Characterization and Pathogenicity of the Porcine Deltacoronavirus Isolated in Southwest China
Abstract
:1. Introduction
2. Material and Methods
2.1. Clinical Samples
2.2. Cells and Antibodies
2.3. Virus Isolation and Propagation
2.4. TCID50 Assay
2.5. Immunofluorescence Assay (IFA)
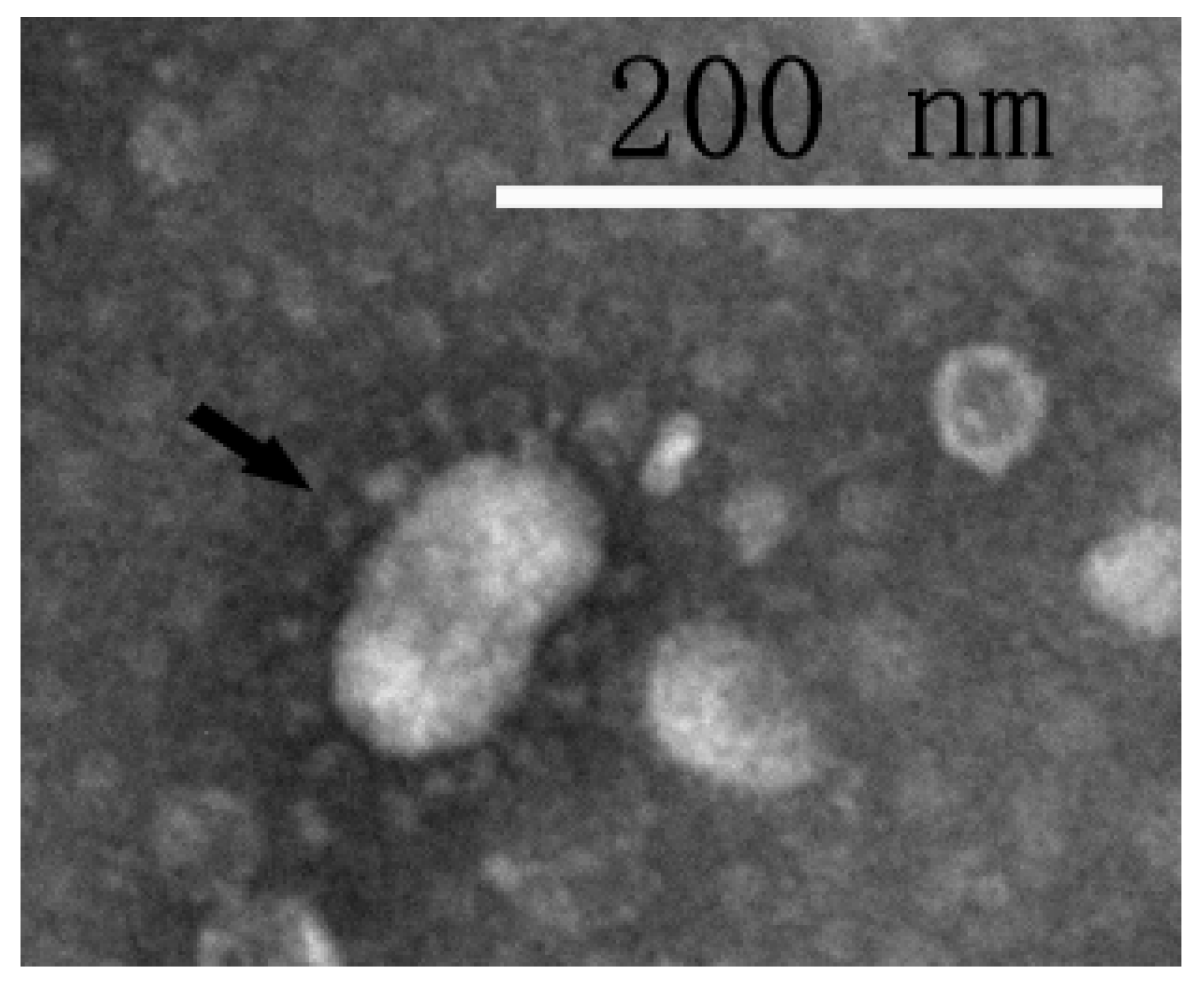
2.6. Electron Microscopy
2.7. RT-PCR/qRT-PCR
2.8. Viral Replication Kinetics
2.9. Genetic Evolution and Mutation Analysis
2.10. Pathogenicity of PDCoV Strain CHN-SC2015 in Piglets
2.11. Statistical Analysis
2.12. Ethics Statement
3. Results
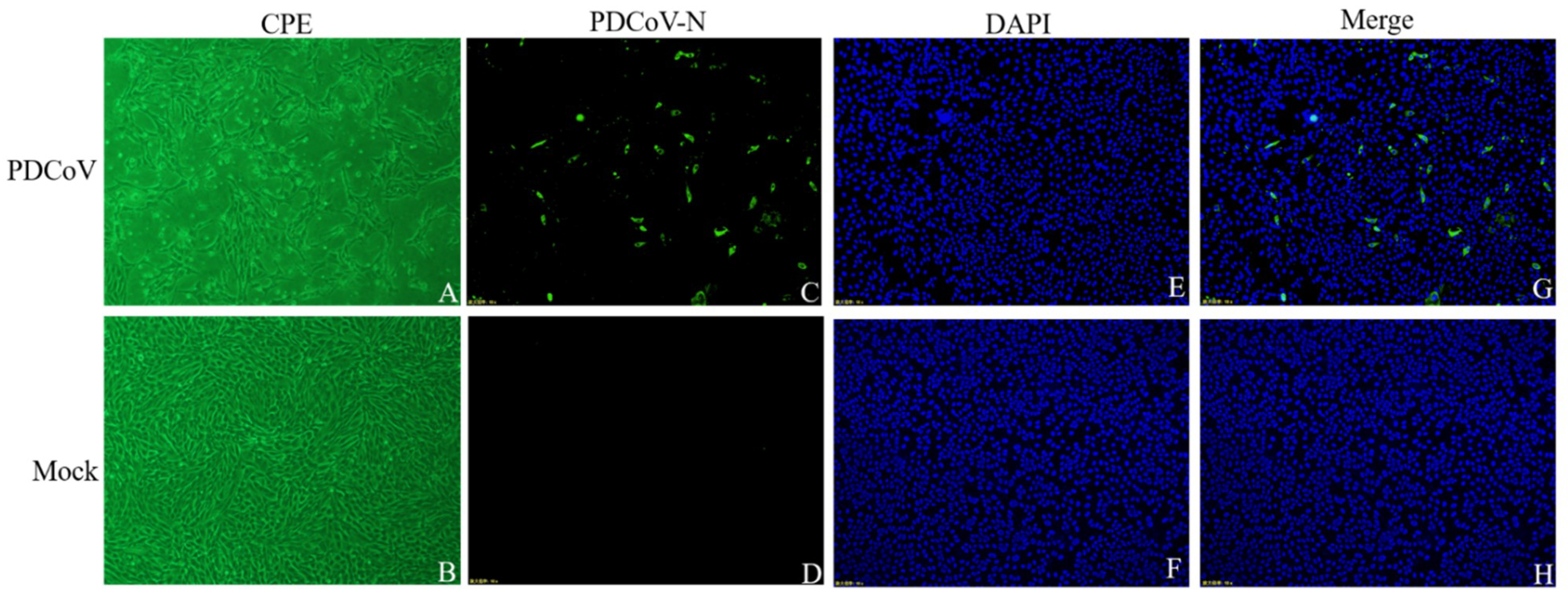
3.1. Virus Isolation and Identification
3.2. Viral Replication
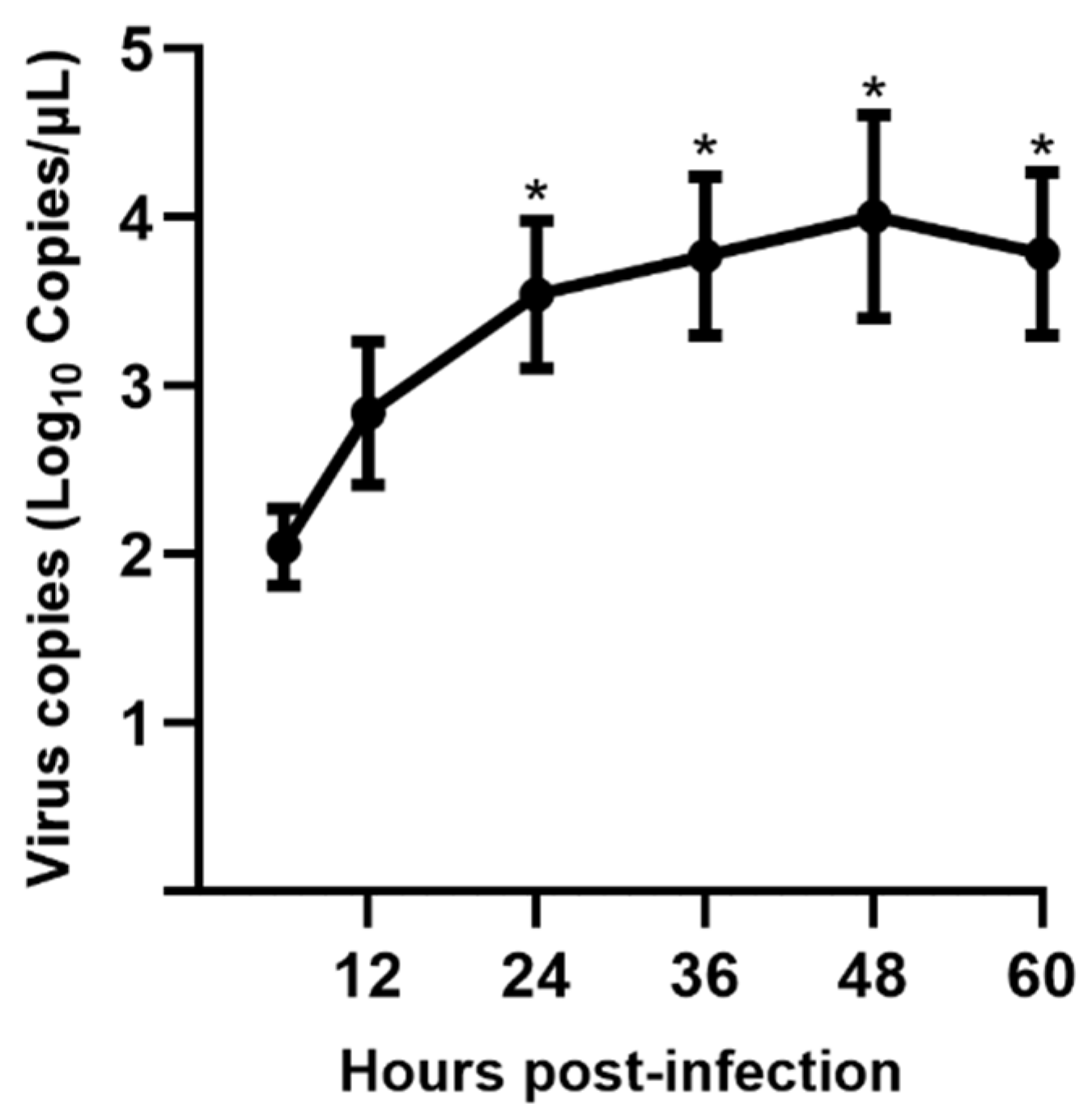
3.3. Viral Replication Kinetics in LLC-PK Cells
3.4. Sequence Variation in the S Gene During Serial Passage
3.5. Genomic Characterization of PDCoV Strain CHN-SC2015
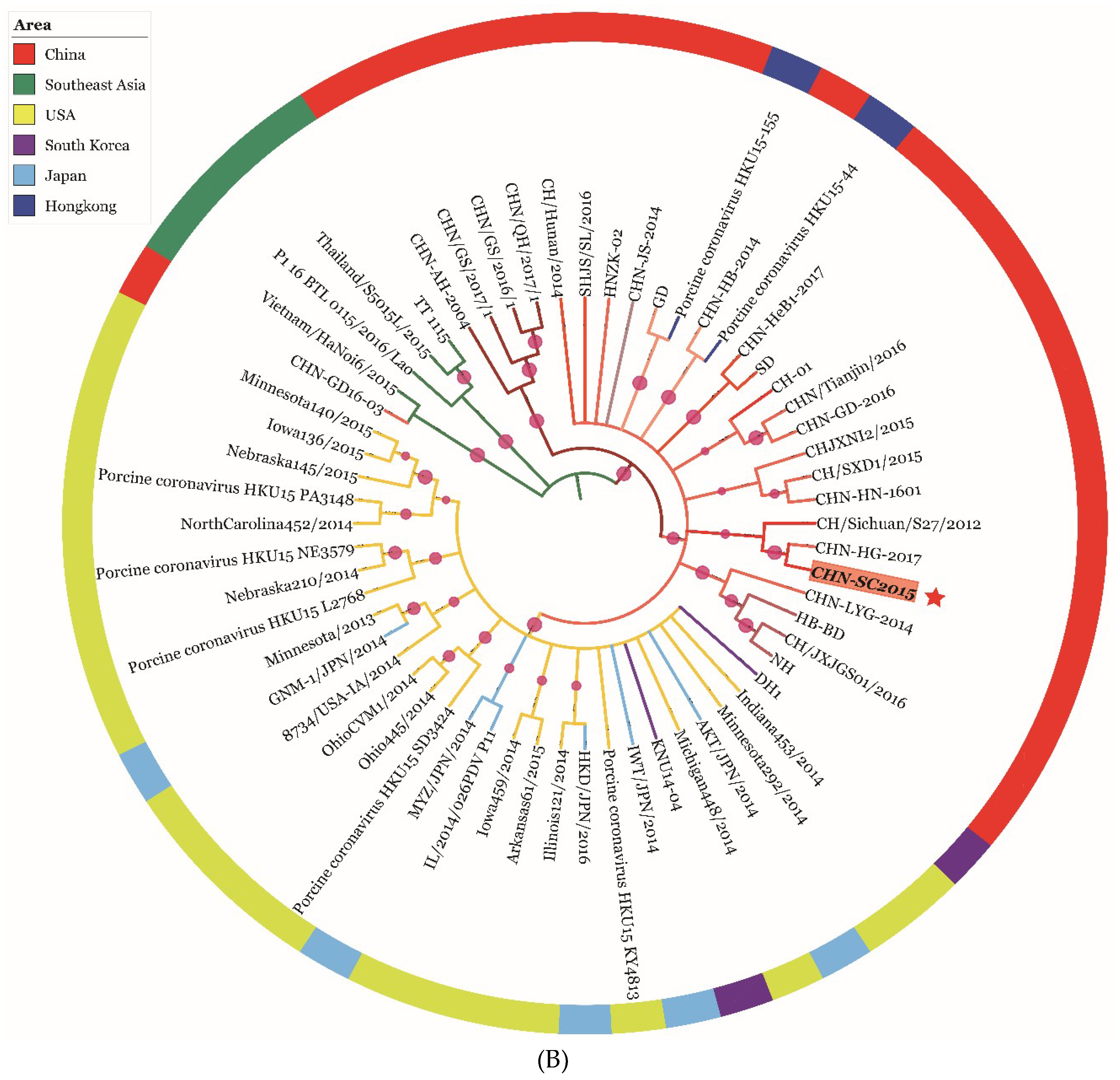
3.6. Phylogenetic and Recombination Analysis of PDCoV Strain CHN-SC2015
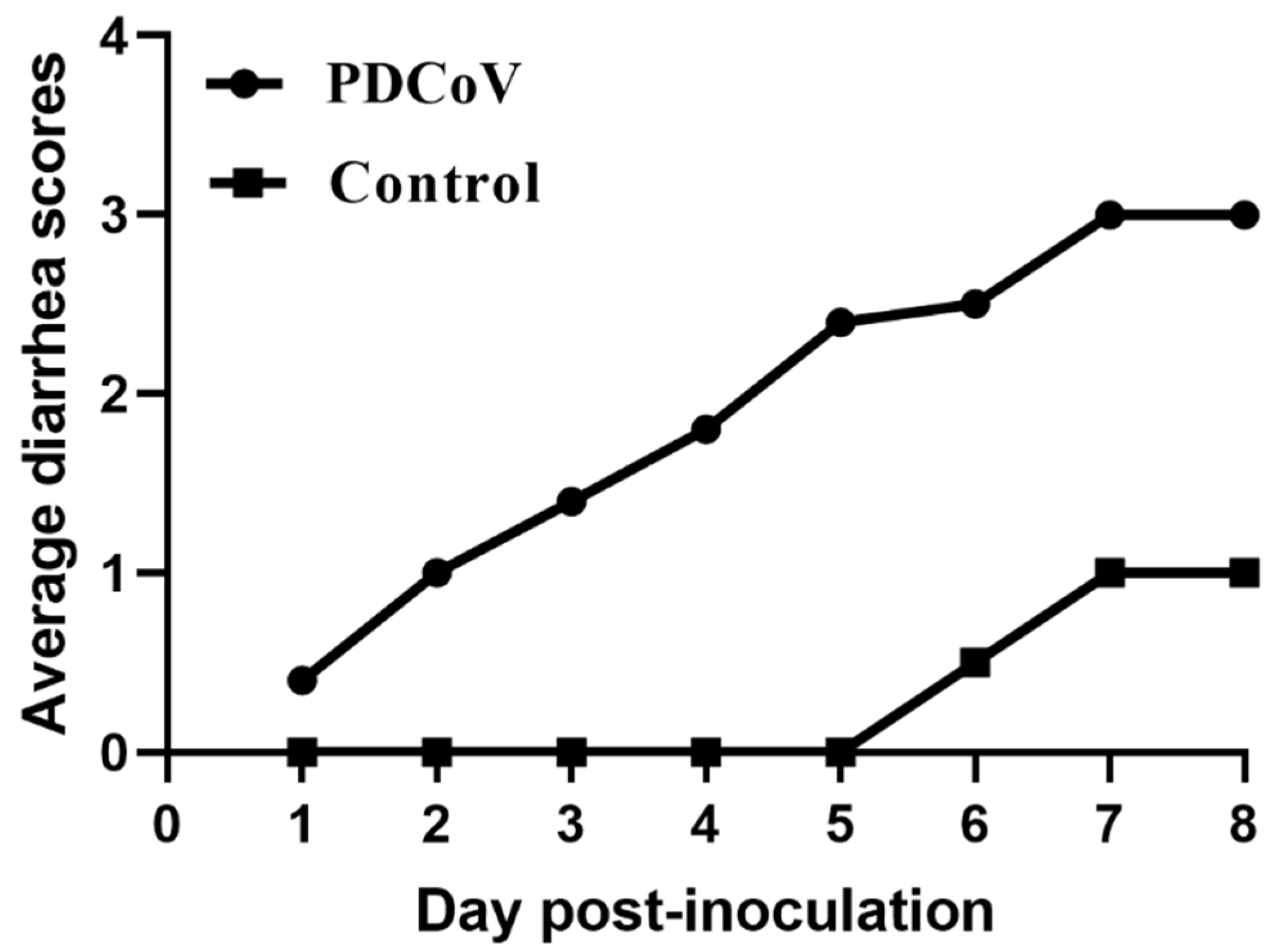
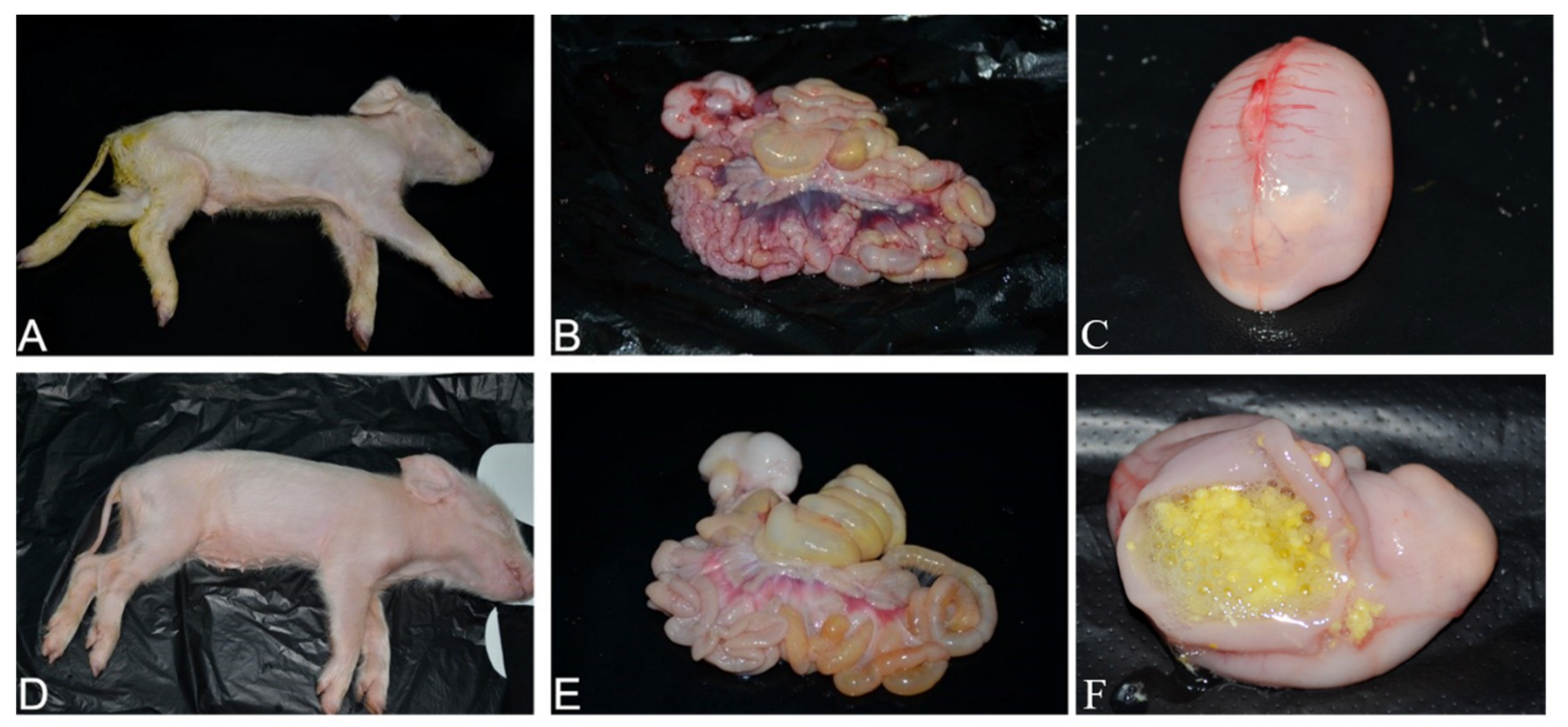
3.7. Pathogenicity of PDCoV Strain CHN-SC2015 in Piglets
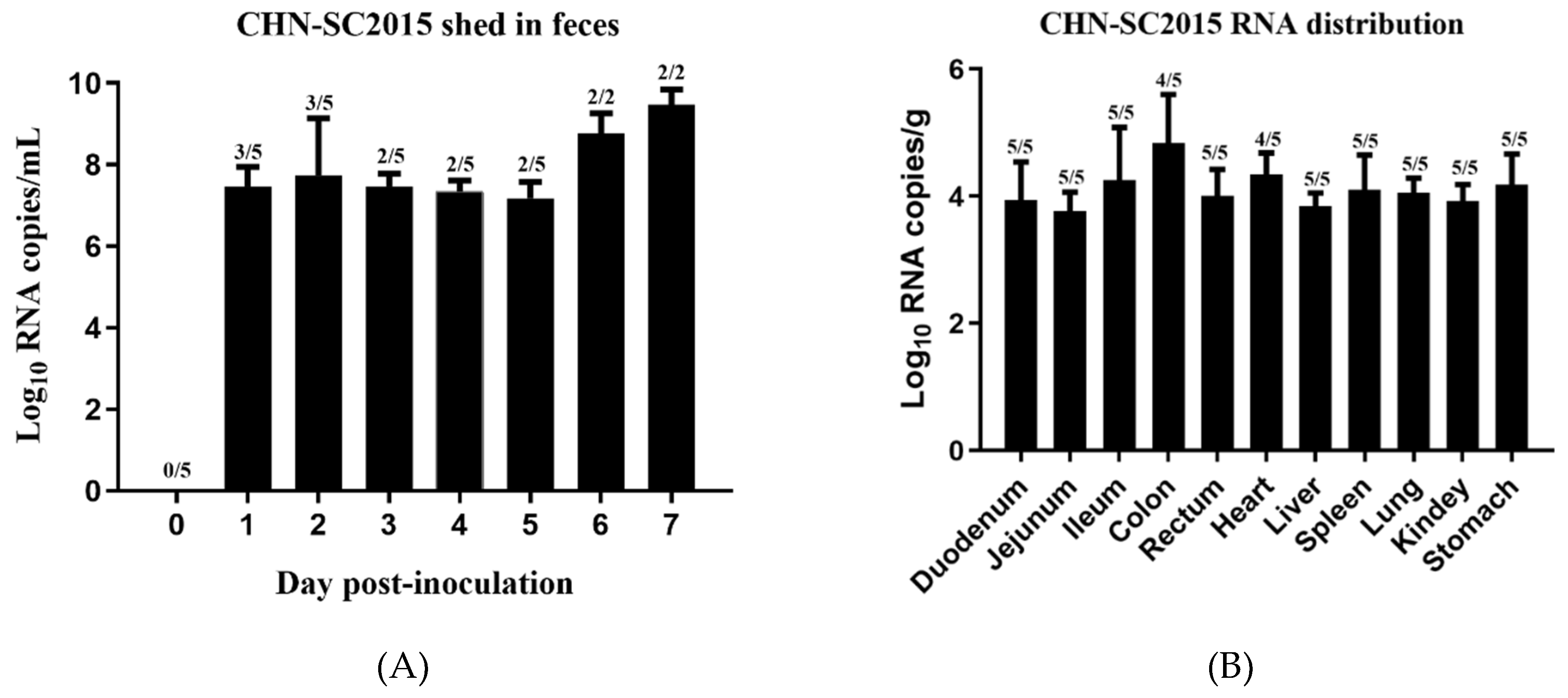
3.8. Viral Shedding and Distribution
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vitosh-Sillman, S.; Loy, J.D.; Brodersen, B.; Kelling, C.; Doster, A.; Topliff, C.; Nelson, E.; Bai, J.; Schirtzinger, E.; Poulsen, E. Experimental infection of conventional nursing pigs and their dams with Porcine deltacoronavirus. J. Vet. Diagn. Investig. 2016, 28, 486–497. [Google Scholar] [CrossRef] [PubMed]
- Saeng-chuto, K.; Jermsutjarit, P.; Stott, C.J.; Vui, D.T.; Tantituvanont, A.; Transboundary, D.N.J.; Diseases, E. Retrospective study, full-length genome characterization and evaluation of viral infectivity and pathogenicity of chimeric porcine deltacoronavirus detected in Vietnam. Transbound. Emerg. Dis. 2019, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Lau, S.K.P.; Lam, C.S.F.; Lau, C.C.Y.; Tsang, A.K.L.; Lau, J.H.N.; Ru, B.; Teng, J.L.L.; Tsang, C.C.C.; Ming, W. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavir. J. Virol. 2012, 86, 3995–4008. [Google Scholar] [CrossRef] [PubMed]
- Leyi, W.; Beverly, B.; Yan, Z. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg. Infect. Dis. 2014, 20, 1227–1230. [Google Scholar]
