978–5984. [Google Scholar] [CrossRef]
- Seto, D.; Chodosh, J.; Brister, J.R.; Jones, M.S.; Members of the Adenovirus Research Community. Using the whole-genome sequence to characterize and name human adenoviruses. J. Virol. 2011, 85, 5701–5702. [Google Scholar] [CrossRef]
- Imperiale, M.J.; Enquist, L.W. What’s in a Name? J. Virol. 2011, 85, 5245. [Google Scholar] [CrossRef][Green Version]
- Gonzalez, G.; Koyanagi, K.O.; Aoki, K.; Kitaichi, N.; Ohno, S.; Kaneko, H.; Ishida, S.; Watanabe, H. Intertypic modular exchanges of genomic segments by homologous recombination at universally conserved segments in human adenovirus species D. Gene 2014, 547, 10–17. [Google Scholar] [CrossRef]
- Crawford-Miksza, L.K.; Schnurr, D.P. Adenovirus serotype evolution is driven by illegitimate recombination in the hypervariable regions of the hexon protein. Virology 1996, 224, 357–367. [Google Scholar] [CrossRef]
- Aoki, K.; Benko, M.; Davison, A.J.; Echavarria, M.; Erdman, D.D.; Harrach, B.; Kajon, A.E.; Schnurr, D.; Wadell, G.; Community, A.R. Toward an Integrated Human Adenovirus Designation System That Utilizes Molecular and Serological Data and Serves both Clinical and Fundamental Virology. J. Virol 2011, 85, 5703–5704. [Google Scholar] [CrossRef] [PubMed]
- Walsh, M.P.; Seto, J.; Liu, E.B.; Dehghan, S.; Hudson, N.R.; Lukashev, A.N.; Ivanova, O.; Chodosh, J.; Dyer, D.W.; Jones, M.S.; et al. Computational analysis of two species C human adenoviruses provides evidence of a novel virus. J. Clin. Microbiol. 2011, 49, 3482–3490. [Google Scholar] [CrossRef] [PubMed]
- Dhingra, A.; Hage, E.; Ganzenmueller, T.; Bottcher, S.; Hofmann, J.; Hamprecht, K.; Obermeier, P.; Rath, B.; Hausmann, F.; Dobner, T.; et al. Molecular Evolution of Human Adenovirus (HAdV) Species C. Sci. Rep. 2019, 9, 1039. [Google Scholar] [CrossRef] [PubMed]
- Tatsumi, C.; Iizuka, S.; Mita, T.; Wada, M.; Hanaoka, N.; Fujimoto, T. First Identification of Human Adenovirus 57 (HAdV-57) in Japan. Jpn. J. Infect. Dis. 2018, 71, 259–263. [Google Scholar] [CrossRef]
- Hong, S.S.; Habib, N.A.; Franqueville, L.; Jensen, S.; Boulanger, P.A. Identification of adenovirus (Ad) penton base neutralizing epitopes by use of sera from patients who had received conditionally replicative ad (Addl1520) for treatment of liver tumors. J. Virol. 2003, 77, 10366–10375. [Google Scholar] [CrossRef]
- Tomita, K.; Sakurai, F.; Iizuka, S.; Hemmi, M.; Wakabayashi, K.; Machitani, M.; Tachibana, M.; Katayama, K.; Kamada, H.; Mizuguchi, H. Antibodies against adenovirus fiber and penton base proteins inhibit adenovirus vector-mediated transduction in the liver following systemic administration. Sci. Rep. 2018, 8, 12315. [Google Scholar] [CrossRef]
- Singh, G.; Robinson, C.M.; Dehghan, S.; Schmidt, T.; Seto, D.; Jones, M.S.; Dyer, D.W.; Chodosh, J. Overreliance on the hexon gene, leading to misclassification of human adenoviruses. J. Virol. 2012, 86, 4693–4695. [Google Scholar] [CrossRef]
- Hashimoto, S.; Gonzalez, G.; Harada, S.; Oosako, H.; Hanaoka, N.; Hinokuma, R.; Fujimoto, T. Recombinant type human mastadenovirus D85 associated with epidemic keratoconjunctivitis since 2015 in Japan. J. Med. Virol. 2018, 90, 881–889. [Google Scholar] [CrossRef]
