Remodeling of the Actin Network Associated with the Non-Structural Protein 1 (NS1) of West Nile Virus and Formation of NS1-Containing Tunneling Nanotubes
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Virus
2.2. Cloning of NS1
2.3. NS1 Production and Purification
2.4. Antibodies
2.5. Protein Gel Electrophoresis and Western Blotting
2.6. Confocal Fluorescence Microscopy
2.7. Flow Cytometry Analysis of Cell Attachment and Uptake of NS1WNV
2.8. Transmission Electron Microscopy
2.9. Transfections
2.10. Kinetics of WNV Replication and Virus Titration
3. Results
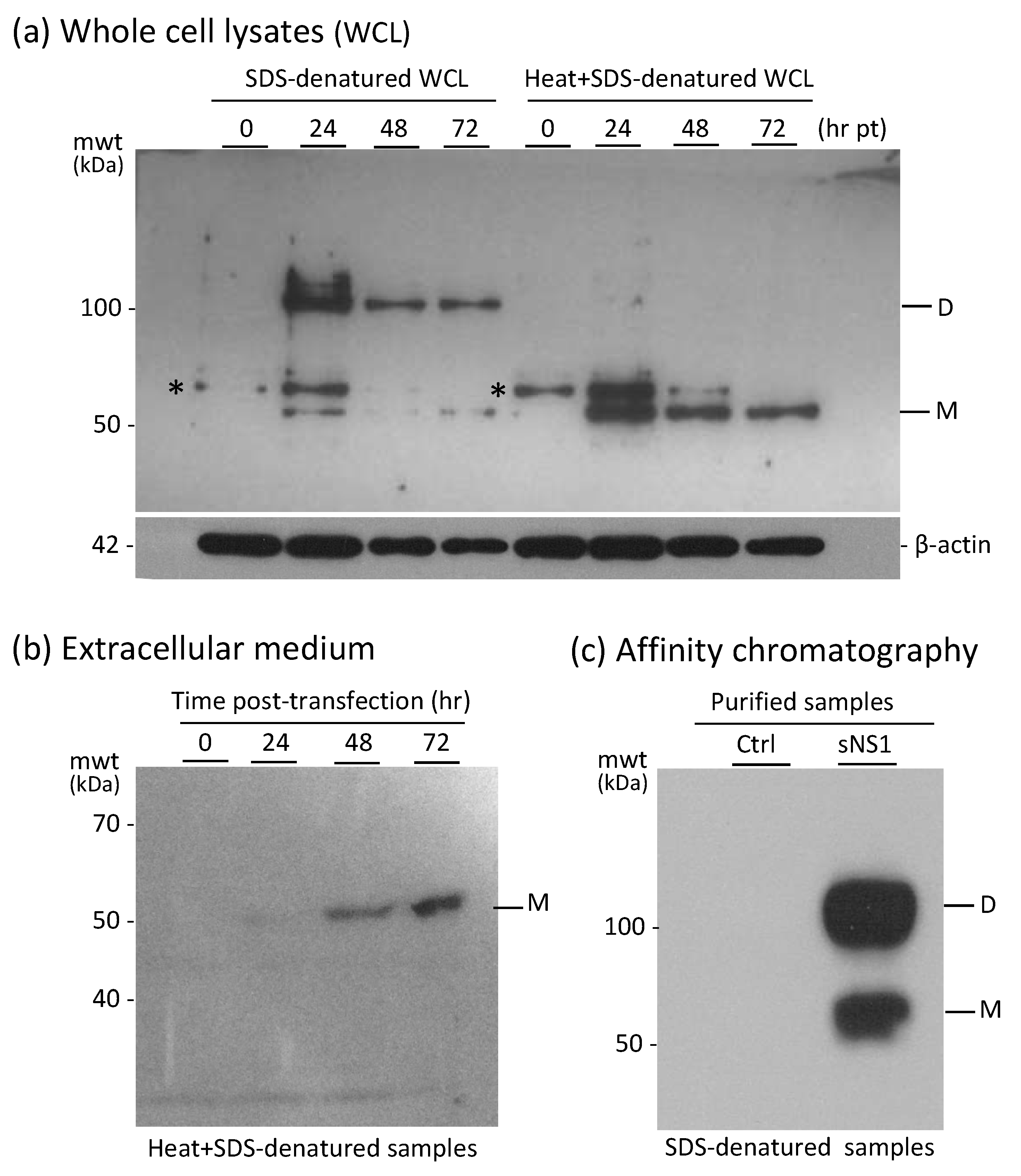
3.1. Expression and Characterization of Intracellular and Secreted Form of NS1WNV in Mammalian Cells
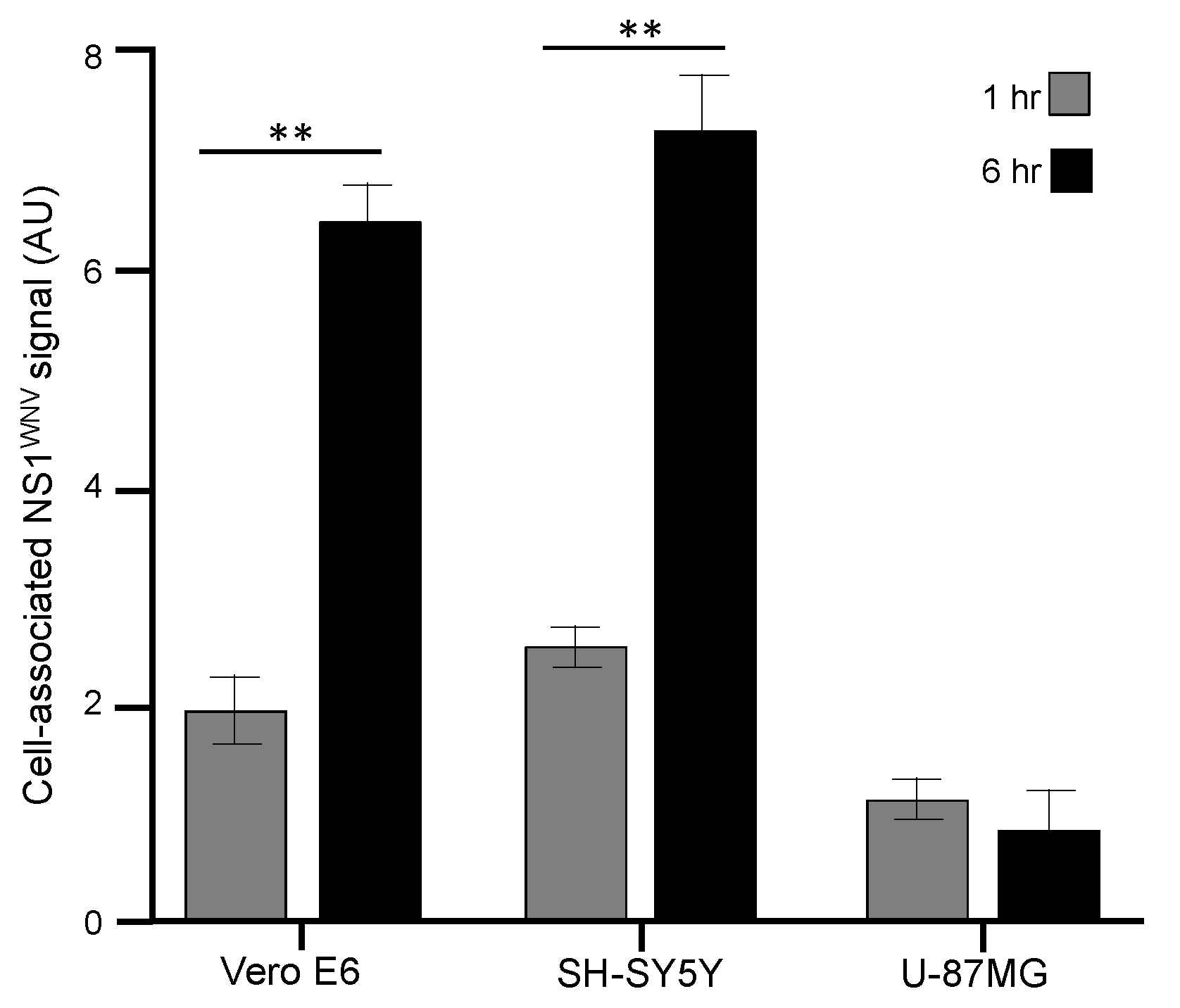
3.2. Cell Attachment and Uptake of sNS1WNV
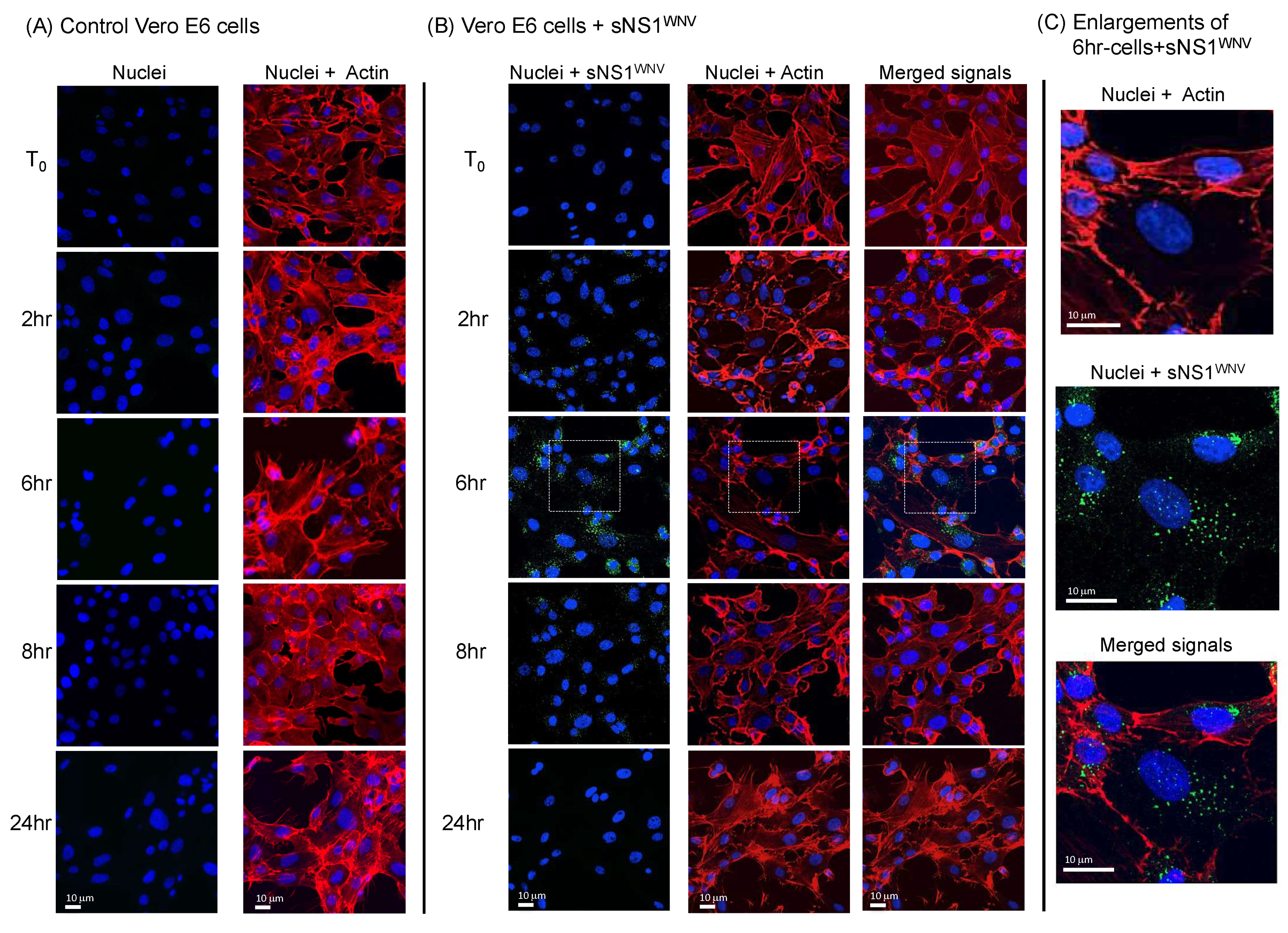
3.3. Cytoskeletal Modifications Associated with sNS1WNV Internalization
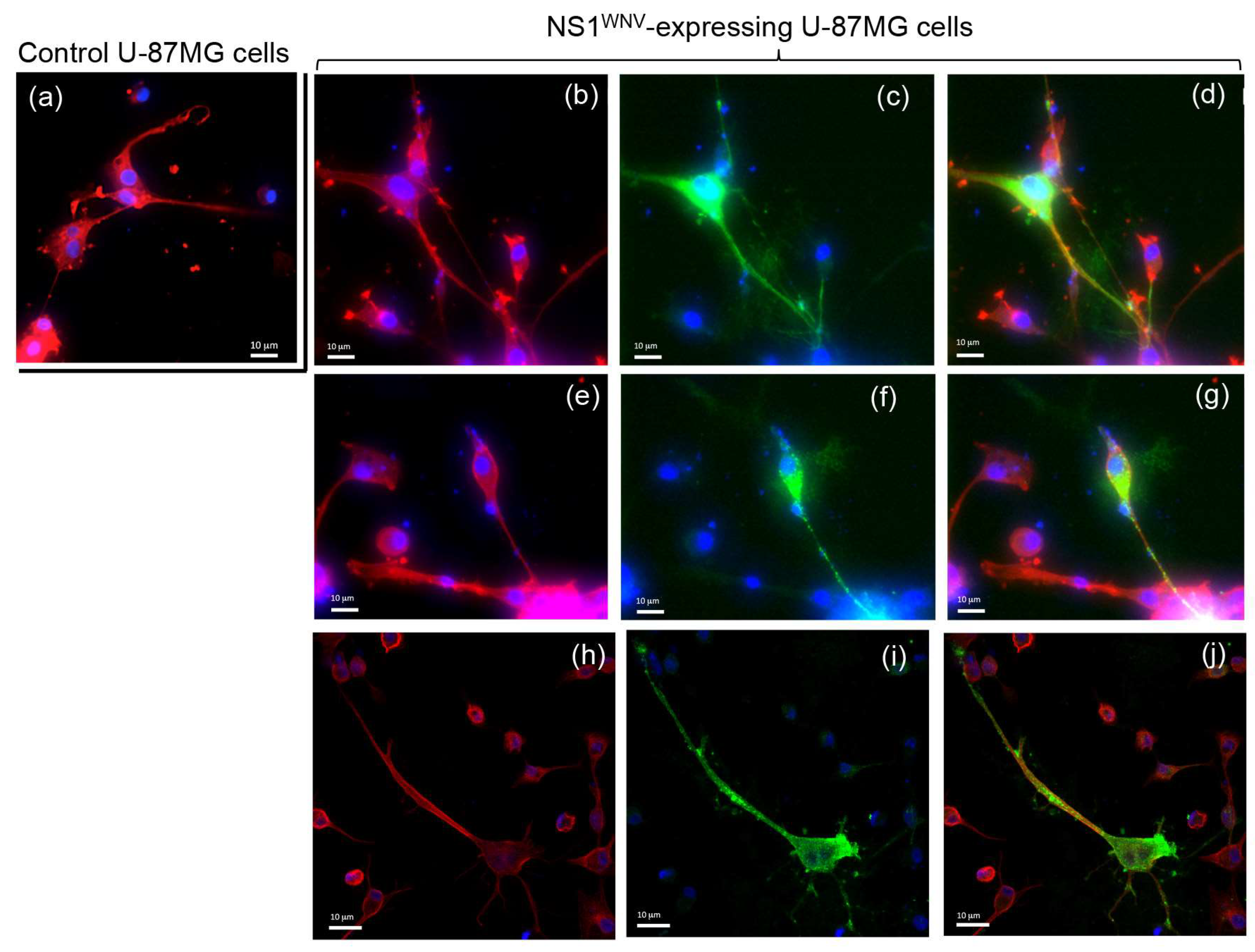
3.4. Presence of NS1WNV Protein in Actin-Containing, TNT-Like Nanofilaments Protruding from NS1WNV-Expressing and WNV-Infected Cells
3.5. NS1WNV and Virus Infectiveness
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Muller, D.A.; Young, P.R. The flavivirus NS1 protein: Molecular and structural biology, immunology, role in pathogenesis and application as a diagnostic biomarker. Antivir. Res. 2013, 98, 192–208. [Google Scholar] [CrossRef] [PubMed]
- Lindenbach, B.D.; Rice, C.M. Molecular biology of flaviviruses. Adv. Virus Res. 2003, 59, 23–61. [Google Scholar] [PubMed]
- Mackenzie, J.M.; Westaway, E.G. Assembly and maturation of the flavivirus Kunjin virus appear to occur in the rough endoplasmic reticulum and along the secreted pathway, respectively. J. Virol. 2001, 75, 10787–10799. [Google Scholar] [CrossRef] [PubMed]
- Gutsche, I.; Coulibaly, F.; Voss, J.E.; Salmon, J.; d’Alayer, J.; Ermonval, M.; Larquet, E.; Charneau, P.; Krey, T.; Megret, F.; et al. Secreted dengue virus nonstructural protein NS1 is an atypical barrel-shaped high-density lipoprotein. Proc. Natl. Acad. Sci. USA 2011, 108, 8003–8008. [Google Scholar] [CrossRef] [PubMed]
- Westaway, E.G.; Goodman, M.R. Variation in distribution of the three flavivirus-specified glycoproteins detected by immunofluorescence in infected Vero cells. Arch. Virol. 1987, 94, 215–228. [Google Scholar] [CrossRef]
- Westaway, E.G.; Mackenzie, J.M.; Kenney, M.T.; Jones, M.K.; Khromykh, A.A. Ultrastructure of Kunjin virus-infected cells: Colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 1997, 71, 6650–6661. [Google Scholar] [PubMed]