- Homwong, N.; Jarvis, M.C.; Lam, H.C.; Diaz, A.; Rovira, A.; Nelson, M.; Marthaler, D. Characterization and evolution of porcine deltacoronavirus in the United States. Prev. Vet. Med. 2016, 123, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Jang, G.; Kim, S.H.; Lee, Y.J.; Kim, S.; Lee, D.S.; Lee, K.K.; Lee, C. Isolation and characterization of a Korean porcine deltacoronavirus strain KNU16-07. J. Vet. Sci. 2018, 19, 577–581. [Google Scholar] [CrossRef]
- Niederwerder, M.C.; Hesse, R.A. Swine enteric coronavirus disease: A review of 4 years with porcine epidemic diarrhoea virus and porcine deltacoronavirus in the United States and Canada. Transbound. Emerg. Dis. 2018, 65, 660–675. [Google Scholar] [CrossRef]
- Suzuki, T.; Shibahara, T.; Imai, N.; Yamamoto, T.; Ohashi, S. Genetic characterization and pathogenicity of Japanese porcine deltacoronavirus. Infect. Genet. Evol. 2018, 61, 176–182. [Google Scholar] [CrossRef]
- Le, V.P.; Song, S.; An, B.H.; Park, G.N.; Pham, N.T.; Le, D.Q.; Nguyen, V.T.; Vu, T.T.H.; Kim, K.S.; Choe, S.E. A novel strain of porcine deltacoronavirus in Vietnam. Arch. Virol. 2018, 163, 203–207. [Google Scholar] [CrossRef]
- Lorsirigool, A.; Saeng-Chuto, K.; Madapong, A.; Temeeyasen, G.; Tripipat, T.; Kaewprommal, P.; Tantituvanont, A.; Piriyapongsa, J.; Nilubol, D. The genetic diversity and complete genome analysis of two novel porcine deltacoronavirus isolates in Thailand in 2015. Virus Genes 2017, 53, 240–248. [Google Scholar] [CrossRef]
- Zhang, H.; Liang, Q.; Li, B.; Cui, X.; Wei, X.; Ding, Q.; Wang, Y.; Hu, H. Prevalence, phylogenetic and evolutionary analysis of porcine deltacoronavirus in Henan province, China. Prev. Vet. Med. 2019, 166, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Mai, K.; Feng, J.; Chen, G.; Li, D.; Zhou, L.; Bai, Y.; Wu, Q.; Ma, J. The detection and phylogenetic analysis of porcine deltacoronavirus from Guangdong Province in Southern China. Transbound. Emerg. Dis. 2018, 65, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Dong, N.; Fang, L.; Yang, H.; Liu, H.; Du, T.; Fang, P.; Wang, D.; Chen, H.; Xiao, S. Isolation, genomic characterization, and pathogenicity of a Chinese porcine deltacoronavirus strain CHN-HN-2014. Vet. Microbiol. 2016, 196, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.J.; Zuo, Y.Z.; Gu, W.Y.; Luo, S.X.; Shi, Q.K.; Hou, L.S.; Zhong, F.; Fan, J.H. Isolation and phylogenetic analysis of porcine deltacoronavirus from pigs with diarrhoea in Hebei province, China. Transbound. Emerg. Dis. 2018, 65, 874–882. