HIV-1 Envelope Glycoprotein Amino Acids Signatures Associated with Clade B Transmitted/Founder and Recent Viruses
Abstract
1. Introduction
2. Materials and Methods
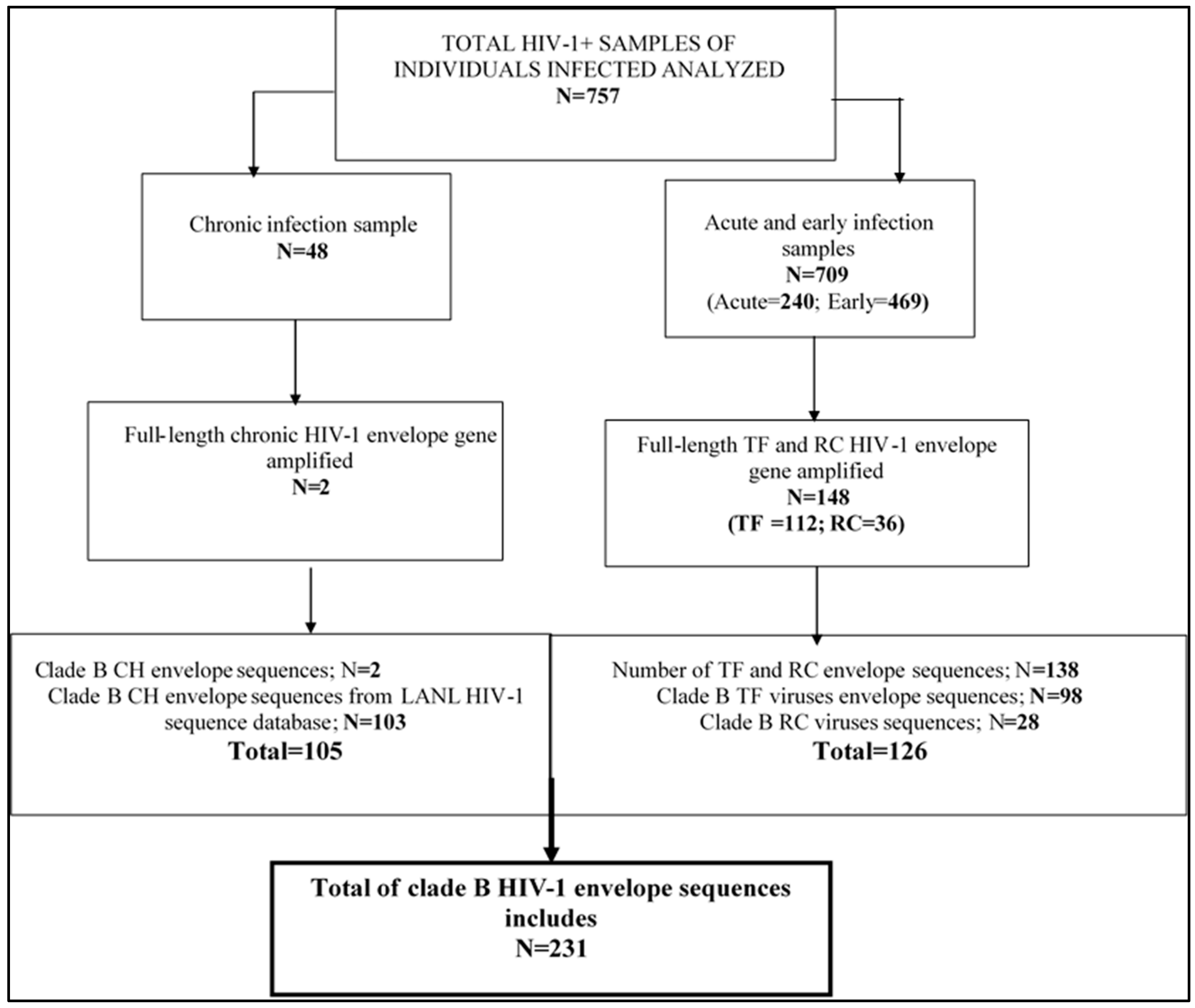
2.1. Description of Specimens
2.2. HIV-1 RNA Extraction
2.3. Reverse Transcription (RT-PCR)
2.4. Second Amplification
2.5. DNA Sequencing and Sequence Assembly
2.6. Data Management and Analysis
2.7. Statistical Analyses
2.8. Ethics Approval and Consent to Participate
3. Results
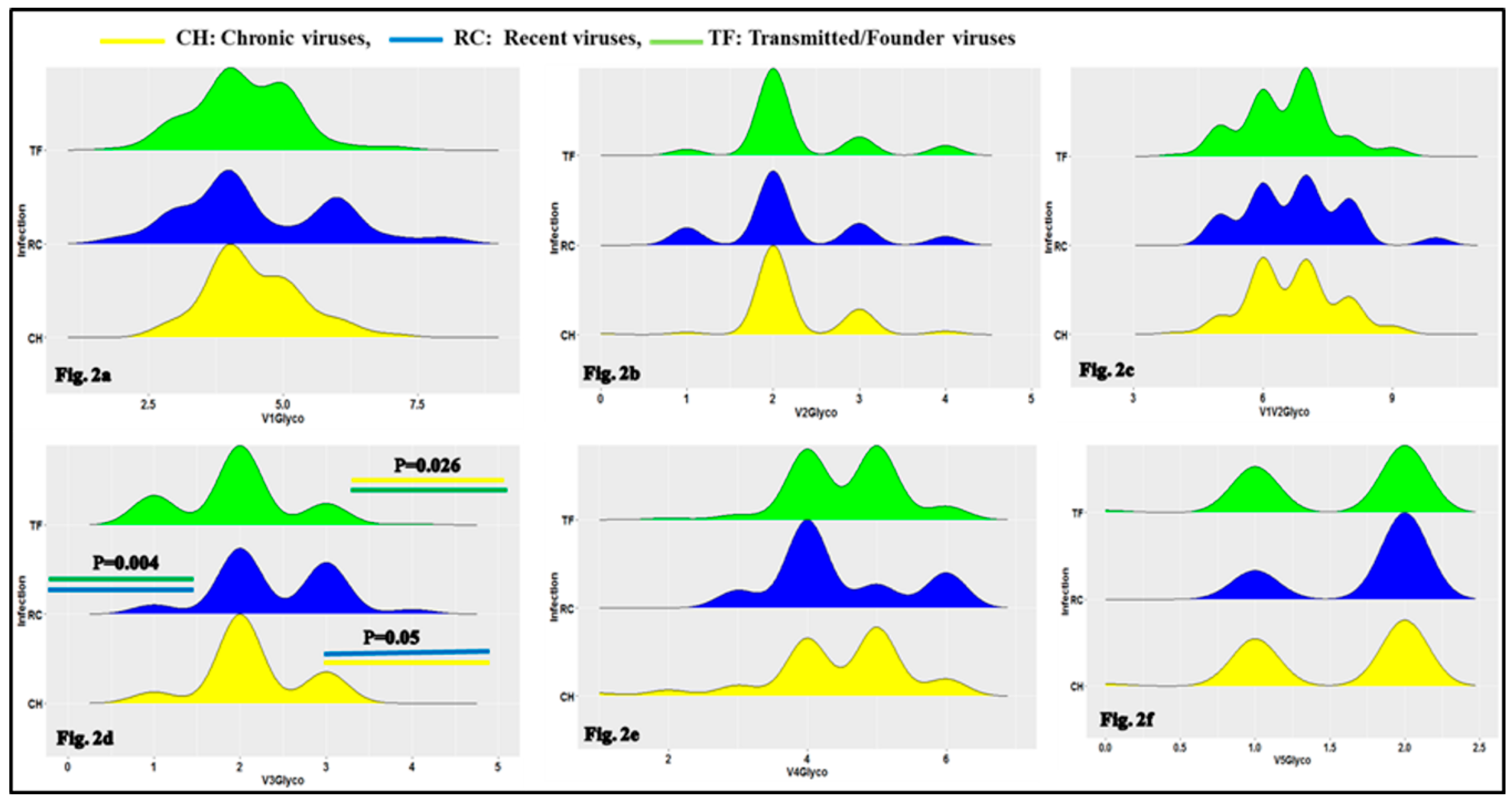
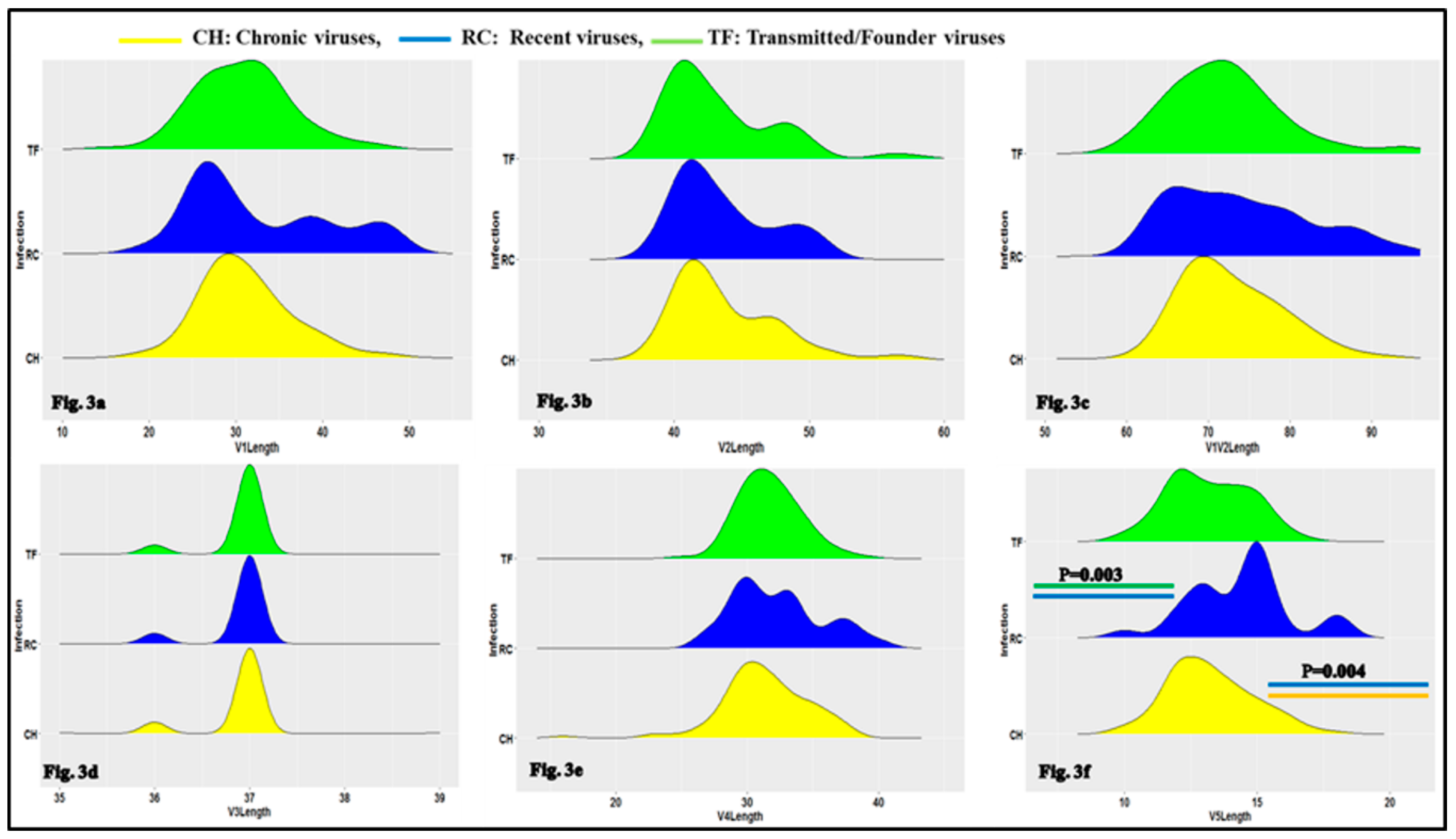
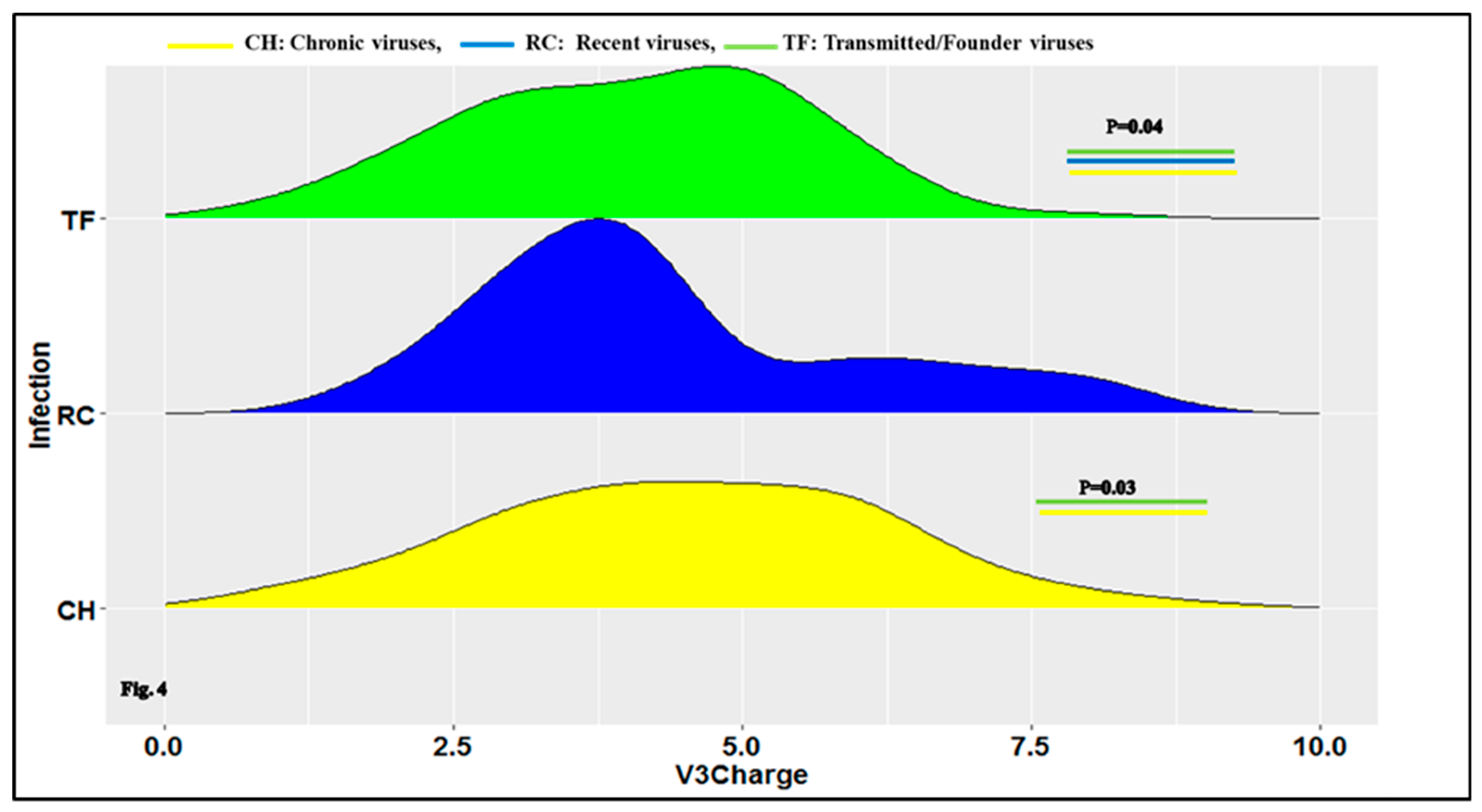
3.1. Characteristics of HIV-1 Envelope Variable Regions
3.2. Clade B HIV-1 Envelope Amino Acids Signatures Associated to Transmitted/Founders and Recent Viruses Compared to Chronic
3.3. HIV-1 Envelope Genetic Signatures Among Transmitted/Founder and Recent Viruses Compared to Chronic
4. Discussion
4.1. Clade B HIV-1 Envelope Variable Loop Characteristics
4.2. Clade B HIV-1 Envelope Amino Acids Signatures Associate to Transmitted/Founder and Recent Viruses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gnanakaran, S.; Bhattacharya, T.; Daniels, M.; Keele, B.F.; Hraber, P.T.; Lapedes, A.S.; Shen, T.; Gaschen, B.; Krishnamoorthy, M.; Li, H.; et al. Recurrent Signature Patterns in HIV-1 B Clade Envelope Glycoproteins Associated with either Early or Chronic Infections. PLoS Pathog. 2011, 7, e1002209. [Google Scholar] [CrossRef]
- Shaw, T.I.; Zhang, M. HIV N-linked glycosylation site analyzer and its further usage in anchored alignment. Nucleic Acids Res. 2013, 41, W454–W458. [Google Scholar] [CrossRef]
- Benjelloun, F.; Lawrence, P.; Verrier, B.; Genin, C.; Paul, S. Role of Human Immunodeficiency Virus Type 1 Envelope Structure in the Induction of Broadly Neutralizing Antibodies. J. Virol. 2012, 86, 13152–13163. [Google Scholar] [CrossRef][Green Version]
- Arrildt, K.T.; Labranche, C.C.; Joseph, S.B.; Dukhovlinova, E.N.; Graham, W.D.; Ping, L.H.; Schnell, G.; Sturdevant, C.B.; Kincer, L.P.; Mallewa, M.; et al. Phenotypic Correlates of HIV-1 Macrophage Tropism. J. Virol. 2015, 89, 11294–11311. [Google Scholar] [CrossRef] [PubMed]
- Pierson, T.; McArthur, J.; Siliciano, R.F. Reservoirs for HIV-1: Mechanisms for Viral Persistence in the Presence of Antiviral Immune Responses and Antiretroviral Therapy. Annu. Rev. Immunol. 2000, 18, 665–708. [Google Scholar] [CrossRef] [PubMed]