- Westaway, E.G.; Mackenzie, J.M.; Khromykh, A.A. Replication and gene function in Kunjin virus. Curr. Top. Microbiol. Immunol. 2002, 267, 323–351. [Google Scholar]
- Fan, W.F.; Mason, P.W. Membrane association and secretion of the Japanese encephalitis virus NS1 protein from cells expressing NS1 cDNA. Virology 1990, 177, 470–476. [Google Scholar] [CrossRef]
- Schlesinger, J.J.; Brandriss, M.W.; Putnak, J.R.; Walsh, E.E. Cell surface expression of yellow fever virus non-structural glycoprotein NS1: Consequences of interaction with antibody. J. Gen. Virol. 1990, 71, 593–599. [Google Scholar] [CrossRef]
- Akey, D.L.; Brown, W.C.; Dutta, S.; Konwerski, J.; Jose, J.; Jurkiw, T.J.; DelProposto, J.; Ogata, C.M.; Skiniotis, G.; Kuhn, R.J.; et al. Flavivirus NS1 structures reveal surfaces for associations with membranes and the immune system. Science 2014, 343, 881–885. [Google Scholar] [CrossRef]
- Crooks, A.J.; Lee, J.M.; Easterbrook, L.M.; Timofeev, A.V.; Stephenson, J.R. The NS1 protein of tick-borne encephalitis virus forms multimeric species upon secretion from the host cell. J. Gen. Virol. 1994, 75, 3453–3460. [Google Scholar] [CrossRef] [PubMed]
- Post, P.R.; Carvalho, R.; Galler, R. Glycosylation and secretion of yellow fever virus nonstructural protein NS1. Virus Res. 1991, 18, 291–302. [Google Scholar] [CrossRef]
- Scaturro, P.; Cortese, M.; Chatel-Chaix, L.; Fischl, W.; Bartenschlager, R. Dengue Virus non-structural protein 1 modulates infectious particle production via interaction with the structural proteins. PLoS Pathog. 2015, 11, e1005277. [Google Scholar] [CrossRef] [PubMed]
- Alcon-LePoder, S.; Drouet, M.T.; Roux, P.; Frenkiel, M.P.; Arborio, M.; Durand-Schneider, A.M.; Maurice, M.; Le Blanc, I.; Gruenberg, J.; Flamand, M. The secreted form of dengue virus nonstructural protein NS1 is endocytosed by hepatocytes and accumulates in late endosomes: Implications for viral infectivity. J. Virol. 2005, 79, 11403–11411. [Google Scholar] [CrossRef] [PubMed]
- Avirutnan, P.; Zhang, L.; Punyadee, N.; Manuyakorn, A.; Puttikhunt, C.; Kasinrerk, W.; Malasit, P.; Atkinson, J.P.; Diamond, M.S. Secreted NS1 of dengue virus attaches to the surface of cells via interactions with heparan sulfate and chondroitin sulfate E. PLoS Pathog. 2007, 3, e183. [Google Scholar] [CrossRef] [PubMed]