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J. Porcine Deltacoronavirus: Overview of Infection Dynamics, Diagnostic Methods, Prevalence and Genetic Evolution. Virus Res. 2016, 226, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Vlasova, A.N.; Kenney, S.P.; Saif, L.J. Emerging and re-emerging coronaviruses in pigs. Curr. Opin. Virol. 2019, 34, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.P.; Wong, E.Y.M.; Tsang, C.C.; Ahmed, S.S.; Au-Yeung, R.K.H.; Yuen, K.Y.; Wernery, U.; Woo, P.C.Y. Discovery and sequence analysis of four deltacoronaviruses from birds in the Middle East suggest interspecies jumping and recombination as potential mechanism for avian-to-avian and avian-to-mammalian transmission. J. Virol. 2018, 92, e00265-18. [Google Scholar] [CrossRef]
- Xu, Z.; Zhong, H.; Zhou, Q.; Du, Y.; Chen, L.; Zhang, Y.; Xue, C.; Cao, Y. A Highly Pathogenic Strain of Porcine Deltacoronavirus Caused Watery Diarrhea in Newborn Piglets. Virol. Sinica 2018, 33, 131–141. [Google Scholar] [CrossRef]
- Song, D.; Zhou, X.; Peng, Q.; Chen, Y.; Zhang, F.; Huang, T.; Zhang, T.; Li, A.; Huang, D.; Wu, Q. Newly Emerged Porcine Deltacoronavirus Associated With Diarrhoea in Swine in China: Identification, Prevalence and Full-Length Genome Sequence Analysis. Transbound. Emerg. Dis. 2015, 62, 575–580. [Google Scholar] [CrossRef]
- Jung, K.; Hu, H.; Saif, L.J. Porcine deltacoronavirus infection: Etiology, cell culture for virus isolation and propagation, molecular epidemiology and pathogenesis. Virus Res. 2016, 226, 50–59. [Google Scholar] [CrossRef]
- Fan, J.H.; Zuo, Y.Z.; Li, J.H.; Pei, L.H. Heterogeneity in membrane protein genes of porcine epidemic diarrhea viruses isolated in China. Virus Genes 2012, 45, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Stodola, J.K.; Dubois, G.; Coupanec, A.L.; Desforges, M.; Talbot, P.J. The OC43 human coronavirus envelope protein is critical for infectious virus production and propagation in neuronal cells and is a determinant of neurovirulence and CNS pathology. Virology 2018, 515, 134–149. [Google Scholar] [CrossRef] [PubMed]
- Hsin, W.C.; Chang, C.H.; Chang, C.Y.; Peng, W.H.; Chien, C.L.; Chang, M.F.; Chang, S.C. Nucleocapsid protein-dependent assembly of the RNA packaging signal of Middle East respiratory syndrome coronavirus. J. Biomed. Sci. 2018, 25, 47. [Google Scholar] [CrossRef] [PubMed]