- Finzi, D.; Blankson, J.; Siliciano, J.D.; Margolick, J.B.; Chadwick, K.; Pierson, T.; Smith, K.; Lisziewicz, J.; Lori, F.; Flexner, C.; et al. Latent infection of CD4(+) T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 1999, 5, 512–517. [Google Scholar] [CrossRef]
- Van Regenmortel, M.H.V. Development of a Preventive HIV Vaccine Requires Solving Inverse Problems Which Is Unattainable by Rational Vaccine Design. Front. Immunol. 2018, 8, 2009. [Google Scholar] [CrossRef] [PubMed]
- Ensoli, B.; Cafaro, A.; Monini, P.; Marcotullio, S.; Ensoli, F. Cna Menges in HIV vaccine research for treatment and prevention. Front. Immunol. 2014, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Keele, B.F.; Giorgi, E.E.; Salazar-Gonzalez, J.F.; Decker, J.M.; Pham, K.T.; Salazar, M.G.; Sun, C.; Grayson, T.; Wang, S.; Li, H.; et al. Identification and characterisation of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. USA 2008, 105, 7552–7557. [Google Scholar] [CrossRef] [PubMed]
- Joseph, S.B.; Swanstrom, R.; Kashuba, A.D.M.; Cohen, M.S. Bottlenecks in HIV-1 transmission: Insights from the study of founder viruses. Nat. Rev. Genet. 2015, 13, 414–425. [Google Scholar] [CrossRef]
- Kariuki, S.M.; Selhorst, P.; Ariën, K.K.; Dorfman, J.R. The HIV-1 transmission bottleneck. Retrovirology 2017, 14, 22. [Google Scholar] [CrossRef] [PubMed]
- Shaw, G.M.; Hunter, E. HIV transmission. Cold Spring Harb. Perspect. Med. 2012, 2, a006965. [Google Scholar] [CrossRef] [PubMed]
- Bar, K.J.; Li, H.; Chamberland, A.; Tremblay, C.; Routy, J.P.; Grayson, T.; Sun, C.; Wang, S.; Learn, G.H.; Morgan, C.J.; et al. Wide Variation in the Multiplicity of HIV-1 Infection among Injection Drug Users. J. Virol. 2010, 84, 6241–6247. [Google Scholar] [CrossRef] [PubMed]
- Frost, S.D.W.; Liu, Y.; Pond, S.L.K.; Chappey, C.; Wrin, T.; Petropoulos, C.J.; Little, S.J.; Richman, D.D. Characterization of Human Immunodeficiency Virus Type 1 (HIV-1) Envelope Variation and Neutralizing Antibody Responses during Transmission of HIV-1 Subtype B. J. Virol. 2005, 79, 6523–6527. [Google Scholar] [CrossRef]
- Checkley, M.A.; Luttge, B.G.; Freed, E.O. HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. J. Mol. Biol. 2011, 410, 582–608. [Google Scholar] [CrossRef]
- Upadhyay, C.; Feyznezhad, R.; Yang, W.; Zhang, H.; Zolla-Pazner, S.; Hioe, C.E. Alterations of HIV-1 envelope phenotype and antibody-mediated neutralization by signal peptide mutations. PLoS Pathog. 2018, 14, e1006812. [Google Scholar] [CrossRef]
- Freed, E.O. HIV-1 replication. Somat. Cell Mol. Genet. 2001, 26, 13–33. [Google Scholar] [CrossRef]
