Genome Analysis of a Novel Clade II.b Alphabaculovirus Obtained from Artaxa digramma
Abstract
1. Introduction
2. Materials and Methods
2.1. Viral DNA Extraction
2.2. Genomic DNA Sequencing and Bioinformatics Analysis
2.3. Phylogenetic Analysis
3. Results and Discussion
3.1. Sequencing and Characterization of ArdiNPV Genome
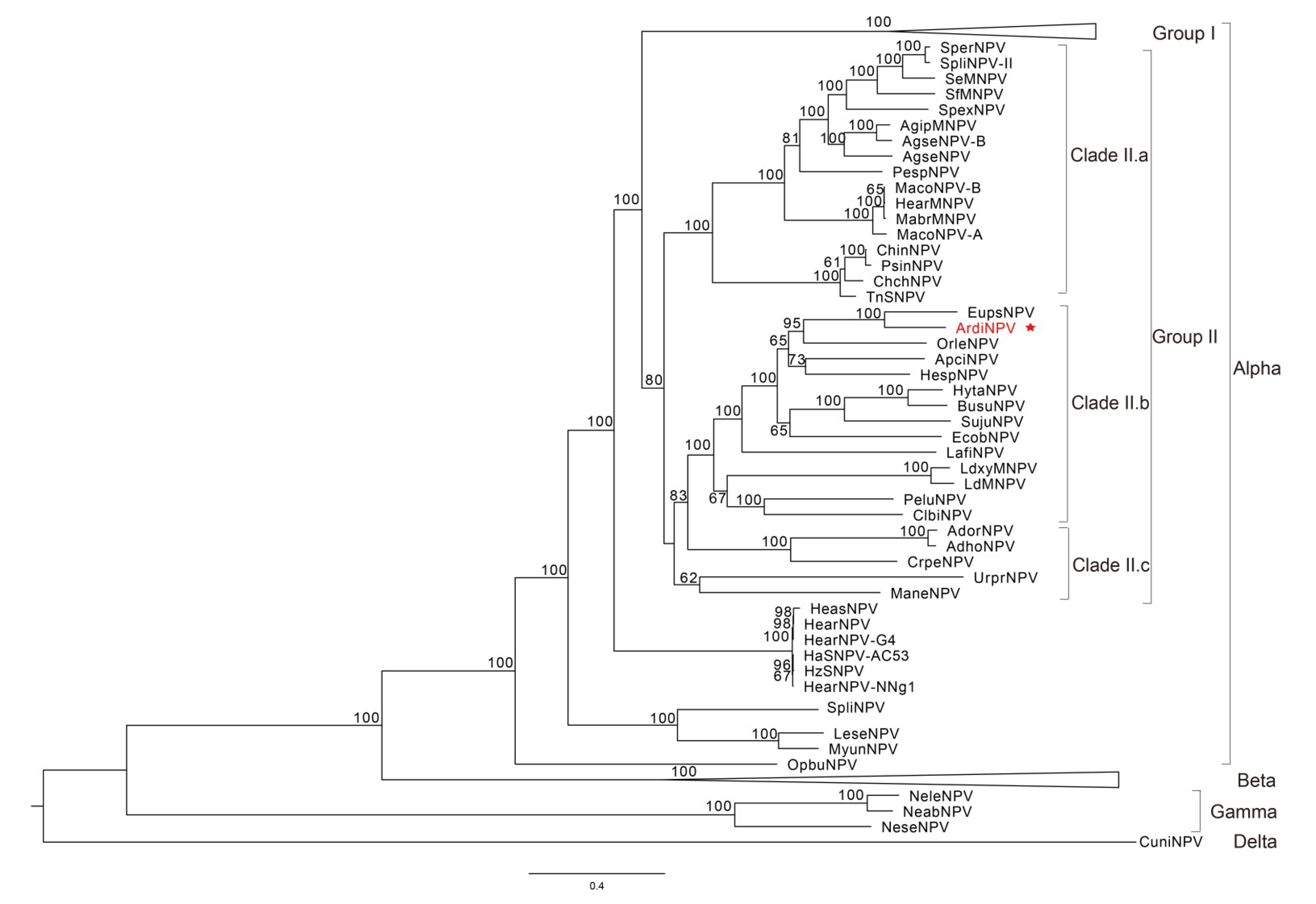
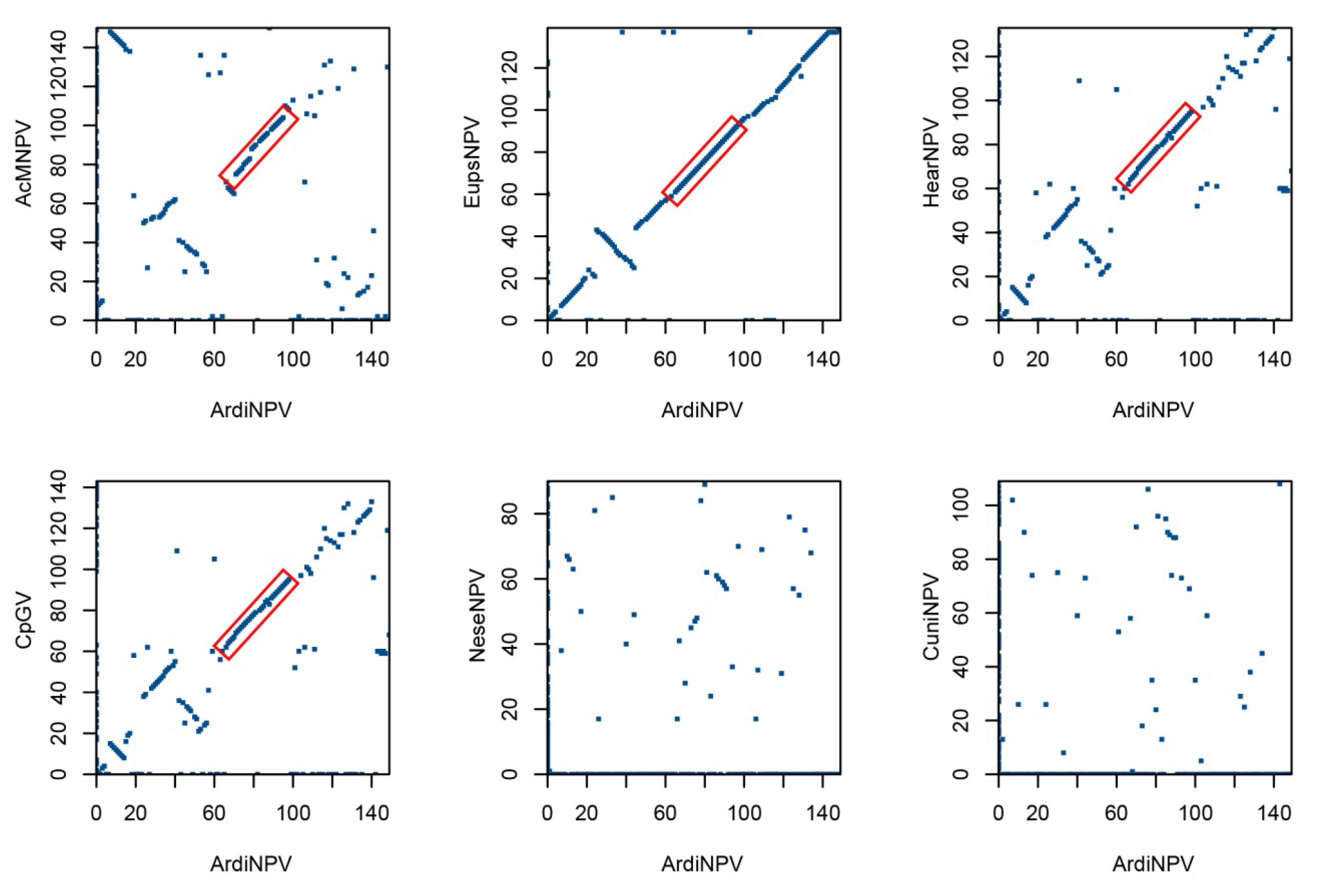
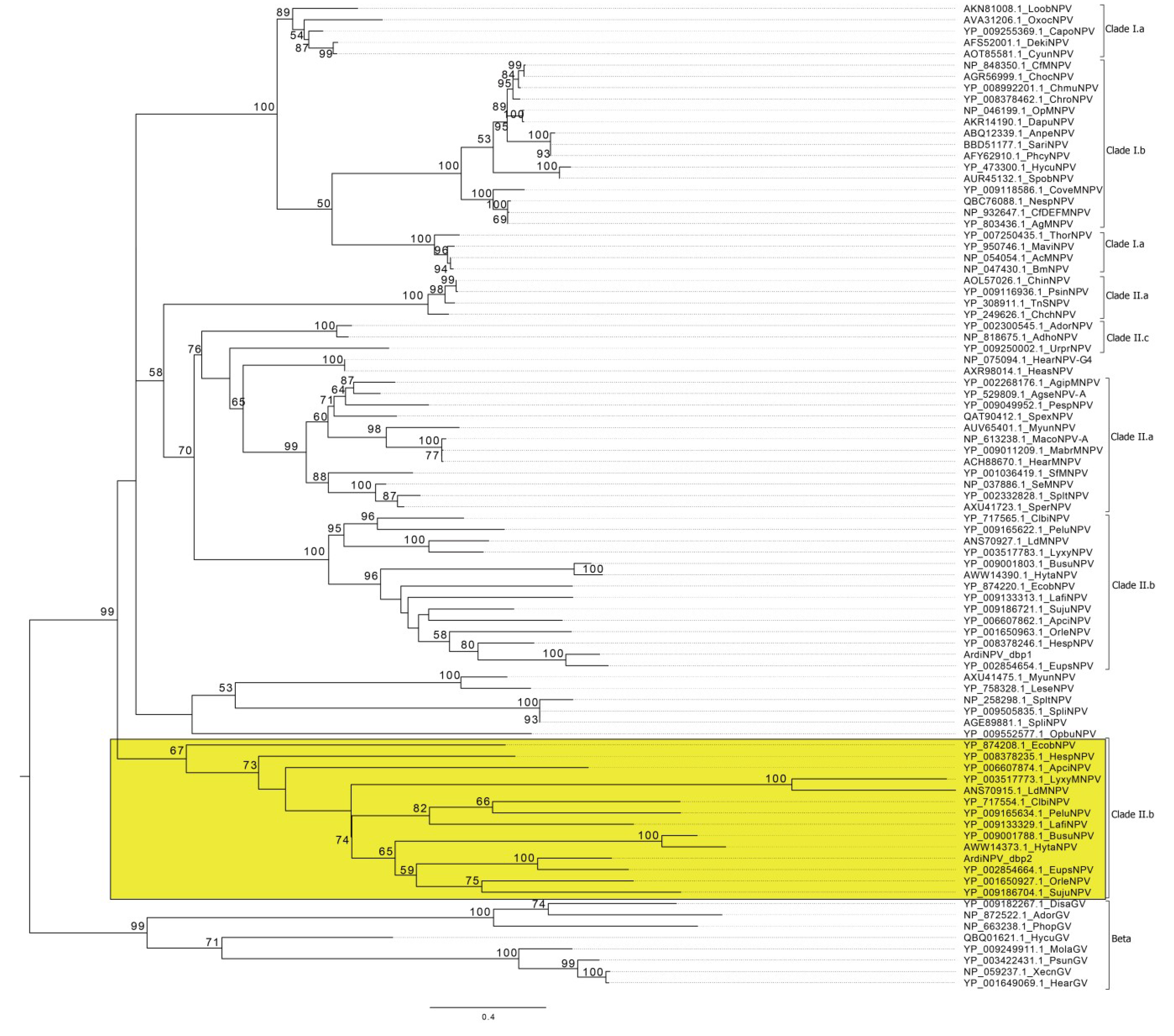
3.2. Phylogenetic Analysis of ArdiNPV
3.3. Classification of ArdiNPV Genes
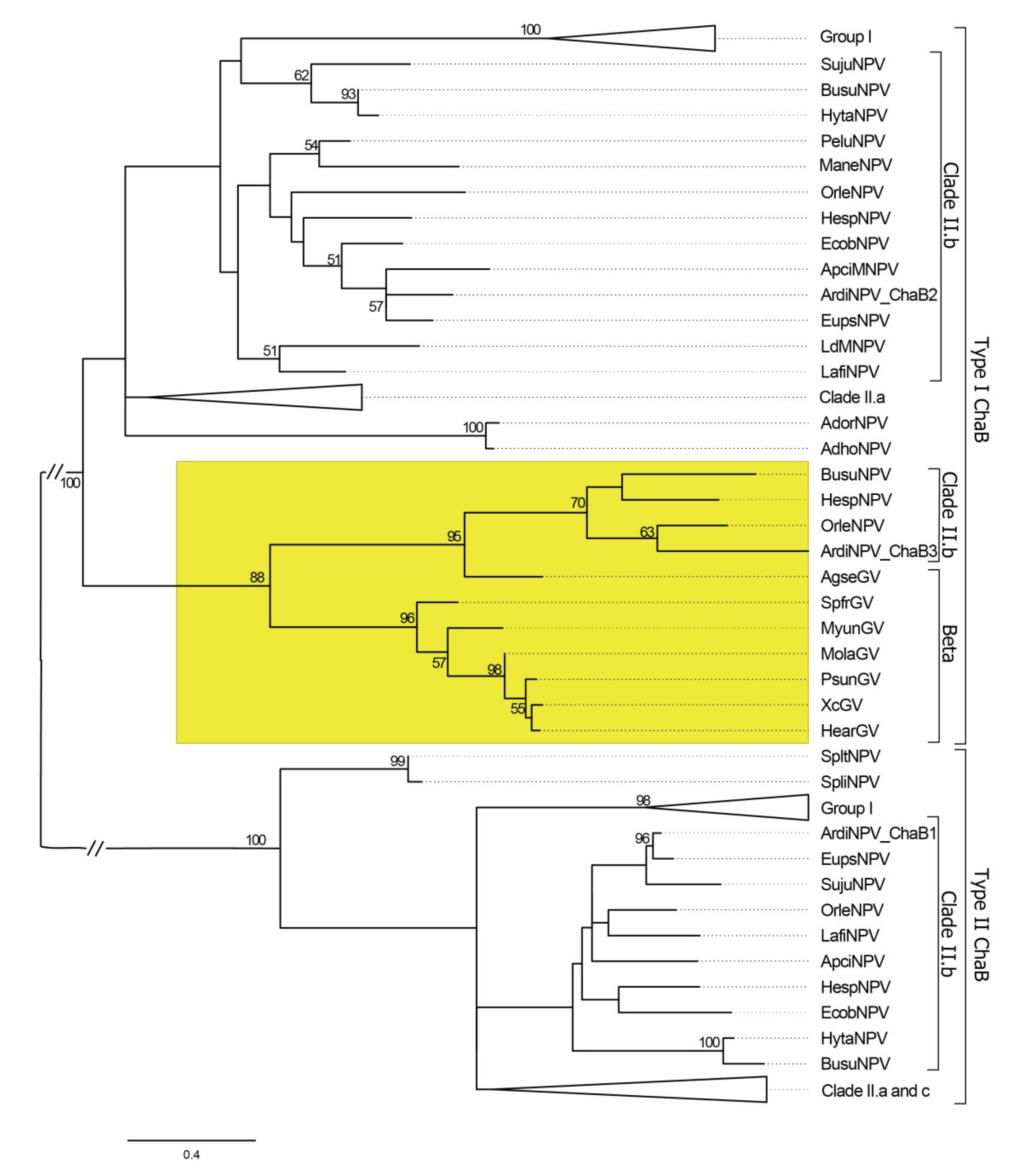
3.4. ArdiNPV Belongs to a Cluster Clade II.b of Group II Alphabaculoviruses Which Contains a Second Copy of dbp Gene