- Crook, K.R.; Miller-Kittrell, M.; Morrison, C.R.; Scholle, F. Modulation of innate immune signaling by the secreted form of the West Nile virus NS1 glycoprotein. Virology 2014, 458, 172–182. [Google Scholar] [CrossRef] [PubMed]
- Modhiran, N.; Watterson, D.; Muller, D.A.; Panetta, A.K.; Sester, D.P.; Liu, L.; Hume, D.A.; Stacey, K.J.; Young, P.R. Dengue virus NS1 protein activates cells via Toll-like receptor 4 and disrupts endothelial cell monolayer integrity. Sci. Transl. Med. 2015, 7, 304ra142. [Google Scholar] [CrossRef] [PubMed]
- Youn, S.; Cho, H.; Fremont, D.H.; Diamond, M.S. A short N-terminal peptide motif on flavivirus nonstructural protein NS1 modulates cellular targeting and immune recognition. J. Virol. 2010, 84, 9516–9532. [Google Scholar] [CrossRef]
- Avirutnan, P.; Fuchs, A.; Hauhart, R.E.; Somnuke, P.; Youn, S.; Diamond, M.S.; Atkinson, J.P. Antagonism of the complement component C4 by flavivirus nonstructural protein NS1. J. Exp. Med. 2010, 207, 793–806. [Google Scholar] [CrossRef]
- Diamond, M.S. Mechanisms of evasion of the type interferon antiviral response by flaviviruses. J. Interferon Cytokine Res. 2009, 29, 521–530. [Google Scholar] [CrossRef]
- Diamond, M.S.; Gale, M. Cell-intrinsic innate immune control of West Nile virus infection. Trends Immunol. 2012, 33, 522–530. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.R.; de Sessions, P.F.; Leon, M.A.; Scholle, F. West Nile virus nonstructural protein 1 inhibits TLR3 signal transduction. J. Virol. 2008, 82, 8262–8271. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-L.; Ye, H.-Q.; Liu, S.-Q.; Deng, C.-L.; Li, X.-D.; Shi, P.-Y.; Zhang, B. West Nile virus NS1 antagonizes interferon-β production by targeting RIG-I and MDA5. J. Virol. 2017, 91, e02396-16. [Google Scholar] [CrossRef] [PubMed]
- Beatty, P.R.; Puerta-Guardo, H.; Killingbeck, S.S.; Glasner, D.R.; Hopkins, K.; Harris, E. Dengue virus NS1 triggers endothelial permeability and vascular leak that is prevented by NS1 vaccination. Sci. Transl. Med. 2015, 7, 304ra141. [Google Scholar] [CrossRef] [PubMed]
- Macdonald, J.; Tonry, J.; Hall, R.A.; Williams, B.; Palacios, G.; Ashok, M.S.; Jabado, O.; Clark, D.; Tesh, R.B.; Briese, T.; et al. NS1 protein secretion during the acute phase of West Nile virus infection. J. Virol. 2005, 79, 13924–13933. [Google Scholar] [CrossRef] [PubMed]
- Sun, E.-C.; Ma, J.-N.; Liu, N.-H.; Yang, T.; Zhao, J.; Geng, H.-W.; Wang, L.-F.; Qin, Y.-L.; Bu, Z.-G.; Yang, Y.-H.; et al. Identification of two linear B-cell epitopes from West Nile virus NS1 by screening a phage-displayed random peptide library. BMC Microbiol. 2011, 11, 160. [Google Scholar] [CrossRef] [PubMed]