- Moore, J.P.; Willey, R.L.; Lewis, G.K.; Robinson, J.; Sodroski, J. Immunological evidence for interactions between the first, 2nd, and 5th conserved domains of the gp120 surface glycoprotein of human-immunodeficiency-virus type-1. J. Virol. 1994, 68, 6836–6847. [Google Scholar]
- Starcich, B.R.; Hahn, B.H.; Shaw, G.M.; McNeely, P.D.; Modrow, S.; Wolf, H.; Parks, E.S.; Parks, W.P.; Josephs, S.F.; Gallo, R.C.; et al. Identification and characterization of conserved and variable regions in the envelope gene of htlv-iii lav, the retrovirus of aids. Cell 1986, 45, 637–648. [Google Scholar] [CrossRef]
- Douglas, N.; Munro, G.; Daniels, R. HIV/SIV glycoproteins: Structure-function relationships. J. Mol. Biol. 1997, 273, 122–149. [Google Scholar] [CrossRef]
- Shang, L.; Yue, L.; Hunter, E. Role of the Membrane-Spanning Domain of Human Immunodeficiency Virus Type 1 Envelope Glycoprotein in Cell-Cell Fusion and Virus Infection. J. Virol. 2008, 82, 5417–5428. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shang, L.; Hunter, E. Residues in the membrane-spanning domain core modulate conformation and fusogenicity of the HIV-1 envelope glycoprotein. Virology 2010, 404, 158–167. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Weiss, C.D. HIV-1 gp41: Mediator of fusion and target for inhibition. AIDS Rev. 2003, 5, 214–221. [Google Scholar] [PubMed]
- Markovic, I.; Clouse, K. Recent Advances in Understanding the Molecular Mechanisms of HIV-1 Entry and Fusion: Revisiting Current Targets and Considering New Options for Therapeutic Intervention. Curr. HIV Res. 2004, 2, 223–234. [Google Scholar] [CrossRef]
- Chen, B.; Chou, J.J. Structure of the transmembrane domain of HIV-1 envelope glycoprotein. FEBS J. 2017, 284, 1171–1177. [Google Scholar] [CrossRef]
- Haffar, O.K.; Dowbenko, D.J.; Berman, W. Topogenic analysis of the human immunodeficiency virus type-1 envelope glycoprotein, gp160, in microsomal-membranes. J. Cell Biol. 1988, 107, 1677–1687. [Google Scholar] [CrossRef]
- Berman, W.; Nunes, W.M.; Haffar, O.K. Expression of membrane-associated and secreted variants of gp160 of human immunodeficiency virus type-1 invitro and in continuous cell-lines. J. Virol. 1988, 62, 3135–3142. [Google Scholar]
- Salzwedel, K.; Johnston, P.B.; Roberts, S.J.; Dubay, J.W.; Hunter, E. Expression and characterization of glycophospholipid-anchored human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 1993, 67, 5279–5288. [Google Scholar]
- Env Feature database: HXB2 Genome Annotation. 2017. Available online: www.hiv.lanl.gov (accessed on 30 October 2019).