- Fang, P.; Fang, L.; Liu, X.; Hong, Y.; Wang, Y.; Dong, N.; Ma, P.; Bi, J.; Wang, D.; Xiao, S. Identification and subcellular localization of porcine deltacoronavirus accessory protein NS6. Virology 2016, 499, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Fang, P.; Fang, L.; Hong, Y.; Liu, X.; Dong, N.; Ma, P.; Bi, J.; Wang, D.; Xiao, S. Discovery of a novel accessory protein NS7a encoded by porcine deltacoronavirus. J. Gen. Virol. 2017, 98, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Fang, P.; Fang, L.; Ren, J.; Hong, Y.; Liu, X.; Zhao, Y.; Wang, D.; Peng, G.; Xiao, S. Porcine deltacoronavirus accessory protein NS6 antagonizes IFN-β production by interfering with the binding of RIG-I/MDA5 to double-stranded RNA. J. Virol. 2018, e00712-18. [Google Scholar]
- Belouzard, S.; Millet, J.K.; Licitra, B.N.; Whittaker, G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 2012, 4, 1011–1033. [Google Scholar] [CrossRef]
- Seongjun, P.; Sanghyun, K.; Daesub, S.; Bongkyun, P. Novel porcine epidemic diarrhea virus variant with large genomic deletion, South Korea. Emerg. Infect. Dis. 2014, 20, 2089–2092. [Google Scholar]
- Sun, J.; Li, Q.; Shao, C.; Ma, Y.; He, H.; Jiang, S.; Zhou, Y.; Wu, Y.; Ba, S.; Shi, L. Isolation and characterization of Chinese porcine epidemic diarrhea virus with novel mutations and deletions in the S gene. Vet. Microbiol. 2018, 221, 81–89. [Google Scholar] [CrossRef]
- Li, C.; Li, W.; Esesarte, E.L.D.; Guo, H.; Elzen, P.V.D.; Aarts, E.; Born, E.V.D.; Rottier, P.J.M.; Bosch, B.J. Cell attachment domains of the PEDV spike protein are key targets of neutralizing antibodies. J. Virol. 2017, 91, e00273-17. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A Simple Method of Estimating Fifty per Cent Endpoints. Am. J. Hyg. 1938, 27, 493–497. [Google Scholar]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef] [PubMed]
- Lole, K.S.; Bollinger, R.C.; Paranjape, R.S.; Gadkari, D.; Kulkarni, S.S.; Novak, N.G.; Ingersoll, R.; Sheppard, H.W.; Ray, S.C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999, 73, 152–160. [Google Scholar] [PubMed]
- Xiong, X.; Tortorici, M.A.; Snijder, J.; Yoshioka, C.; Walls, A.C.; Li, W.; McGuire, A.T.; Rey, F.A.; Bosch, B.J.; Veesler, D. Glycan shield and fusion activation of a deltacoronavirus spike glycoprotein fine-tuned for enteric infections. J. Virol. 2018, 92, e01628-17. [Google Scholar] [CrossRef]