- Fujimoto, T.; Yamane, S.; Ogawa, T.; Hanaoka, N.; Ogura, A.; Hotta, C.; Niwa, T.; Chiba, Y.; Gonzalez, G.; Aoki, K. A novel complex recombinant form of type 48-related human adenovirus species D isolated in Japan. Jpn. J. Infect. Dis. 2014, 67, 282–287. [Google Scholar] [CrossRef]
- Fujimoto, T.; Okafuji, T.; Okafuji, T.; Ito, M.; Nukuzuma, S.; Chikahira, M.; Nishio, O. Evaluation of a bedside immunochromatographic test for detection of adenovirus in respiratory samples, by comparison to virus isolation, PCR, and real-time PCR. J. Clin. Microbiol. 2004, 42, 5489–5492. [Google Scholar] [CrossRef]
- Okada, M.; Ogawa, T.; Kubonoya, H.; Yoshizumi, H.; Shinozaki, K. Detection and sequence-based typing of human adenoviruses using sensitive universal primer sets for the hexon gene. Arch. Virol. 2007, 152, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Matsushima, Y.; Nakajima, E.; Ishikawa, M.; Kano, A.; Komane, A.; Fujimoto, T.; Hanaoka, N.; Okabe, N.; Shimizu, H. Construction of New Primer Sets for Corresponding to Genetic Evolution of Human Adenoviruses in Major Capsid Genes through Frequent Recombination. Jpn. J. Infect. Dis. 2014, 67, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics 2016, 32, 1933–1942. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Lole, K.S.; Bollinger, R.C.; Paranjape, R.S.; Gadkari, D.; Kulkarni, S.S.; Novak, N.G.; Ingersoll, R.; Sheppard, H.W.; Ray, S.C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999, 73, 152–160. [Google Scholar]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus. Evol. 2015, 1, vev003. [Google Scholar] [CrossRef]
- Gahery-Segard, H.; Farace, F.; Godfrin, D.; Gaston, J.; Lengagne, R.; Tursz, T.; Boulanger, P.; Guillet, J.G. Immune response to recombinant capsid proteins of adenovirus in humans: Antifiber and anti-penton base antibodies have a synergistic effect on neutralizing activity. J. Virol. 1998, 72, 2388–2397. [Google Scholar]
- Larranaga, C.; Kajon, A.; Villagra, E.; Avendano, L.F. Adenovirus surveillance on children hospitalized for acute lower respiratory infections in Chile (1988-1996). J. Med. Virol. 2000, 60, 342–346. [Google Scholar] [CrossRef]
- Binder, A.M.; Biggs, H.M.; Haynes, A.K.; Chommanard, C.; Lu, X.Y.; Erdman, D.D.; Watson, J.T.; Gerber, S.I. Human Adenovirus Surveillance - United States, 2003-2016. Mmwr-Morb. Mortal. W. 2017, 66, 1039–1042. [Google Scholar] [CrossRef]
- Gonzalez, G.; Koyanagi, K.O.; Aoki, K.; Watanabe, H. Interregional Coevolution Analysis Revealing Functional and Structural Interrelatedness between different genomic regions in human mastadenovirus D. J. Virol. 2015, 89, 6209–6217. [Google Scholar] [CrossRef] [PubMed]
- Yoshitomi, H.; Sera, N.; Gonzalez, G.; Hanaoka, N.; Fujimoto, T. First isolation of a new type of human adenovirus (genotype 79), species Human mastadenovirus B (B2) from sewage water in Japan. J. Med. Virol. 2017, 89, 1192–1200. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.L.; Chi, C.Y.; Kuo, P.H.; Tsai, H.P.; Wang, S.M.; Liu, C.C.; Su, I.J.; Wang, J.R. High-incidence of human adenoviral co-infections in taiwan. PLoS ONE 2013, 8, e75208. [Google Scholar] [CrossRef] [PubMed]
- Kalu, S.U.; Loeffelholz, M.; Beck, E.; Patel, J.A.; Revai, K.; Fan, J.; Henrickson, K.J.; Chonmaitree, T. Persistence of adenovirus nucleic acids in nasopharyngeal secretions: A diagnostic conundrum. Pediatr. Infect. Dis J. 2010, 29, 746–750. [Google Scholar] [CrossRef]