- Postler, T.S.; Desrosiers, R.C. The tale of the long tail: The cytoplasmic domain of HIV-1 gp41. J. Virol. 2013, 87, 2–15. [Google Scholar] [CrossRef]
- Yang, P.; Ai, L.S.; Huang, S.C.; Li, H.F.; Chan, W.E.; Chang, C.W.; Ko, C.Y.; Chen, S.S. The Cytoplasmic Domain of Human Immunodeficiency Virus Type 1 Transmembrane Protein gp41 Harbors Lipid Raft Association Determinants. J. Virol. 2010, 84, 59–75. [Google Scholar] [CrossRef]
- Edwards, T.G.; Wyss, S.; Reeves, J.D.; Zolla-Pazner, S.; Hoxie, J.A.; Doms, R.W.; Baribaud, F. Truncation of the Cytoplasmic Domain Induces Exposure of Conserved Regions in the Ectodomain of Human Immunodeficiency Virus Type 1 Envelope Protein. J. Virol. 2002, 76, 2683–2691. [Google Scholar] [CrossRef] [PubMed]
- Bültmann, A.; Muranyi, W.; Seed, B.; Haas, J. Identification of Two Sequences in the Cytoplasmic Tail of the Human Immunodeficiency Virus Type 1 Envelope Glycoprotein That Inhibit Cell Surface Expression. J. Virol. 2001, 75, 5263–5276. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Murakami, T.; Freed, E.O. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc. Natl. Acad. Sci. USA 2000, 97, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Piller, S.C.; Dubay, J.W.; Derdeyn, C.A.; Hunter, E. Mutational Analysis of Conserved Domains within the Cytoplasmic Tail of gp41 from Human Immunodeficiency Virus Type 1: Effects on Glycoprotein Incorporation and Infectivity. J. Virol. 2000, 74, 11717–11723. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Aiken, C. Maturation-Dependent Human Immunodeficiency Virus Type 1 Particle Fusion Requires a Carboxyl-Terminal Region of the gp41 Cytoplasmic Tail. J. Virol. 2007, 81, 9999–10008. [Google Scholar] [CrossRef]
- Kalia, V.; Sarkar, S.; Gupta, P.; Montelaro, R.C. Rational site-directed mutations of the LLP-1 and LLP-2 lentivirus lytic peptide domains in the intracytoplasmic tail of human immunodeficiency virus type 1 gp4l indicate common functions in cell-cell fusion but distinct roles in virion envelope incorporation. J. Virol. 2003, 77, 3634–3646. [Google Scholar]
- Cohen, M.S.; Shaw, G.M.; McMichael, A.J.; Haynes, B.F. Acute HIV-1 Infection. N. Engl. J. Med. 2011, 364, 1943–1954. [Google Scholar] [CrossRef]
- Serhir, B.; Hamel, D.; Doualla-Bell, F.; Routy, J.P.; Beaulac, S.-N.; Legault, M.; Fauvel, M.; Tremblay, C. Quebec Primary HIV infection study group Performance of Bio-Rad and Limiting Antigen Avidity Assays in Detecting Recent HIV Infections Using the Quebec Primary HIV-1 Infection Cohort. PLoS ONE 2016, 11, e0156023. [Google Scholar] [CrossRef]
- Brian, R.; Wood, M.; David, H.; Spach, M.D. Acute and Recent HIV Infection. Section 1: Screening and Diagnosis, Topic 4: Acute and Recent HIV Infection 2019 April 24th. 2019. Available online: https://www.hiv.uw.edu/go/screening-diagnosis/acute-recent-early-hiv/core-concept/all#tables (accessed on 8 July 2019).
- Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the Use of Antiretroviral Agents in Adults and Adolescents with HIV. Considerations for Antiretroviral Use in Special Patient Populations: Acute and Recent (early) HIV Infection. In National Instute of health (NIH), Department of Health and Human Services; AIDSinfo, 2018. Available online: https://aidsinfo.nih.gov/guidelines (accessed on 3 October 2019).
- Revilla, A.; Delgado, E.; Christian, E.C.; Dalrymple, J.; Vega, Y.; Carrera, C.; González-Galeano, M.; Ocampo, A.; de Castro, R.O.; Lezaún, M.J.; et al. Construction and phenotypic characterization of HIV type 1 functional envelope clones of subtypes G and F. AIDS Res. Hum. Retrovir. 2011, 27, 889–901. [Google Scholar] [CrossRef]