- Pan, Y. Isolation and characterization of a variant porcine epidemic diarrhea virus in China. Virol. J. 2012, 9, 195. [Google Scholar] [CrossRef] [Green Version]
- Xia, L.; Yang, Y.; Wang, J.; Jing, Y.; Yang, Q. Impact of TGEV infection on the pig small intestine. Virol. J. 2018, 15, 102. [Google Scholar] [CrossRef]
- Beatrice, B.M.; Alice, P.; Antonio, L.; Giovanni, A.; Enrica, S.; Chiara, C.; Silvia, F.; Paolo, B.; Paolo, C.; Douglas, M. Porcine Epidemic Diarrhea Virus and Discovery of a Recombinant Swine Enteric Coronavirus, Italy. Emerg. Infect. Dis. 2016, 22, 83–87. [Google Scholar]
- Gong, L.; Li, J.; Zhou, Q.; Xu, Z.; Chen, L.; Zhang, Y.; Xue, C.; Wen, Z.; Cao, Y. A New Bat-HKU2–like Coronavirus in Swine, China, 2017. Emerg. Infect. Dis. 2017, 23, 1607–1609. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.; Tian, X.; Qin, P.; Wang, B.; Zhao, P.; Yang, Y.L.; Wang, L.; Wang, D.; Song, Y.; Zhang, X.; et al. Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China. Vet. Microbiol. 2017, 211, 15–21. [Google Scholar] [CrossRef]
- Zhou, P.; Fan, H.; Lan, T.; Yang, X.L.; Shi, W.F.; Zhang, W.; Zhu, Y.; Zhang, Y.W.; Xie, Q.M.; Mani, S. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 2018, 556, 255–258. [Google Scholar] [CrossRef]
- Jang, G.; Lee, K.K.; Kim, S.H.; Lee, C. Prevalence, complete genome sequencing and phylogenetic analysis of porcine deltacoronavirus in South Korea, 2014–2016. Transbound. Emerg. Dis. 2017, 64, 1364–1370. [Google Scholar] [CrossRef] [PubMed]
- Wicht, O.; Li, W.; Willems, L.; Meuleman, T.J.; Wubbolts, R.W.; van Kuppeveld, F.J.; Rottier, P.J.; Bosch, B.J. Proteolytic activation of the porcine epidemic diarrhea coronavirus spike fusion protein by trypsin in cell culture. J. Virol. 2014, 88, 7952–7961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, K.; Miyazaki, A.; Hui, H.; Saif, L.J. Susceptibility of porcine IPEC-J2 intestinal epithelial cells to infection with porcine deltacoronavirus (PDCoV) and serum cytokine responses of gnotobiotic pigs to acute infection with IPEC-J2 cell culture-passaged PDCoV. Vet. Microbiol. 2018, 221, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Hui, H.; Kwonil, J.; Vlasova, A.N.; Juliet, C.; Zhongyan, L.; Qiuhong, W.; Saif, L.J. Isolation and characterization of porcine deltacoronavirus from pigs with diarrhea in the United States. J. Clin. Microbiol. 