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Harrison, R.L.; Herniou, E.A.; Jehle, J.A.; Theilmann, D.A.; Burand, J.P.; Becnel, J.J.; Krell, P.J.; van Oers, M.M.; Mowery, J.D.; Bauchan, G.R.; et al. ICTV Virus Taxonomy Profile: Baculoviridae. J. Gen. Virol. 2018, 99, 1185–1186. [Google Scholar] [CrossRef] [PubMed]
- Kelly, B.J.; King, L.A.; Possee, R.D. Introduction to Baculovirus Molecular Biology. Methods Mol. Biol. 2016, 338, 25–53. [Google Scholar]
- Possee, R.D. Baculovirus as expression vectors. Curr. Opin. Biotechnol. 1997, 8, 569–572. [Google Scholar] [CrossRef]
- Summers, M.D. Milestones leading to the genetic engineering of baculoviruses as expression vector systems and viral pesticides. Adv. Virus Res. 2006, 68, 3–73. [Google Scholar] [PubMed]
- Ono, C.; Okamoto, T.; Abe, T.; Matsuura, Y. Baculovirus as a tool for gene delivery and gene therapy. Viruses 2018, 10, 510. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.J.; Lefkowitz, E.J.; King, A.M.Q.; Harrach, B.; Harrison, R.L.; Knowles, N.J.; Kropinski, A.M.; Krupovic, M.; Kuhn, J.H.; Mushegian, A.R.; et al. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2016). Arch. Virol. 2016, 161, 2921–2949. [Google Scholar] [CrossRef] [PubMed]
- Jehle, J.A.; Blissard, G.W.; Bonning, B.C.; Cory, J.S.; Herniou, E.A.; Rohrmann, G.F.; Theilmann, D.A.; Thiem, S.M.; Vlak, J.M. On the classification and nomenclature of baculoviruses: A proposal for revision. Arch. Virol. 2006, 151, 1257–1266. [Google Scholar] [CrossRef]
- Thézé, J.; Lopez-Vaamonde, C.; Cory, J.S.; Herniou, E. Biodiversity, Evolution and Ecological Specialization of Baculoviruses: A Treasure Trove for Future Applied Research. Viruses 2018, 10, 366. [Google Scholar] [CrossRef]
- Species 2000: Naturalis, Leiden, The Netherlands. Available online: www.catalogueoflife.org/annual-checklist/2019 (accessed on 8 September 2019).