- Watterson, D.; Modhiran, N.; Young, P.R. The many faces of the flavivirus NS1 protein offer a multitude of options for inhibitor design. Antivir. Res. 2016, 130, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Foo, K.Y.; Chee, H.Y. Interaction between flavivirus and cytoskeleton during virus replication. Biomed. Res. Int. 2015, 2015, 427814. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.P.; Koyuncu, O.O.; Enquist, L.W. Subversion of the actin cytoskeleton during viral infection. Nat. Rev. Microbiol. 2011, 9, 427–439. [Google Scholar] [CrossRef] [PubMed]
- Chu, J.J.; Choo, B.G.; Lee, J.W.; Ng, M.L. Actin filaments participate in West Nile (Sarafend) virus maturation process. J. Med. Virol. 2003, 71, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Hackett, B.A.; Cherry, S. Flavivirus internalization is regulated by a size-dependent endocytic pathway. Proc. Natl. Acad. Sci. USA 2018, 115, 4246–4251. [Google Scholar] [CrossRef] [PubMed]
- Sum, M.S.H. The involvement of microtubules and actin during the infection of Japanese encephalitis virus in neuroblastoma cell line, IMR32. Biomed. Res. Int. 2015, 2015, 695283. [Google Scholar]
- Ng, M.L.; Chu, J.H. Interaction of West Nile and Kunjin viruses with cellular components during morphogenesis. Curr. Top. Microbiol. Immunol. 2002, 267, 353–372. [Google Scholar] [PubMed]
- Ng, M.L.; Hong, S.S. Flavivirus infection: Essential ultrastructural changes and association of Kunjin virus NS3 protein with microtubules. Arch. Virol. 1989, 106, 103–120. [Google Scholar] [CrossRef] [PubMed]
- Poungsawai, J.; Kanlaya, R.; Pattanakitsakul, S.N.; Thongboonkerd, V. Subcellular localizations and time-course expression of dengue envelope and non-structural 1 proteins in human endothelial cells. Microb. Pathog. 2011, 51, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Růzek, D.; Vancová, M.; Tesarová, M.; Ahantarig, A.; Kopecký, J.; Grubhoffer, L. Morphological changes in human neural cells following tick-borne encephalitis virus infection. J. Gen. Virol. 2009, 90, 1649–1658. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zou, L.; Hu, Z.; Chen, W.; Zhang, J.; Zhu, J.; Fang, X.; Yuan, W.; Hu, X.; Hu, F.; et al. Identification and characterization of a 43 kDa actin protein involved in the DENV-2 binding and infection of ECV304 cells. Microbes Infect. 2013, 15, 310–318. [Google Scholar] [CrossRef]