- Shcherbakova, N.S.; Shalamova, L.A.; Delgado, E.; Fernández-García, A.; Vega, Y.; Karpenko, L.I.; Ilyichev, A.A.; Sokolov, Y.V.; Shcherbakov, D.N.; Pérez-Álvarez, L.; et al. Short communication: Molecular epidemiology, phylogeny, and phylodynamics of CRF63_02A1, a recently originated HIV-1 circulating recombinant form spreading in Siberia. AIDS Res. Hum. Retrovir. 2014, 30, 912–919. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Schneider, T.D.; Stephens, R. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990, 18, 6097–6100. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.A.; Diaz-Rohrer, B.; Saminathan, P.; Jacobs, A. The Membrane Proximal External Regions of gp41 from HIV-1 Strains HXB2 and JRFL Have Different Sensitivities to Alanine Mutation. Biochemistry 2015, 54, 1681–1693. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Lu, L.; Du, L.; Zhu, X.; Debnath, A.K.; Jiang, S. Approaches for Identification of HIV-1 Entry Inhibitors Targeting gp41 Pocket. Viruses 2013, 5, 127–149. [Google Scholar] [CrossRef]
- Hunt, R. Virology—Chapter Seven Part Nine Human Immunodeficiency Virus and Aids Structure: The Genome and Proteins of HIV. In Microbiology and Immunology On-line; Hunt, R.C., Ed.; University of South Carolina School of Medicine: Columbia, CA, USA, 2016. [Google Scholar]
- Archary, D.; Gordon, M.L.; Green, T.N.; Coovadia, H.M.; Goulder, P.J.; Ndung’U, T. HIV-1 subtype C envelope characteristics associated with divergent rates of chronic disease progression. Retrovirology 2010, 7, 92. [Google Scholar] [CrossRef]
- McClure, P.; Curran, R.; Boneham, S.; Ball, J.K. A polymerase chain reaction method for the amplification of full-length envelope genes of HIV-1 from DNA samples containing single molecules of HIV-1 provirus. J. Virol. Methods 2000, 88, 73–80. [Google Scholar] [CrossRef]
- Cornelissen, M.; Gall, A.; Vink, M.; Zorgdrager, F.; Binter, Š.; Edwards, S.; Jurriaans, S.; Bakker, M.; Ong, S.H.; Gras, L.; et al. From clinical sample to complete genome: Comparing methods for the extraction of HIV-1 RNA for high-throughput deep sequencing. Virus Res. 2017, 239, 10–16. [Google Scholar] [CrossRef]
- Balasubramanian, C.; Chillemi, G.; Abbate, I.; Capobianchi, M.R.; Rozera, G.; Desideri, A. Importance of V3 Loop Flexibility and Net Charge in the Context of Co-Receptor Recognition. A Molecular Dynamics Study on HIV gp120. J. Biomol. Struct. Dyn. 2012, 29, 879–891. [Google Scholar] [CrossRef]
- De Wolf, F.; Hogervorst, E.; Goudsmit, J.; Fenyö, E.-M.; Rübsamen-Waigmann, H.; Holmes, H.; Galvão-Castro, B.; Karita, E.; Wasi, C.; Sempala, S.; et al. Syncytium-Inducing and Non-Syncytium-Inducing Capacity of Human Immunodeficiency Virus Type 1 Subtypes Other Than B: Phenotypic and Genotypic Characteristics. AIDS Res. Hum. Retrovir. 1994, 10, 1387–1400. [Google Scholar] [CrossRef]
- Kaleebu, P.; Nankya, I.L.; Yirrell, D.L.; Shafer, L.A.; Kyosiimire-Lugemwa, J.; Lule, D.B.; Morgan, D.; Beddows, S.; Weber, J.; Whitworth, J.A. Relation between chemokine receptor use, disease stage, and HIV-1 subtypes A and D—Results from a rural Ugandan cohort. Jaids-J. Acquir. Immune Defic. Syndr. 2007, 45, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Baalwa, J.; Wang, S.; Parrish, N.F.; Decker, J.M.; Keele, B.F.; Learn, G.H.; Yue, L.; Ruzagira, E.; Ssemwanga, D.; Kamali, A.; et al. Molecular identification, cloning and characterization of transmitted/founder HIV-1 subtype A, D and A/D infectious molecular clones. Virology 2013, 436, 33–48. [Google Scholar] [CrossRef] [PubMed]