- Zhao, Z. Fauna Sinica Insecta, Lepidoptera: Lymantriidae; Science Press: Beijing, China, 2003; Volume 30, p. 484. (In Chinese) [Google Scholar]
- Zheng, M.; Dai, C.; Chen, L.; Peng, Y.; Liang, D.; Zhao, K.; Zhang, Q. Some insect viruses discovered in Guizhou. J. Northwestern Coll. For. 1988, 3, 55–63. [Google Scholar]
- Tang, X.-D.; Xiao, Q.; Ma, X.-C.; Zhu, Z.-R.; Zhang, C.-X. Morphology and genome of Euproctis pseudoconspersa nucleopolyhedrovirus. Virus Genes 2009, 38, 495–506. [Google Scholar] [CrossRef]
- Volkman, L.E.; Summeers, M.D.; Hsieh, C.-H. Occluded and nonoccluded nuclear polyhedrosis virus grown in Trichoplusia ni: Comparative neutralization comparative infectivity, and in vitro growth studies. J. Virol. 1976, 19, 820–832. [Google Scholar] [PubMed]
- Hu, Z.H.; Arif, B.M.; Sun, J.S.; Chen, X.W.; Zuidema, D.; Goldbach, R.W.; Vlak, J.M. Genetic organization of the HindIII-I region of the single-nucleocapsid nucleopolyhedrovirus of Buzura suppressaria. Virus Res. 1998, 55, 71–82. [Google Scholar] [CrossRef]
- EMBOSS Stretcher. Available online: www.bioinformatics.nl/cgi-bin/emboss/stretcher (accessed on 8 July 2019).
- Tandem Repeats Finder. Available online: http://tandem.bu.edu/trf/trf.html (accessed on 10 July 2019).
- BLAST. Available online: http://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 8 July 2019).
- FGENESV0. Available online: http://linux1.softberry.com/berry.phtml (accessed on 6 July 2019).
- ORF Finder. Available online: https://www.ncbi.nlm.nih.gov/orffinder/ (accessed on 20 July 2019).
- Hu, Z.H.; Arif, B.M.; Jin, F.; Martens, J.W.M.; Chen, X.W.; Sun, J.S.; Zuidema, D.; Goldbach, R.W.; Vlak, J.M. Distinct gene arrangement in the Buzura suppressaria single-nucleocapsid nucleopolyhedrovirus genome. J. Gen. Virol. 1998, 79, 2841–2851. [Google Scholar] [CrossRef]
- Garavaglia, M.J.; Miele, S.A.; Iserte, J.A.; Belaich, M.N.; Ghiringhelli, P.D. The ac53, ac78, ac101, and ac103 genes are newly discovered core genes in the family Baculoviridae. J. Virol. 2012, 86, 12069–12079. [Google Scholar] [CrossRef] [PubMed]
- Javed, M.A.; Biswas, S.; Willis, L.G.; Harris, S.; Pritchard, C.; van Oers, M.M.; Donly, B.C.; Erlandson, M.A.; Hegedus, D.D.; Theilmann, D.A. Autographa californica multiple nucleopolyhedrovirus AC83 is a per os infectivity factor (pif) protein required for occlusion-derived Virus (ODV) and budded virus nucleocapsid assembly as well as assembly of the pif complex in ODV envelopes. J. Virol. 2017, 91, 5. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using clustalW and clustalX. Curr. Protoc. Bioinform. 2003, 2, 2.3.1–2.3.22. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Sanderson, M.J.; Wojciechowski, M.F. Improved bootstrap confidence limits in large-scale phylogenies, with an example from Neo-Astragalus (Leguminosae). Syst. Biol. 2000, 49, 671–685. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef]
- Leisy, D.J.; Rohrmann, G.F. Characterization of the replication of plasmids containing hr sequences in baculovirus-infected Spodoptera frugiperda cells. Virology 1993, 196, 722–730. [Google Scholar] [CrossRef]