- Hafirassou, M.L.; Meertens, L.; Umaña-Diaz, C.; Labeau, A.; Dejarnac, O.; Bonnet-Madin, L.; Kümmerer, B.M.; Delaugerre, C.; Roingeard, P.; Vidalain, P.O.; et al. A global interactome map of the Dengue virus NS1 identifies virus restriction and dependency host factors. Cell Rep. 2017, 21, 3900–3913. [Google Scholar] [CrossRef]
- Dechtawewat, T.; Paemanee, A.; Roytrakul, S.; Songprakhon, P.; Limjindaporn, T.; Yenchitsomanus, P.T.; Saitornuang, S.; Puttikhunt, C.; Kasinrerk, W.; Malasit, P.; et al. Mass spectrometric analysis of host cell proteins interacting with dengue virus nonstructural protein 1 in dengue virus-infected HepG2 cells. Biochim. Biophys. Acta 2016, 1864, 1270–1280. [Google Scholar] [CrossRef]
- Gerdes, H.H.; Carvalho, R.N. Intercellular transfer mediated by tunneling nanotubes. Curr. Opin. Cell Biol. 2008, 20, 470–475. [Google Scholar] [CrossRef]
- Gerdes, H.H.; Rustom, A.; Wang, X. Tunneling nanotubes, an emerging intercellular communication route in development. Mech. Dev. 2013, 130, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Gurke, S.; Barroso, J.F.; Gerdes, H.H. The art of cellular communication: Tunneling nanotubes bridge the divide. Histochem. Cell Biol. 2008, 129, 539–550. [Google Scholar] [CrossRef] [PubMed]
- Gurke, S.; Barroso, J.F.; Hodneland, E.; Bukoreshtliev, N.V.; Schlicker, O.; Gerdes, H.H. Tunneling nanotube (TNT)-like structures facilitate a constitutive, actomyosin-dependent exchange of endocytic organelles between normal rat kidney cells. Exp. Cell Res. 2008, 314, 3669–3683. [Google Scholar] [CrossRef] [PubMed]
- Rustom, A.; Saffrich, R.; Markovic, I.; Walther, P.; Gerdes, H.H. Nanotubular highways for intercellular organelle transport. Science 2004, 303, 1007–1010. [Google Scholar] [CrossRef] [PubMed]
- Sartori-Rupp, A.; Cordero Cervantes, D.; Pepe, A.; Gousset, K.; Delage, E.; Corroyer-Dulmont, S.; Schmitt, C.; Krijnse-Locker, J.; Zurzolo, C. Correlative cryo-electron microscopy reveals the structure of TNTs in neuronal cells. Nat. Commun. 2019, 10, 342. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.; Baginski, J.; Hassan, S.F.; Volin, M.; Shukla, D.; Tiwari, V. Filopodia and viruses: An analysis of membrane processes in entry mechanisms. Front. Microbiol. 2016, 7, 300. [Google Scholar] [CrossRef] [PubMed]
- Eugenin, E.A.; Gaskill, P.J.; Berman, J.W. Tunneling nanotubes (TNT): A potential mechanism for intercellular HIV trafficking. Commun. Integr. Biol. 2009, 2, 243–244. [Google Scholar] [CrossRef] [PubMed]