2015, 53, 1537–1548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X.; Liu, S.; Wang, X.; Luo, Z.; Shi, Y.; Wang, D.; Peng, G.; Chen, H.; Fang, L.; Xiao, S. Contribution of porcine aminopeptidase N to porcine deltacoronavirus infection. Emerg. Microb. Infect. 2018, 7, 65. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Hulswit, R.J.G.; Kenney, S.P.; Widjaja, I.; Bosch, B.-J. Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility. Proc. Natl. Acad. Sci. USA 2018, 115, E5135–E5143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef] [Green Version]
- Fang, L. Receptor recognition mechanisms of coronaviruses: A decade of structural studies. J. Virol. 2015, 89, 1954–1964. [Google Scholar]
- Shang, J.; Zheng, Y.; Yang, Y.; Liu, C.; Geng, Q.; Tai, W.; Du, L.; Zhou, Y.; Zhang, W.; Li, F. Cryo-EM structure of porcine delta coronavirus spike protein in the pre-fusion state. J. Virol. 2018, 92, e01556-17. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Wang, Y.; Baloch, A.R.; Pan, Y.; Tian, L.; Xu, F.; Shivaramu, S.; Chen, S.; Zeng, Q. Detection and genetic characterization of porcine deltacoronavirus in Tibetan pigs surrounding the Qinghai-Tibet Plateau of China. Transbound. Emerg. Dis. 2018, 65, 363–369. [Google Scholar] [CrossRef]
- Lau, S.K.P.; Li, K.S.M.; Yi, H.; Chung-Tong, S.; Herman, T.; Ming, W.; Choi, G.K.Y.; Huifang, X.; Lam, C.S.F.; Rongtong, G. Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related Rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self-limiting infection that allows recombination events. J. Virol. 2010, 84, 2808–2819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nefedeva, M.; Titov, I.; Malogolovkin, A. Molecular characteristics of a novel recombinant of porcine epidemic diarrhea virus. Arch. Virol. 2019, 164, 1199–1204. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Wong, G.; Shi, W.; Liu, J.; Lai, A.C.K.; Zhou, J.; Liu, W.; Bi, Y.; Gao, G.F. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016, 24, 490–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.-J.; Liu, D.-J.; Liu, X.-L.; Ge, X.-Y.; Jongkaewwattana, A.; He, Q.-G.; Luo, R. Genomic characterization and pathogenicity of porcine deltacoronavirus strain CHN-HG-2017 from China. Arch. Virol. 2019, 164, 413–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, H.; Jung, K.; Vlasova, A.; Saif, L. Experimental infection of gnotobiotic pigs with the cell-culture-adapted porcine deltacoronavirus strain OH-FD22. Arch. Virol. 