- Kool, M.; Ahrens, C.H.; Vlak, J.M.; Rohrmann, G.F. Replication of baculovirus DNA. J. Gen. Virol. 1995, 76, 2103–2118. [Google Scholar] [CrossRef] [PubMed]
- Theilmann, D.A.; Stewart, S. Tandemly repeated sequence at the 3′end of the baculovirus Orygia pseudotsugata multicapsid nuclear polyhedrosis virus is an enhancer element. Virology 1992, 187, 97–106. [Google Scholar] [CrossRef]
- Van Oers, M.M.; Abma-Henkens, M.H.; Herniou, E.A.; de Groot, J.C.; Peters, S.; Vlak, J.M. Genome sequence of Chrysodeixis chalcites nucleopolyhedrovirus, a baculovirus with two DNA photolyase genes. J. Gen. Virol 2005, 86, 2069–2080. [Google Scholar] [CrossRef] [PubMed]
- Craveiro, S.R.; Inglis, P.W.; Togawa, R.C.; Grynberg, P.; Melo, F.L.; Ribeiro, Z.M.; Ribeiro, B.M.; Bao, S.N.; Castro, M.E. The genome sequence of Pseudoplusia includens single nucleopolyhedrovirus and an analysis of p26 gene evolution in the baculoviruses. BMC Genom. 2015, 16, 127. [Google Scholar] [CrossRef] [PubMed]
- Willis, L.G.; Seipp, R.; Stewart, T.M.; Erlandson, M.A.; Theilmann, D.A. Sequence analysis of the complete genome of Trichoplusia ni single nucleopolyhedrovirus and the identification of a baculoviral photolyase gene. Virology 2005, 338, 209–226. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Yin, F.; Liu, X.; Hou, D.; Wang, J.; Zhang, L.; Arif, B.; Wang, H.; Deng, F.; Hu, Z. Genome sequence and analysis of Buzura suppressaria Nucleopolyhedrovirus: A Group II Alphabaculovirus. PLoS ONE 2014, 9, e86450. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhu, S.Y.; Yi, J.P.; Shen, W.D.; Wang, L.Q.; He, H.G.; Wang, Y.; Li, B.; Wang, W.B. Genomic sequence, organization and characteristics of a new nucleopolyhedrovirus isolated from Clanis bilineata larva. BMC Genom. 2009, 10, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Rohrmann, G.F.; Erlandson, M.A.; Theilmann, D.A. The genome of a baculovirus isolated from Hemileuca sp. encodes a serpin ortholog. Virus Genes 2013, 47, 357–364. [Google Scholar] [CrossRef]
- Ma, X.-C.; Shang, J.-Y.; Yang, Z.-N.; Bao, Y.-Y.; Xiao, Q.; Zhang, C.-X. Genome sequence and organization of a nucleopolyhedrovirus that infects the tea looper caterpillar, Ectropis obliqua. Virology 2007, 360, 235–246. [Google Scholar] [CrossRef]
- Thumbi, D.K.; Eveleigh, R.J.; Lucarotti, C.J.; Lapointe, R.; Graham, R.I.; Pavlik, L.; Lauzon, H.A.; Arif, B.M. Complete sequence, analysis and organization of the Orgyia leucostigma nucleopolyhedrovirus genome. Viruses 2011, 3, 2301–2327. [Google Scholar] [CrossRef]
- Liu, X.; Yin, F.; Zhu, Z.; Hou, D.; Wang, J.; Zhang, L.; Wang, M.; Wang, H.; Hu, Z.; Deng, F. Genomic sequencing and analysis of Sucra jujuba Nucleopolyhedrovirus. PLoS ONE 2014, 9, e110023. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.; Suryamohan, K.; Kuriakose, B.; Janakiraman, V.; Reichelt, M.; Chaudhuri, S.; Guillory, J.; Divakaran, N.; Rabins, P.E.; Goel, R.; et al. Comprehensive analysis of single molecule sequencing-derived complete genome and whole transcriptome of Hyposidra talaca nuclear polyhedrosis virus. Sci. Rep. 2018, 8, 8924–8935. [Google Scholar] [CrossRef] [PubMed]