- Eugenin, E.A.; Gaskill, P.J.; Berman, J.W. Tunneling nanotubes (TNT) are induced by HIV-infection of macrophages: A potential mechanism for intercellular HIV trafficking. Cell. Immunol. 2009, 254, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Sherer, N.M. Long-distance relationships: Do membrane nanotubes regulate cell-cell communication and disease progression? Mol. Biol. Cell 2013, 24, 1095–1098. [Google Scholar] [CrossRef] [PubMed]
- Sherer, N.M.; Lehmann, M.J.; Jimenez-Soto, L.F.; Horensavitz, C.; Pypaert, M.; Mothes, W. Retroviruses can establish filopodial bridges for efficient cell-to-cell transmission. Nat. Cell Biol. 2007, 9, 310–315. [Google Scholar] [CrossRef] [PubMed]
- Sherer, N.M.; Mothes, W. Cytonemes and tunneling nanotubules in cell-cell communication and viral pathogenesis. Trends Cell Biol. 2008, 18, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Sowinski, S.; Jolly, C.; Berninghausen, O.; Purbhoo, M.A.; Chauveau, A.; Kohler, K.; Oddos, S.; Eissmann, P.; Brodsky, F.M.; Hopkins, C.; et al. Membrane nanotubes physically connect T cells over long distances presenting a novel route for HIV-1 transmission. Nat. Cell Biol. 2008, 10, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Kim, J.H.; Ranjan, P.; Metcalfe, M.G.; Cao, W.; Mishina, M.; Gangappa, S.; Guo, Z.; Boyden, E.S.; Zaki, S.; et al. Influenza virus exploits tunneling nanotubes for cell-to-cell spread. Sci. Rep. 2017, 7, 40360. [Google Scholar] [CrossRef] [PubMed]
- Cudmore, S.; Cossart, P.; Griffiths, G.; Way, M. Actin-based motility of vaccinia virus. Nature 1995, 378, 636–638. [Google Scholar] [CrossRef] [PubMed]
- Cudmore, S.; Reckmann, I.; Griffiths, G.; Way, M. Vaccinia virus: A model system for actin-membrane interactions. J. Cell Sci. 1996, 109, 1739–1747. [Google Scholar] [PubMed]
- Favoreel, H.W.; Van Minnebruggen, G.; Adriaensen, D.; Nauwynck, H.J. Cytoskeletal rearrangements and cell extensions induced by the US3 kinase of an alphaherpesvirus are associated with enhanced spread. Proc. Natl. Acad. Sci. USA 2005, 102, 8990–8995. [Google Scholar] [CrossRef]
- Favoreel, H.W.; Van Minnebruggen, G.; Van de Walle, G.R.; Ficinska, J.; Nauwynck, H. Herpesvirus interference with virus-specific antibodies: Bridging antibodies, internalizing antibodies, and hiding from antibodies. Vet. Microbiol. 2006, 113, 257–263. [Google Scholar] [CrossRef]