- Wilen, C.B.; Parrish, N.F.; Pfaff, J.M.; Decker, J.M.; Henning, E.A.; Haim, H.; Petersen, J.E.; Wojcechowskyj, J.A.; Sodroski, J.; Haynes, B.F.; et al. Phenotypic and Immunologic Comparison of Clade B Transmitted/Founder and Chronic HIV-1 Envelope Glycoproteins. J. Virol. 2011, 85, 8514–8527. [Google Scholar] [CrossRef] [PubMed]
- Finzi, A.; Pacheco, B.; Xiang, S.-H.; Pancera, M.; Herschhorn, A.; Wang, L.; Zeng, X.; Desormeaux, A.; Kwong, P.D.; Sodroski, J. Lineage-Specific Differences between Human and Simian Immunodeficiency Virus Regulation of gp120 Trimer Association and CD4 Binding. J. Virol. 2012, 86, 8974–8986. [Google Scholar] [CrossRef][Green Version]
- Wyatt, R. The HIV-1 Envelope Glycoproteins: Fusogens, Antigens, and Immunogens. Science 1998, 280, 1884–1888. [Google Scholar] [CrossRef]
- Chohan, B.; Lang, D.; Sagar, M.; Korber, B.; Lavreys, L.; Richardson, B.; Overbaugh, J. Selection for Human Immunodeficiency Virus Type 1 Envelope Glycosylation Variants with Shorter V1-V2 Loop Sequences Occurs during Transmission of Certain Genetic Subtypes and May Impact Viral RNA Levels. J. Virol. 2005, 79, 6528–6531. [Google Scholar] [CrossRef]
- Helseth, E.; Olshevsky, U.; Furman, C.; Sodroski, J. Human-Immunodeficiency-Virus Type-1 Gp120 Envelope Glycoprotein Regions Important For Association With The Gp41 Transmembrane Glycoprotein. J. Virol. 1991, 65, 2119–2123. [Google Scholar]
- Yuan, T.; Li, J.; Zhang, M.-Y. HIV-1 Envelope Glycoprotein Variable Loops Are Indispensable for Envelope Structural Integrity and Virus Entry. PLoS ONE 2013, 8, e69789. [Google Scholar] [CrossRef]
- Pejchal, R.; Doores, K.J.; Walker, L.M.; Khayat, R.; Huang, P.-S.; Wang, S.-K.; Stanfield, R.L.; Julien, J.-P.; Ramos, A.; Crispin, M.; et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 2011, 334, 1097–1103. [Google Scholar] [CrossRef]
- Zhou, T.; Georgiev, I.; Wu, X.; Yang, Z.Y.; Dai, K.; Finzi, A.; Kwon, Y.D.; Scheid, J.F.; Shi, W.; Xu, L.; et al. Structural Basis for Broad and Potent Neutralization of HIV-1 by Antibody VRC01. Sci. 2010, 329, 811–817. [Google Scholar] [CrossRef]
- Zolla-Pazner, S.; Cardozo, T. Structure-function relationships of HIV-1 envelope sequence-variable regions refocus vaccine design. Nat. Rev. Immunol. 2010, 10, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.S.; Gay, C.L.; Busch, M.P.; Hecht, F.M. The Detection of Acute HIV Infection. J. Infect. Dis. 2010, 202, S270–S277. [Google Scholar] [CrossRef] [PubMed]
- Fiebig, E.W.; Wright, D.J.; Rawal, B.D.; E Garrett, P.; Schumacher, R.T.; Peddada, L.; Heldebrant, C.; Smith, R.; Conrad, A.; Kleinman, S.H.; et al. Dynamics of HIV viremia and antibody seroconversion in plasma donors: Implications for diagnosis and staging of primary HIV infection. AIDS 2003, 17, 1871–1879. [Google Scholar] [CrossRef] [PubMed]
- Venable, R.M.; Pastor, R.W.; Brooks, B.R.; Carson, F.W. Theoretically Determined Three-Dimensional Structures for Amphipathic Segments of the HIV-1 gp41 Envelope Protein. AIDS Res. Hum. Retrovir. 1989, 5, 7–22. [Google Scholar] [CrossRef]
- Eisenberg, D.; Wesson, M. The most highly amphiphilic alpha-helices include two amino acid segments in human immunodeficiency virus glycoprotein 41. Biopolymers 1990, 29, 171–177. [Google Scholar] [CrossRef]
- Fernandez, M.V.; Freed, E.O. Meeting Review: 2018 International Workshop on Structure and Function of the Lentiviral gp41 Cytoplasmic Tail. Viruses 2018, 10, 613. [Google Scholar] [CrossRef]
- Kao, S.M.; Miller, E.D.; Su, L. A leucine zipper motif in the cytoplasmic domain of gp41 is required for HIV-1 replication and pathogenesis in vivo. Virology 2001, 289, 208–217. [Google Scholar] [CrossRef]