- Rohrmann, G.F.; Erlandson, M.A.; Theilmann, D.A. Genome Sequence of an Alphabaculovirus Isolated from the Oak Looper, Lambdina fiscellaria, Contains a Putative 2-Kilobase-Pair Transposable Element Encoding a Transposase and a FLYWCH Domain-Containing Protein. Genome Announc. 2015, 3, e00186-15. [Google Scholar] [CrossRef] [PubMed]
- Ardisson-Araujo, D.M.; Lima, R.N.; Melo, F.L.; Clem, R.J.; Huang, N.; Bao, S.N.; Sosa-Gomez, D.R.; Ribeiro, B.M. Genome sequence of Perigonia lusca single nucleopolyhedrovirus: Insights into the evolution of a nucleotide metabolism enzyme in the family baculoviridae. Sci. Rep. 2016, 6, 24612–24626. [Google Scholar] [CrossRef] [PubMed]
- Kuzio, J.; Pearson, M.N.; Harwood, S.H.; Funk, C.J.; Evans, J.T.; Slavicek, J.M.; Rohrmann, G.F. Sequence and Analysis of the Genome of a Baculovirus Pathogenic for Lymantria dispar. Virology 1999, 253, 17–34. [Google Scholar] [CrossRef] [PubMed]
- Nai, Y.S.; Wu, C.Y.; Wang, T.C.; Chen, Y.R.; Lau, W.H.; Lo, C.F.; Tsai, M.F.; Wang, C.H. Genomic sequencing and analyses of Lymantria xylina multiple nucleopolyhedrovirus. BMC Genom. 2010, 11, 116–130. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hou, D.; Wang, Q.; Kuang, W.; Zhang, L.; Li, J.; Shen, S.; Deng, F.; Wang, H.; Hu, Z.; et al. Genome analysis of a novel Group I alphabaculovirus obtained from Oxyplax ochracea. PLoS ONE 2018, 13, e0192279. [Google Scholar] [CrossRef]
- Zhu, Z.; Wang, J.; Wang, Q.; Yin, F.; Liu, X.; Hou, D.; Zhang, L.; Liu, H.; Li, J.; Arif, B.M.; et al. Genome characteristics of the Cyclophragma Undans nucleopolyhedrovirus: A distinct species in Group I of Alphabaculovirus. Virol. Sin. 2018, 33, 359–368. [Google Scholar] [CrossRef]
- Rollie, J. Clem Viral IAPs, then and now. Semin. Cell Dev. Biol. 2015, 39, 72–77. [Google Scholar]
- Zheng, F.; Huang, Y.; Long, G.; Sun, X.; Wang, H. Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus ORF51 is a ChaB homologous gene involved in budded virus production and DNA replication. Virus Res. 2011, 155, 203–212. [Google Scholar] [CrossRef]
- Mikhailov, V.S.; Vanarsdall, A.L.; Rohrmann, G.F. Isolation and characterization of the DNA-binding protein (DBP) of the Autographa californica multiple nucleopolyhedrovirus. Virology 2008, 370, 415–429. [Google Scholar] [CrossRef] [PubMed]
- Vanarsdall, A.L.; Mikhailov, V.S.; Rohrmann, G.F. Characterization of a baculovirus lacking the DBP (DNA-binding protein) gene. Virology 2007, 364, 475–485. [Google Scholar] [CrossRef] [PubMed]
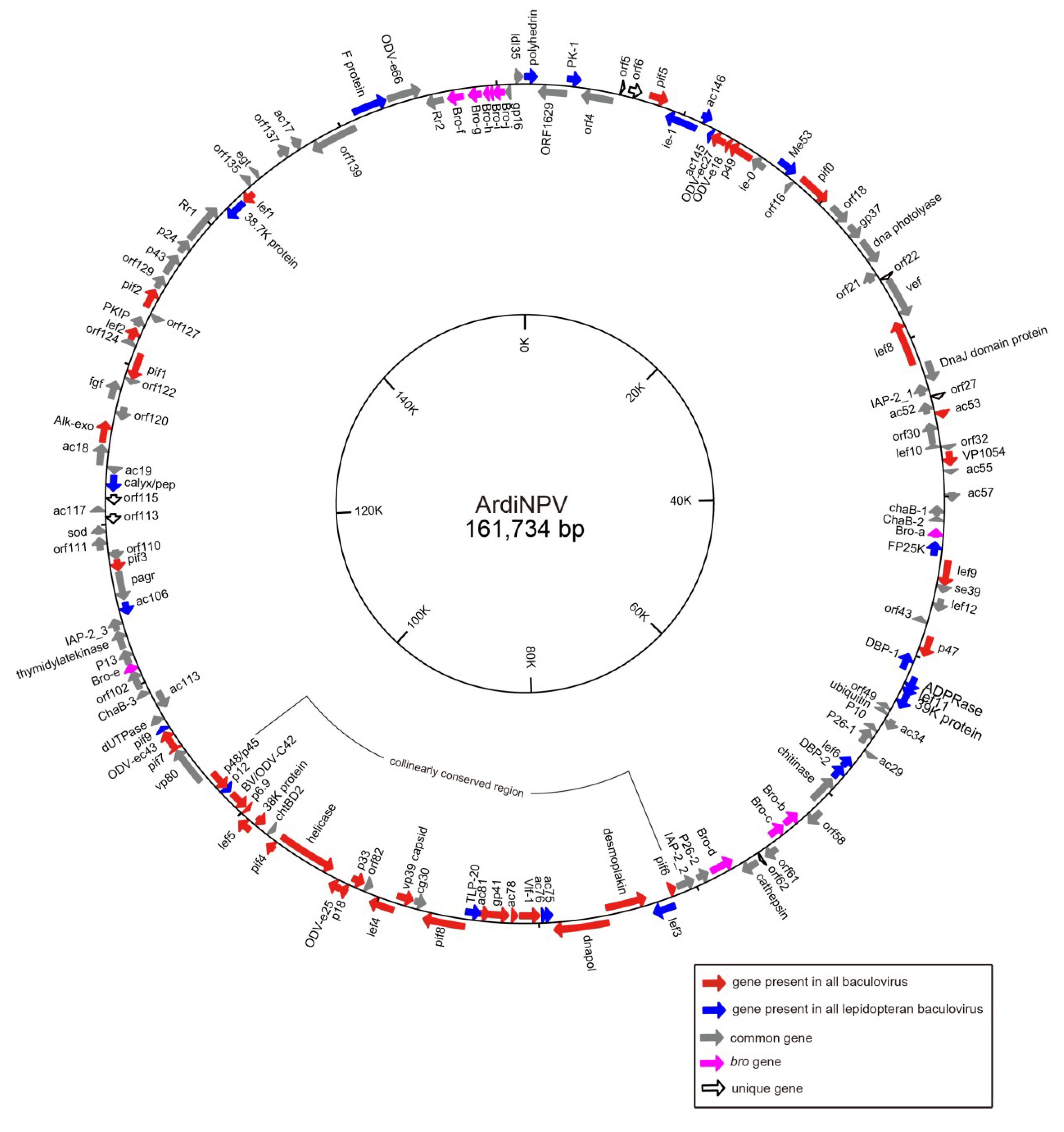
| Gene Type | Core Genes | Lepidoptera Baculovirus Conserved Genes | Other Baculovirus Genes |
|---|---|---|---|
| Replication | dna-pol(orf70), helicase(orf86), Alk-exo(ORF119), lef2(orf125), lef1(orf134) | ie-1 (orf8), me53 (orf15), lef11 (orf47), dbp-1 (orf45), dbp-2(orf56), lef3 (orf68) | dna photolyase(orf20), dUTPase(orf99), rr1(orf132), rr2(orf142) |