- Panasiuk, M.; Rychłowski, M.; Derewonko, N.; Bienkowska-Szewczyk, K. Tunneling nanotubes as a novel route of cell-to-cell spread of herpesviruses. J. Virol. 2018, 92, e00090-18. [Google Scholar] [CrossRef]
- Abounit, S.; Wu, J.W.; Duff, K.; Victoria, G.S.; Zurzolo, C. Tunneling nanotubes: A possible highway in the spreading of tau and other prion-like proteins in neurodegenerative diseases. Prion 2016, 10, 344–351. [Google Scholar] [CrossRef]
- Gousset, K.; Schiff, E.; Langevin, C.; Marijanovic, Z.; Caputo, A.; Browman, D.T.; Chenouard, N.; deChaumont, F.; Martino, A.; Enninga, J.; et al. Prions hijack tunnelling nanotubes for intercellular spread. Nat.Cell Biol. 2009, 11, 328–336. [Google Scholar] [CrossRef]
- Tardivel, M.; Bégard, S.; Bousset, L.; Dujardin, S.; Coens, A.; Melki, R.; Buée, L.; Colin, M. Tunneling nanotube (TNT)-mediated neuron-to neuron transfer of pathological Tau protein assemblies. Acta Neuropathol. Commun. 2016, 4, 117. [Google Scholar] [CrossRef] [PubMed]
- Malkinson, M.; Banet, C.; Weisman, Y.; Pokamunski, S.; King, R.; Drouet, M.T.; Deubel, V. Introduction of West Nile virus in the Middle East by migrating white storks. Emerg. Infect. Dis. 2002, 8, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Brien, J.D.; Lazear, H.M.; Diamond, M.S. Propagation, quantification, detection, and storage of West Nile virus. Curr. Protoc. Microbiol. 2013, 31, 15D.3. [Google Scholar] [PubMed]
- Chen, H.R.; Chao, C.H.; Liu, C.C.; Ho, T.S.; Tsai, H.P.; Perng, G.C.; Lin, Y.S.; Wang, J.R.; Yeh, T.M. Macrophage migration inhibitory factor is critical for dengue NS1-induced endothelial glycocalyx degradation and hyperpermeability. PLoS Pathog. 2018, 14, e1007033. [Google Scholar] [CrossRef] [PubMed]
- Muller, D.A.; Landsberg, M.J.; Bletchly, C.; Rothnagel, R.; Waddington, L.; Hankamer, B.; Young, P.R. Structure of the dengue virus glycoprotein non-structural protein 1 by electron microscopy and single-particle analysis. J. Gen. Virol. 2012, 93, 771–779. [Google Scholar] [CrossRef]
- Lee, J.W.M.; Ng, M.L. A nano-view of West Nile virus-induced cellular changes during infection. J. Nanobiotechnol. 2004, 2, 6. [Google Scholar] [CrossRef][Green Version]
- Donadieu, E.; Lowenski, S.; Servely, J.L.; Laloy, E.; Lilin, T.; Nowotny, N.; Richardson, J.; Zientara, S.; Lecollinet, S.; Coulpier, M. Comparison of the neuropathology induced by two West Nile virus strains. PLoS ONE 2013, 8, e84473. [Google Scholar] [CrossRef]