- Gonzalez, G.; Yawata, N.; Aoki, K.; Kitaichi, N. Challenges in management of epidemic keratoconjunctivitis with emerging recombinant human adenoviruses. J. Clin. Virol. 2019, 112, 1–9. [Google Scholar] [CrossRef]
- Aoki, K.; Gonzalez, G.; Hinokuma, R.; Yawata, N.; Tsutsumi, M.; Ohno, S.; Kitaichi, N. Assessment of clinical signs associated with adenoviral epidemic keratoconjunctivitis cases in southern Japan between 2011 and 2014. Diagn. Microbiol. Infect. Dis. 2019, 95, 114885. [Google Scholar] [CrossRef]
- Wickham, T.J.; Mathias, P.; Cheresh, D.A.; Nemerow, G.R. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell 1993, 73, 309–319. [Google Scholar] [CrossRef]

| Strain * | Anti-Ad-1 | Anti-Ad-2 | Anti-Ad-5 | Anti-Ad-6 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 U | 10 U | 20 U | 5 U | 10 U | 20 U | 5 U | 10 U | 20 U | 5 U | 10 U | 20 U | |
| K19-085-2012 | ++++ | +++ | + | - | - | - | ++++ | +++ | + | +++ | + | ± |
| K27-108-2013 | ++++ | +++ | ++ | - | - | - | ++++ | +++ | + | +++ | + | - |
| K40-191-2014 | +++ | ++ | + | - | - | - | +++ | ++ | + | +++ | + | ± |
| K55-255-2014 | +++ | ++ | ± | - | - | - | ++++ | +++ | + | +++ | + | ± |
| K67-339-2016 | ++++ | +++ | + | - | - | - | ++++ | ++++ | ++ | +++ | + | ± |
| K85-422-2017 | ++++ | +++ | + | - | - | - | ++++ | +++ | ++ | +++ | ++ | + |
| Sequence | No. | Start | End | Minor Parent | Detection Method Support (p-Value) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| RDP | GENECONV | Bootscan | Maxchi | Chimaera | SiSscan | 3Seq | |||||
| K19-85 2012 | 1 | 18,725 | 21,251 | Ad-2 (J01917) | NS | 5 × 10−9 | NS | 1 × 10−2 | 1 × 10−10 | 4 × 10−12 | 1 × 10−2 |
| 2 | 27,376 | 31,060 | Ad-57 (HQ003817) | 2 × 10−31 | 6 × 10−101 | 9 × 10−100 | 3 × 10−36 | 2 × 10−32 | 4 × 10−54 | 3 × 10−15 | |
| 3 | 31,229 | 32,755 | Ad-2 (J01917) | 5 × 10−33 | 2 × 10−82 | 5 × 10−80 | 5 × 10−40 | 2 × 10−39 | 1 × 10−55 | 3 × 10−15 | |
| 4 | 33,123 | 34,253 | Ad-2 (J01917) | NS | 2 × 10−7 | 2 × 10−3 | 2 × 10−7 | 1 × 10−5 | 3 × 10−4 | 2 × 10−3 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takahashi, K.; Gonzalez, G.; Kobayashi, M.; Hanaoka, N.; Carr, M.J.; Konagaya, M.; Nojiri, N.; Ogi, M.; Fujimoto, T. Pediatric Infections by Human mastadenovirus C Types 2, 89, and a Recombinant Type Detected in Japan between 2011 and 2018. Viruses 2019, 11, 1131. https://doi.org/10.3390/v11121131
Takahashi K, Gonzalez G, Kobayashi M, Hanaoka N, Carr MJ, Konagaya M, Nojiri N, Ogi M, Fujimoto T. Pediatric Infections by Human mastadenovirus C Types 2, 89, and a Recombinant Type Detected in Japan between 2011 and 2018. Viruses. 2019; 11(12):1131. https://doi.org/10.3390/v11121131
Chicago/Turabian StyleTakahashi, Kenichiro, Gabriel Gonzalez, Masaaki Kobayashi, Nozomu Hanaoka, Michael J. Carr, Masami Konagaya, Naomi Nojiri, Miki Ogi, and Tsuguto Fujimoto. 2019. "Pediatric Infections by Human mastadenovirus C Types 2, 89, and a Recombinant Type Detected in Japan between 2011 and 2018" Viruses 11, no. 12: 1131. https://doi.org/10.3390/v11121131
APA StyleTakahashi, K., Gonzalez, G., Kobayashi, M., Hanaoka, N., Carr, M. J., Konagaya, M., Nojiri, N., Ogi, M., & Fujimoto, T. (2019). Pediatric Infections by Human mastadenovirus C Types 2, 89, and a Recombinant Type Detected in Japan between 2011 and 2018. Viruses, 11(12), 1131. https://doi.org/10.3390/v11121131