| Transcription | lef8(orf24), lef9(orf40), p47(orf44), vlf-1(orf73), lef4(orf81), lef5(orf90) | pk-1 (orf3), 39k (orf48), lef6 (orf55) | lef10 (orf32), lef12 (orf42),ie-0(orf14) |
| Structure | odv-ec27 (orf11), odv-e18 (orf12), 49k (orf13), ac53(orf29), vp1054(orf33), desmoplakin (orf69), ac78 (orf74), gp41 (orf75), ac81 (orf76), vp39 (orf80), p33 (orf83), p18 (orf84), odv-e25 (orf85), 38k (orf89), p6.9 (orf91), BV/ODV-C42 (orf92), p48/p45 (orf94), odv-ec43 (orf97) | polyhedrin (orf1), fp25k (orf39), tlp-20 (orf77), p12 (orf93), F (orf140), calyx/pep (orf116) | p10 (orf52), vp80 (orf95), pkip (orf126), p24 (orf131), gp16 (orf148), ORF1629 (orf2), cg30 (orf79) |
| Oral infection | pif5 (orf7), pif0 (orf17), pif6 (orf67), pif8 (orf78), pif4 (orf87), pif7 (orf96), pif3 (orf109), pif1 (orf123), pif2 (orf128) | pif9 (orf98) | odv-e66 (orf141) |
| Auxiliary | ADPRase (orf46), 38.7k (orf133) | iap-2_1 (orf26), ubiquitin (orf50), p26-1 (orf53), chitinase (orf57), cathepsin (orf63), gp37 (orf19), p26-2 (orf65), iap-2_2 (orf66), iap-2_3 (orf106), sod (orf112), fgf (orf121), egt(orf136), bro-a(orf38), bro-b(orf59), bro-c(orf60), bro-d(orf64), bro-e(orf103), bro-f(orf143), bro-g(orf144), bro-h(orf145), bro-i(orf146), bro-j(orf147), vef(orf23), ring finger protein(orf82), p13(orf104) | |
| Unknown | ac146 (orf9), ac145 (orf10), ac75 (orf71), ac76 (orf72), ac106 (orf107) | ac52 (orf28), orf31, ac55 (orf34), ac57 (orf35), chaB-1 (orf36), chaB-2 (orf37), ac34 (orf51), ac29 (orf54), chtBD2 (orf88), ac113 (orf100), ac117 (orf114), ac19 (orf117), ac18 (orf118), ac17 (orf138), DnaJ domain protein (orf25), p43(orf130), hoar(orf4), orf16, orf18, orf21, orf30, orf41, orf43, orf49, orf58, orf61, chaB-3(orf101),orf102, orf110,HE65(orf111), orf120, orf122, orf124, orf127, orf129, orf135, orf137, ldl35(orf149), thymidylate kinase(orf105), pagr(orf108), peptidase MA superfamily(orf139) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Duan, X.; Wang, Q.; Zhang, L.; Deng, F.; Wang, H.; Hu, Z.; Wang, M.; Wang, J. Genome Analysis of a Novel Clade II.b Alphabaculovirus Obtained from Artaxa digramma. Viruses 2019, 11, 925. https://doi.org/10.3390/v11100925
Li J, Duan X, Wang Q, Zhang L, Deng F, Wang H, Hu Z, Wang M, Wang J. Genome Analysis of a Novel Clade II.b Alphabaculovirus Obtained from Artaxa digramma. Viruses. 2019; 11(10):925. https://doi.org/10.3390/v11100925
Chicago/Turabian StyleLi, Jiang, Xiaoyan Duan, Qianran Wang, Lei Zhang, Fei Deng, Hualin Wang, Zhihong Hu, Manli Wang, and Jun Wang. 2019. "Genome Analysis of a Novel Clade II.b Alphabaculovirus Obtained from Artaxa digramma" Viruses 11, no. 10: 925. https://doi.org/10.3390/v11100925
APA StyleLi, J., Duan, X., Wang, Q., Zhang, L., Deng, F., Wang, H., Hu, Z., Wang, M., & Wang, J. (2019). Genome Analysis of a Novel Clade II.b Alphabaculovirus Obtained from Artaxa digramma. Viruses, 11(10), 925. https://doi.org/10.3390/v11100925