- Kalia, V.; Sarkar, S.; Gupta, P.; Montelaro, R.C. Antibody Neutralization Escape Mediated by Point Mutations in the Intracytoplasmic Tail of Human Immunodeficiency Virus Type 1 gp41. J. Virol. 2005, 79, 2097–2107. [Google Scholar] [CrossRef]
- Newman, J.T.; Sturgeon, T.J.; Gupta, P.; Montelaro, R.C. Differential functional phenotypes of two primary HIV-1 strains resulting from homologous point mutations in the LLP domains of the envelope gp41 intracytoplasmic domain. Virology 2007, 367, 102–116. [Google Scholar] [CrossRef]
- Lee, S.F.; Ko, C.Y.; Wang, C.T.; Chen, S.S.L. Effect of Point Mutations in the N Terminus of the Lentivirus Lytic Peptide-1 Sequence of Human Immunodeficiency Virus Type 1 Transmembrane Protein gp41 on Env Stability. J. Biol. Chem. 2002, 277, 15363–15375. [Google Scholar] [CrossRef]
- Asmal, M.; Hellmann, I.; Liu, W.; Keele, B.F.; Perelson, A.S.; Bhattacharya, T.; Gnanakaran, S.; Daniels, M.; Haynes, B.F.; Korber, B.T.; et al. A Signature in HIV-1 Envelope Leader Peptide Associated with Transition from Acute to Chronic Infection Impacts Envelope Processing and Infectivity. PLoS ONE 2011, 6, e23673. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gonzalez, M.W.; DeVico, A.L.; Lewis, G.K.; Spouge, J.L. Conserved Molecular Signatures in gp120 Are Associated with the Genetic Bottleneck during Simian Immunodeficiency Virus (SIV), SIV-Human Immunodeficiency Virus (SHIV), and HIV Type 1 (HIV-1) Transmission. J. Virol. 2015, 89, 3619–3629. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, T.; Pisch, T.; Devitt, G.; Holtkotte, D.; Bösch, V. Effects of signal peptide exchange on HIV-1 glycoprotein expression and viral infectivity in mammalian cells. FEBS Lett. 2006, 580, 3775–3778. [Google Scholar] [CrossRef] [PubMed]
- Yolitz, J.; Schwing, C.; Chang, J.; Van Ryk, D.; Nawaz, F.; Wei, D.; Cicala, C.; Arthos, J.; Fauci, A.S. Signal peptide of HIV envelope protein impacts glycosylation and antigenicity of gp120. Proc. Natl. Acad. Sci. USA 2018, 115, 2443–2448. [Google Scholar] [CrossRef] [PubMed]

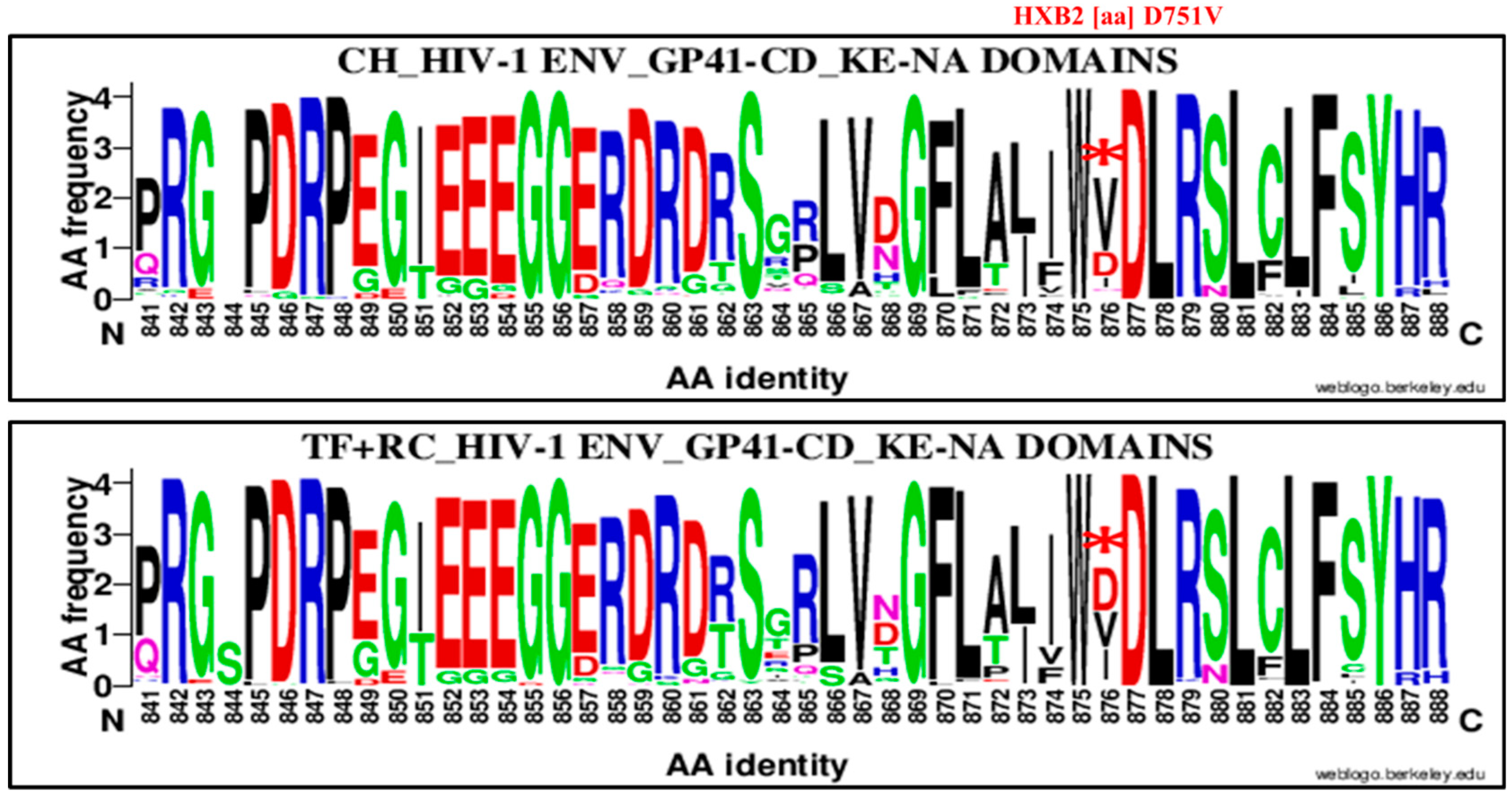
| HXB2 | Env | Amino Acids | CH | TF | Chi2 Test | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Position | Amino Acids | Subregion/Domain | Alignment Position | Change | Genetic Signature | YES | NO | Total | YES | NO | Total | OR | 95% CI | B-H adjusted p-values |
| 841 | R | LLP-1 | 959 | I | R841I | 10 | 95 | 105 | 32 | 63 | 95 | 0.2 | 0.09–0.44 | 0.00001 |
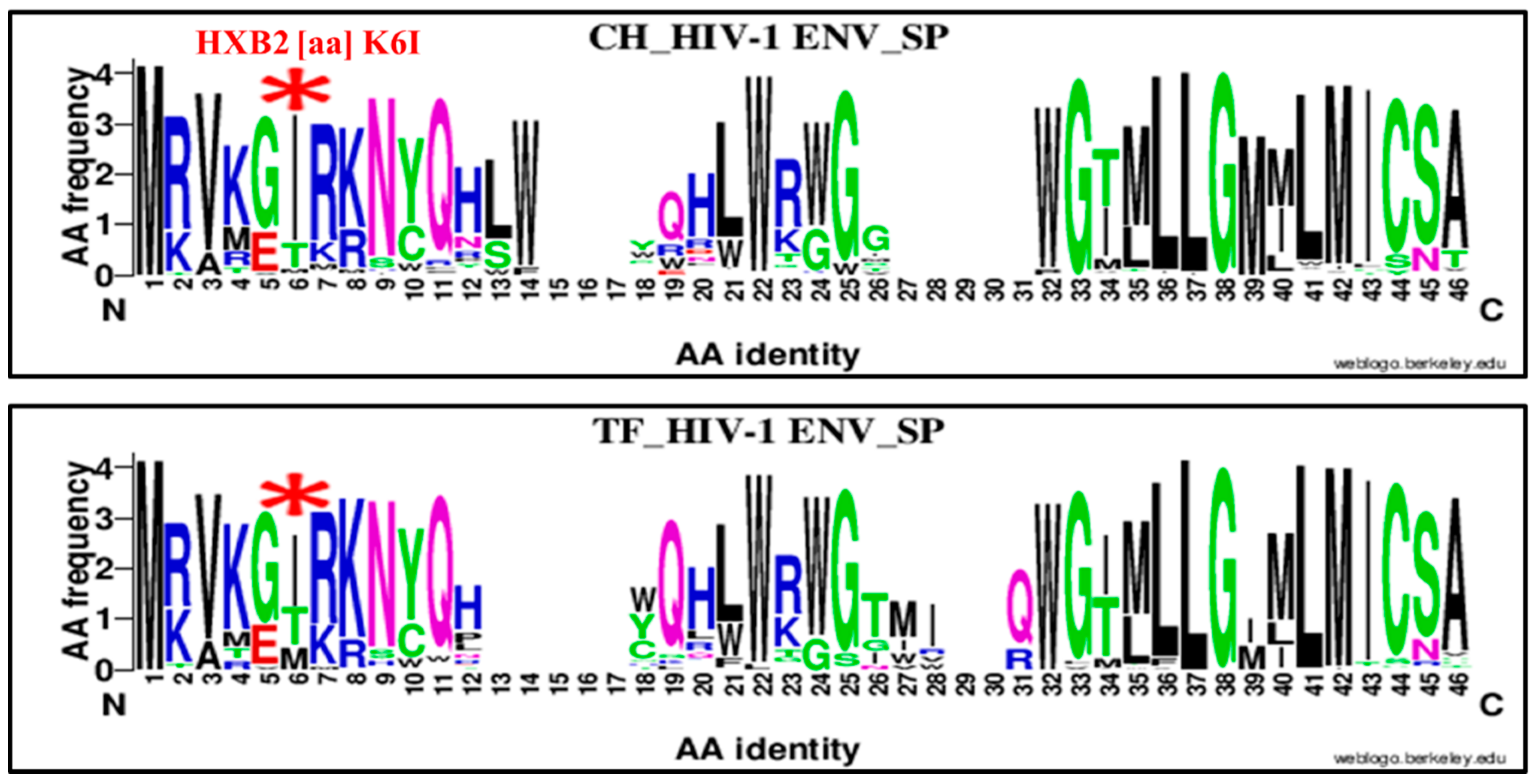
| 6 | K | SP | 6 | I | K6I | 83 | 22 | 105 | 52 | 45 | 97 | 3.26 | 1.76–6.02 | 0.0001 |
| 62 | D | C1 | 81 | E | D62E | 20 | 85 | 105 | 4 | 94 | 98 | 5.52 | 1.89–16.04 | 0.006 |
| 514 | G | ECD | 628 | T | G514T | 9 | 22 | 31 | 26 | 12 | 38 | 0.18 | 0.06–0.52 | 0.006 |
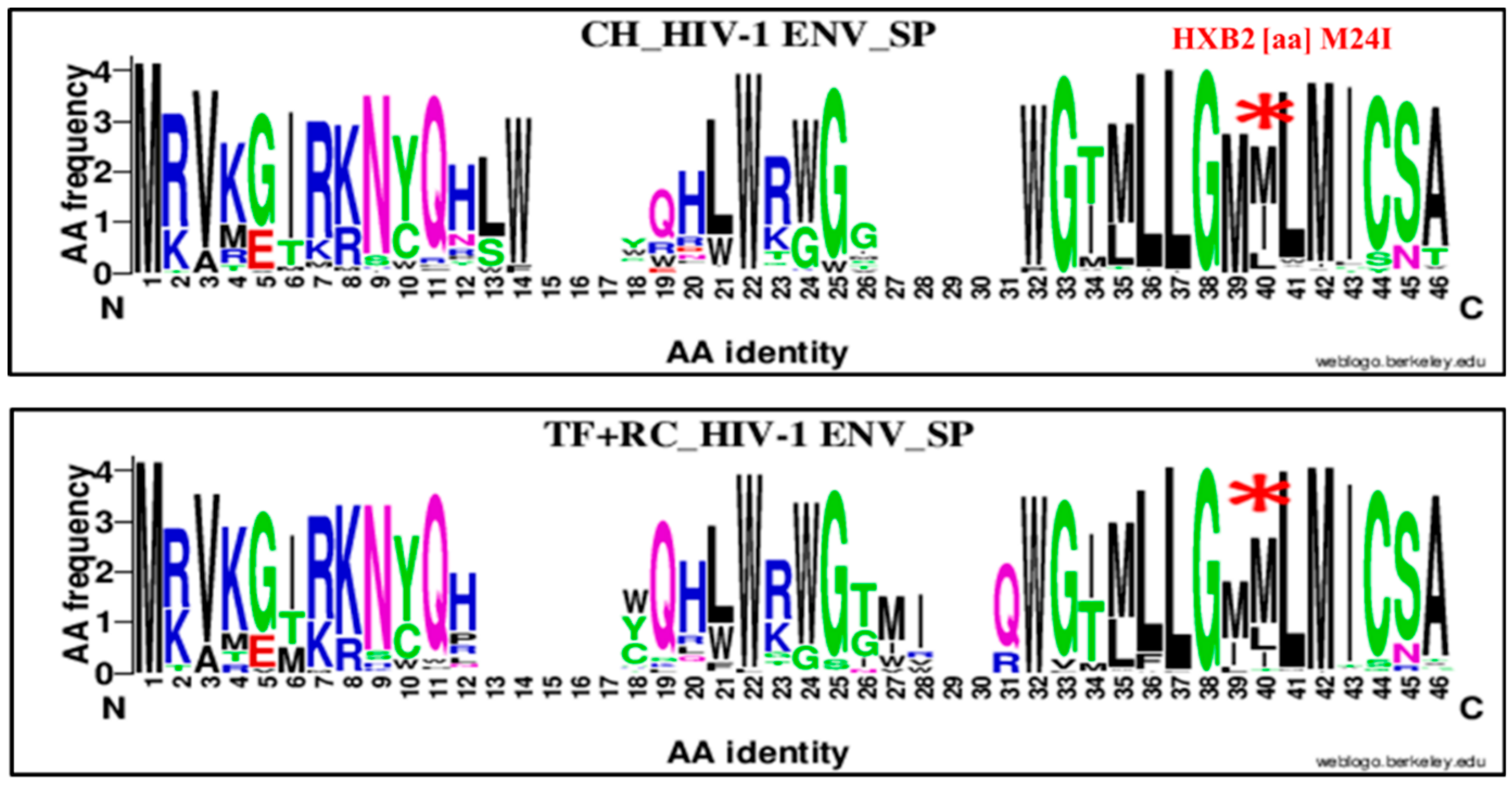
| 24 | M | SP | 40 | I | M24I | 38 | 66 | 104 | 16 | 81 | 97 | 2.91 | 1.50–5.64 | 0.006 |
| 743 | R | HIR/KE | 865 | R | 743R | 45 | 60 | 105 | 62 | 35 | 97 | 0.42 | 0.24–0.74 | 0.008 |
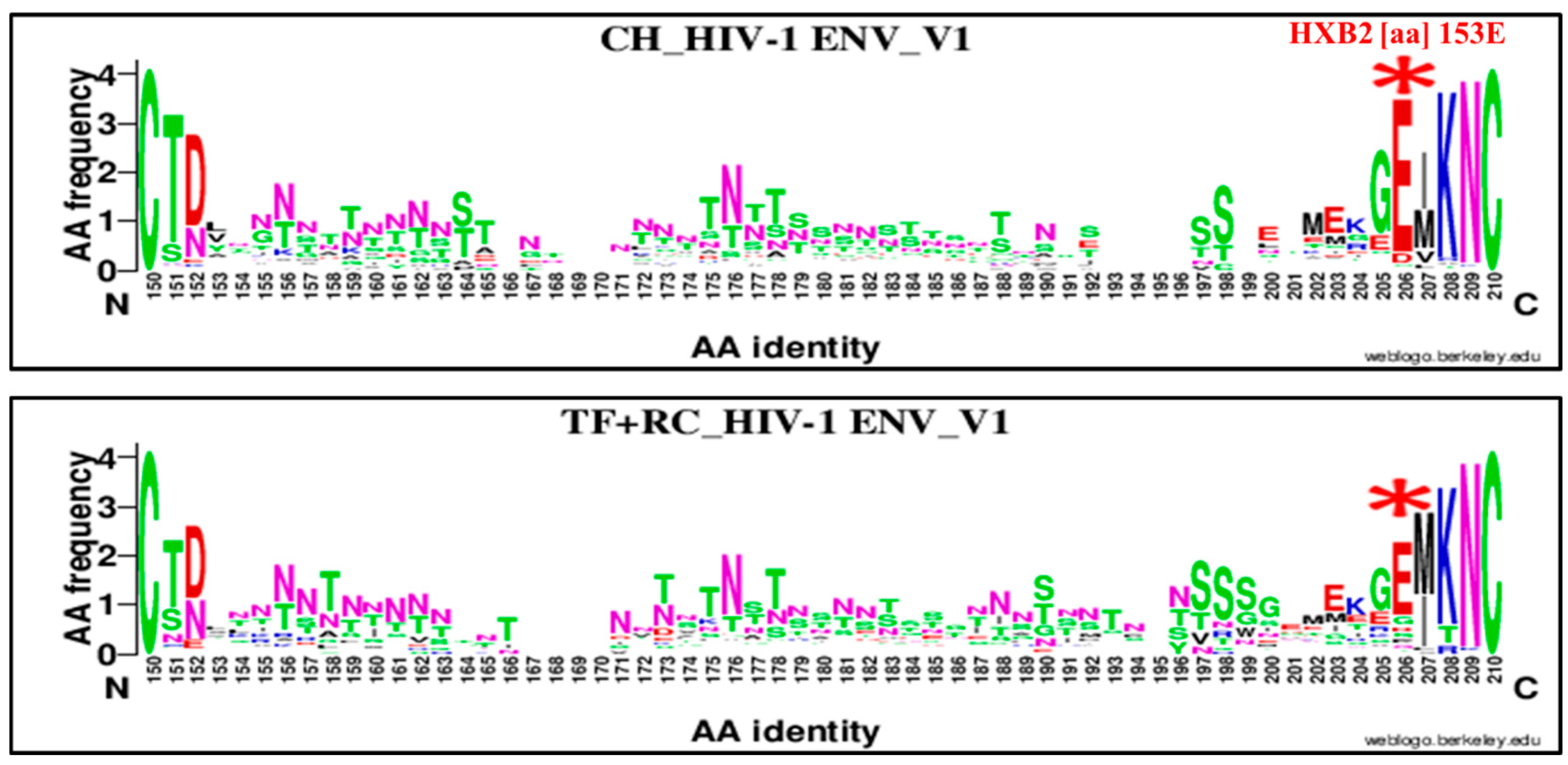
| 153 | E | V1 | 206 | E | 153E | 93 | 11 | 104 | 71 | 26 | 97 | 3.09 | 1.44–6.59 | 0.008 |
| 744 | R | HIR/KE | 862 | R | 744R | 78 | 27 | 105 | 53 | 44 | 97 | 2.39 | 1.32–4.32 | 0.008 |
| 717 | F | HIR/KE | 834 | F | 717F | 86 | 19 | 105 | 62 | 35 | 97 | 2.55 | 1.34–4.85 | 0.008 |
| 717 | F | HIR/KE | 834 | L | F717L | 19 | 86 | 105 | 35 | 62 | 97 | 0.39 | 0.20–0.74 | 0.008 |
| 154 | I | V1 | 207 | M | I154M | 35 | 70 | 105 | 52 | 46 | 98 | 0.44 | 0.25–0.77 | 0.008 |
| 744 | R | HIR/KE | 862 | T | R744T | 15 | 90 | 105 | 30 | 67 | 97 | 0.37 | 0.18–0.74 | 0.008 |
| 841 | R | LLP-1 | 959 | L | R841L | 81 | 24 | 105 | 56 | 39 | 95 | 2.35 | 1.27–4.31 | 0.009 |