2016, 161, 3421–3434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, K.; Hu, H.; Saif, L.J. Porcine deltacoronavirus induces apoptosis in swine testicular and LLC porcine kidney cell lines in vitro but not in infected intestinal enterocytes in vivo. Vet. Microbiol. 2016, 182, 57–63. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, Y.; Liang, X.; Lou, F.; Oglesbee, M.; Krakowka, S.; Li, J. Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. MBio 2014, 6, e00064-15. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Wu, Q.; Huang, L.; Yuan, C.; Wang, J.; Yang, Q. An alternative pathway of enteric PEDV dissemination from nasal cavity to intestinal mucosa in swine. Nat. Commun. 2018, 9, 3811. [Google Scholar] [CrossRef]
- Liang, Q.; Zhang, H.; Li, B.; Ding, Q.; Wang, Y.; Gao, W.; Guo, D.; Wei, Z.; Hu, H. Susceptibility of Chickens to Porcine Deltacoronavirus Infection. Viruses 2019, 11, 573. [Google Scholar] [CrossRef] [Green Version]







| Isolate | Parameter | Results of Different Passage | ||||||
|---|---|---|---|---|---|---|---|---|
| P1 | P5 | P10 | P15 | P20 | P25 | P30 | ||
| CHN-SC2015 | CPEs | + | + | + | + | + | + | + |
| qRT-PCR CT | ND | 22.09 | 24.01 | 18.71 | 18.44 | 18.15 | 20.02 | |
| Virus RNA Copies (Log10 Copies/μL) | ND | 1.75 | 1.19 | 2.74 | 2.82 | 2.90 | 2.36 | |
| Infectious titer (Log10 TCID50/mL) | 5.26 | 4.31 | 7.00 | 7.53 | 8.22 | 7.53 | 8.11 | |
| Passage | Nucleotide Position | Amino Acid Position | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 484 | 500 | 578 | 787 | 946 | 1188 | 1443 | 1450 | 1587 | 1922 | 2636 | 162 | 167 | 193 | 263 | 316 | 396 | 484 | 641 | 879 | |
| parent | T | A | A | G | A | T | A | G | C | A | T | Y | H | D | V | N | N | E | Q | L |
| P5 | G | A | A | G | G | T | A | G | T | G | T | D | H | D | V | D | N | E | R | L |
| P10 | T | C | A | G | G | T | A | G | T | G | C | Y | P | D | V | D | N | E | R | S |
| P15 | G | A | A | G | G | T | A | C | T | G | C | D | H | D | V | D | N | Q | R | S |
| P20 | T | C | A | G | G | T | A | C | T | G | C | Y | P | D | V | D | N | Q | R | S |
| P25 | T | C | A | T | G | T | A | C | T | G | C | Y | P | D | L | D | N | Q | R | S |
| P30 | T | C | C | T | G | A | G | C | T | G | C | Y | P | A | L | D | K | Q | R | S |
| Area | Nucleotide and Amino Acid Identity (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Complete Genome | ORF1a | ORF1ab | S | E | M | NS6 | N | NS7 | |