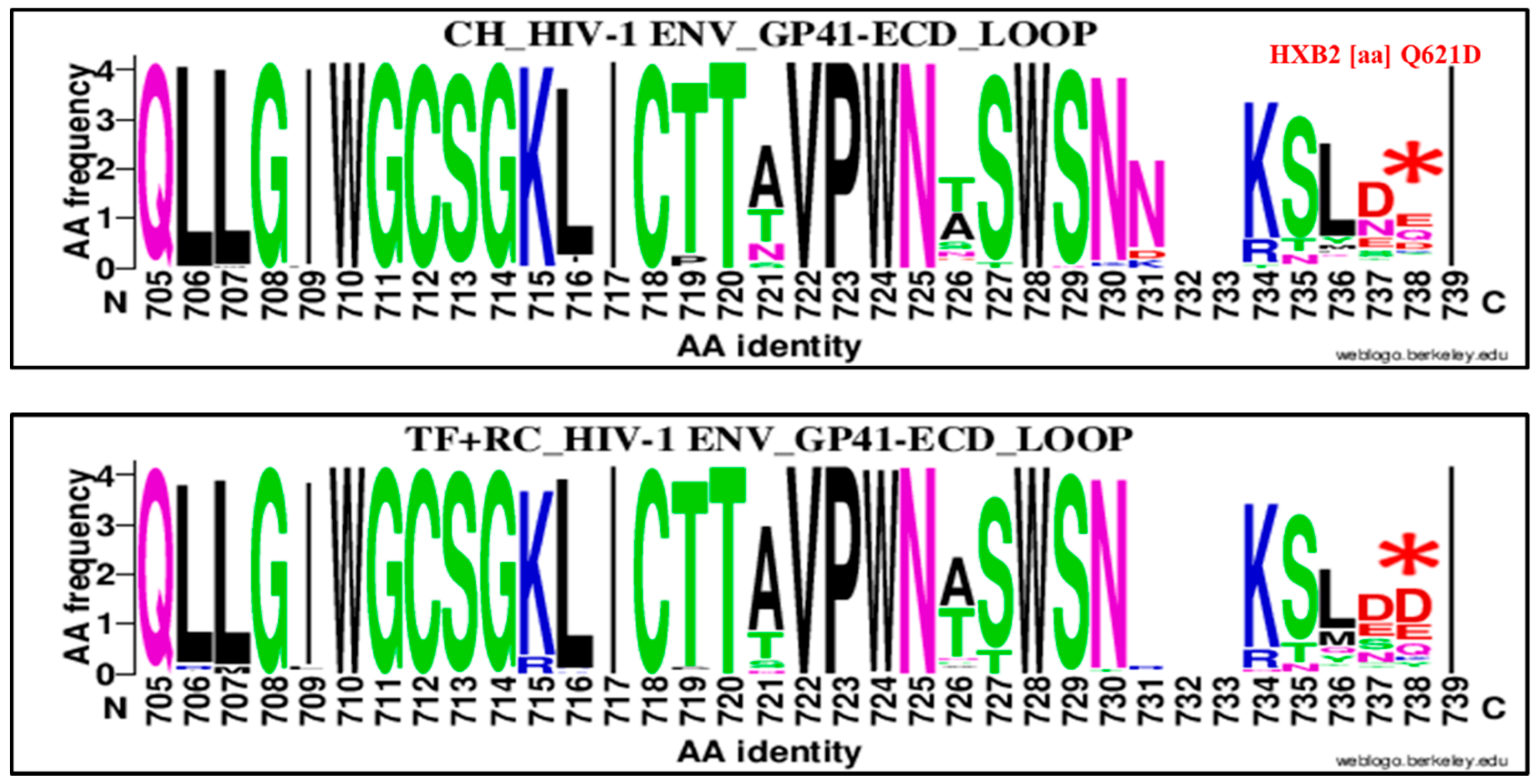
| 621 | Q | GP 41 Loop | 738 | D | Q621D | 16 | 88 | 104 | 31 | 67 | 98 | 0.39 | 0.20–0.77 | 0.009 |
| 464 | L | V5 | 566 | N | L464N | 24 | 45 | 69 | 42 | 31 | 73 | 0.39 | 0.20–0.77 | 0.009 |
| 543 | Q | FPPR | 657 | Q | 543Q | 63 | 42 | 105 | 76 | 22 | 98 | 0.43 | 0.23–0.79 | 0.009 |
| HXB2 | Env | Amino Acid | CH | TF + RC | Chi2 test | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Position | Amino acid | Subregion/Domain | Alignment position | Change | Genetic signature | YES | NO | TOTAL | YES | NO | TOTAL | OR | 95% CI | B-H Adjusted p value |
| 153 | E | V1 | 206 | E | 153E | 93 | 11 | 104 | 82 | 43 | 125 | 4.43 | 2.16, 9.05 | 0.000001 |
| 24 | M | SP | 40 | I | M24I | 38 | 66 | 104 | 16 | 109 | 125 | 3.92 | 2.04, 7.53 | 0.00001 |
| 621 | Q | GP 41 CT Loop | 738 | D | Q621D | 16 | 88 | 104 | 52 | 73 | 125 | 0.25 | 0.13, 0.48 | 0.00001 |
| 751 | D | CT (HIR/KE) | 876 | V | D751V | 64 | 41 | 105 | 39 | 86 | 125 | 3.44 | 1.99, 5.92 | 0.00001 |
| 6 | K | SP | 6 | I | K6I | 83 | 22 | 105 | 69 | 56 | 125 | 3.06 | 1.70, 5.48 | 0.0003 |
| 33 | K | C1 | 49 | Q | K33Q | 39 | 55 | 94 | 22 | 102 | 124 | 3.28 | 1.78, 6.06 | 0.0003 |
| 717 | F | EC (YSPL) and HIR/KE | 834 | F | F717F | 86 | 19 | 105 | 73 | 52 | 125 | 3.22 | 1.75, 5 | 0.0003 |
| 717 | F | EC (YSPL) and HIR/KE | 834 | L | F717L | 19 | 86 | 105 | 52 | 73 | 125 | 0.31 | 0.16, 0.56 | 0.0003 |
| 747 | R | HIR/KE NA | 865 | R | R747R | 45 | 60 | 105 | 85 | 40 | 125 | 0.35 | 0.20, 0.60 | 0.0003 |
| 132 | T | V2 | 151 | T | T132T | 86 | 19 | 105 | 75 | 51 | 126 | 3.07 | 1.67, 5.64 | 0.0004 |
| 154 | I | V1 | 207 | M | I154M | 35 | 70 | 105 | 73 | 53 | 126 | 0.36 | 0.21, 0.62 | 0.0004 |
| 360 | I | C3 | 441 | V | I360V | 52 | 52 | 104 | 33 | 93 | 126 | 2.81 | 1.62, 4.88 | 0.0004 |
| 737 | R | HIR/KE | 862 | T | R737T | 15 | 90 | 105 | 44 | 81 | 125 | 0.3 | 0.16, 0.58 | 0.0006 |
| 841 | R | LLP-1 | 959 | I | R841I | 10 | 95 | 105 | 35 | 87 | 122 | 0.26 | 0.12, 0.55 | 0.0006 |
| 744 | R | HIR/KE | 862 | R | R744R | 78 | 27 | 105 | 65 | 60 | 125 | 2.66 | 1.52, 4.65 | 0.0009 |
| 236 | T | Loop D | 312 | S | T236T | 9 | 95 | 104 | 33 | 93 | 126 | 0.26 | 0.12, 0.58 | 0.001 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kafando, A.; Martineau, C.; El-Far, M.; Fournier, E.; Doualla-Bell, F.; Serhir, B.; Kazienga, A.; Sangaré, M.N.; Sylla, M.; Chamberland, A.; et al. HIV-1 Envelope Glycoprotein Amino Acids Signatures Associated with Clade B Transmitted/Founder and Recent Viruses. Viruses 2019, 11, 1012. https://doi.org/10.3390/v11111012
Kafando A, Martineau C, El-Far M, Fournier E, Doualla-Bell F, Serhir B, Kazienga A, Sangaré MN, Sylla M, Chamberland A, et al. HIV-1 Envelope Glycoprotein Amino Acids Signatures Associated with Clade B Transmitted/Founder and Recent Viruses. Viruses. 2019; 11(11):1012. https://doi.org/10.3390/v11111012
Chicago/Turabian StyleKafando, Alexis, Christine Martineau, Mohamed El-Far, Eric Fournier, Florence Doualla-Bell, Bouchra Serhir, Adama Kazienga, Mohamed Ndongo Sangaré, Mohamed Sylla, Annie Chamberland, and et al. 2019. "HIV-1 Envelope Glycoprotein Amino Acids Signatures Associated with Clade B Transmitted/Founder and Recent Viruses" Viruses 11, no. 11: 1012. https://doi.org/10.3390/v11111012
APA StyleKafando, A., Martineau, C., El-Far, M., Fournier, E., Doualla-Bell, F., Serhir, B., Kazienga, A., Sangaré, M. N., Sylla, M., Chamberland, A., Charest, H., & Tremblay, C. L. (2019). HIV-1 Envelope Glycoprotein Amino Acids Signatures Associated with Clade B Transmitted/Founder and Recent Viruses. Viruses, 11(11), 1012. https://doi.org/10.3390/v11111012