| China | 97.7–99.1 | 98.0–99.1/ 98.0–99.4 | 98.1–99.1/ 98.0–99.6 | 97.0–99.3/ 96.0–99.5 | 96.4–100/ 95.2–100 | 97.7–99.8/ 96.8–99.5 | 96.5–100/ 92.6–100 | 97.1–99.9/ 95.3–100 | 97.5–100/ 94.0–100 |
| Japan | 97.9–98.4 | 98.0–98.5/ 92.6–99.2 | 98.3–98.7/ 95.5–99.4 | 97.6–98.3/ 97.8–98.6 | 99.2–99.6/ 100 | 98.5–98.8/ 99.1 | 98.2–98.9/ 96.8–97.9 | 98.7–99.2/ 99.7–100 | 99.2–99.5/ 97.5–98.5 |
| South Korea | 98.5–98.6 | 98.4–98.5/ 99.1–99.2 | 98.6/ 99.3–99.4 | 98.1–98.3/ 98.2–98.6 | 99.6/100 | 98.6/99.1 | 98.9/ 97.9–98.9 | 99.0/100 | 99.2–99.3/ 97.5–98 |
| USA | 98.1–98.6 | 98.4–98.5/ 99.0–99.2 | 98.6–98.7/ 99.3–99.4 | 98.1–98.3/ 98.1–98.6 | 99.2–100/ 100 | 98.3–98.8/ 98.6–99.1 | 98.6–99.3/ 97.9–98.9 | 98.9–99.2/ 99.4–100 | 99.2–99.5/ 97.5–99.0 |
| Southeast | 97.5–98 | 97.8–98.0/ 98.2–98.6 | 97.9–98.1/ 98.7–99 | 96.0–96.7/ 96.8–97.8 | 99.6–100/ 100 | 98.2–99.1/ 98.6–99.1 | 98.2–98.9/ 98.9 | 97.6–99.1/ 98.8–99.7 | 97.5–99.2/ 93.5–97.5 |
| DPI | Lethargy and Anorexia | Fecal Consistency | ||
|---|---|---|---|---|
| Normal | Mild Diarrhea | Watery Diarrhea | ||
| 1 | 3/5 | 4/5 | 1/5 | 0/5 |
| 2 | 5/5 | 2/5 | 2/5 | 1/5 |
| 3 | 5/5 | 1/5 | 2/5 | 2/5 |
| 4 | 5/5 | 0/5 | 3/5 | 2/5 |
| 5 | 5/5 | 0/5 | 1/5 | 4/5 |
| 6 | 2/2 | 0/2 | 0/2 | 2/2 |
| 7 | 2/2 | 0/2 | 0/2 | 2/2 |
| 8 | 2/2 | 0/2 | 0/2 | 2/2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Qu, H.; Hu, J.; Fu, J.; Chen, R.; Li, C.; Cao, S.; Wen, Y.; Wu, R.; Zhao, Q.; et al. Characterization and Pathogenicity of the Porcine Deltacoronavirus Isolated in Southwest China. Viruses 2019, 11, 1074. https://doi.org/10.3390/v11111074
Zhao Y, Qu H, Hu J, Fu J, Chen R, Li C, Cao S, Wen Y, Wu R, Zhao Q, et al. Characterization and Pathogenicity of the Porcine Deltacoronavirus Isolated in Southwest China. Viruses. 2019; 11(11):1074. https://doi.org/10.3390/v11111074
Chicago/Turabian StyleZhao, Yujia, Huan Qu, Jingfei Hu, Jiayu Fu, Rui Chen, Cheng Li, Sanjie Cao, Yiping Wen, Rui Wu, Qin Zhao, and et al. 2019. "Characterization and Pathogenicity of the Porcine Deltacoronavirus Isolated in Southwest China" Viruses 11, no. 11: 1074. https://doi.org/10.3390/v11111074
APA StyleZhao, Y., Qu, H., Hu, J., Fu, J., Chen, R., Li, C., Cao, S., Wen, Y., Wu, R., Zhao, Q., Yan, Q., Wen, X., & Huang, X. (2019). Characterization and Pathogenicity of the Porcine Deltacoronavirus Isolated in Southwest China. Viruses, 11(11), 1074. https://doi.org/10.3390/v11111074