- Laassri, M.; Bidzhieva, B.; Speicher, J.; Pletnev, A.G.; Chumakov, K. Microarray hybridization for assessment of the genetic stability of chimeric West Nile/dengue 4 virus. J. Med. Virol. 2011, 83, 910–920. [Google Scholar] [CrossRef]
- Daniels, B.P.; Jujjavarapu, H.; Durrant, D.M.; Williams, J.L.; Green, R.G.; White, J.P.; Lazear, H.M.; Gale, M.J.; Diamond, M.S.; Klein, R.S. Regional astrocyte IFN signaling restricts pathogenesis during neurotropic viral infection. J. Clin. Investig. 2017, 127, 843–856. [Google Scholar] [CrossRef]
- Puerta-Guardo, H.; Glasner, D.R.; Espinosa, D.A.; Biering, S.B.; Patana, M.; Ratnasiri, K.; Wang, C.; Beatty, P.R.; Harris, E. Flavivirus NS1 triggers tissue-specific vascular endothelial dysfunction reflecting disease tropism. Cell Rep. 2019, 26, 1598–1613. [Google Scholar] [CrossRef]
- Singh, S.; Anupriya, M.G.; Modak, A.; Sreekumat, E. Dengue virus or NS1 protein induces trans-endothelial cell permeability associated with VE-Cadherin and RhoA phosphorylation in HMEC-1 cells preventable by angiopoietin-1. J. Gen. Virol. 2018, 99, 1658–1670. [Google Scholar] [CrossRef]
- Fraisier, C.; Camoin, L.; Lim, S.M.; Bakli, M.; Belghazi, M.; Fourquet, P.; Granjeaud, S.; Osterhaus, A.D.; Koraka, P.; Martina, B.; et al. Altered protein networks and cellular pathways in severe West Nile disease in mice. PLoS ONE 2013, 8, e68318. [Google Scholar] [CrossRef]
- Nobile, C.; Rudnicka, D.; Hasan, M.; Aulner, N.; Porrot, F.; Machu, C.; Renaud, O.; Prévost, M.C.; Hivroz, C.; Schwartz, O.; et al. HIV-1 Nef inhibits ruffles, induces filopodia, and modulates migration of infected lymphocytes. J. Virol. 2010, 84, 2282–2293. [Google Scholar] [CrossRef]
- Stolp, B.; Abraham, L.; Rudolph, J.M.; Fackler, O.T. Lentiviral Nef proteins utilize PAK2-mediated deregulation of cofilin as a general strategy to interfere with actin remodeling. J. Virol. 2010, 84, 3935–3948. [Google Scholar] [CrossRef]
- Wang, C.; Puerta-Guardo, H.; Biering, S.B.; Glasner, D.R.; Tran, E.B.; Patana, M.; Gomberg, T.A.; Malvar, C.; Lo, N.T.N.; Espinosa, D.A.; et al. Endocytosis of flavivirus NS1 is required for NS1-mediated endothelial hyperpermeability and is abolished by a single N-glycosylation site mutation. PLoS Pathog. 2019, 15, e1007938. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Furnon, W.; Fender, P.; Confort, M.-P.; Desloire, S.; Nangola, S.; Kitidee, K.; Leroux, C.; Ratinier, M.; Arnaud, F.; Lecollinet, S.; et al. Remodeling of the Actin Network Associated with the Non-Structural Protein 1 (NS1) of West Nile Virus and Formation of NS1-Containing Tunneling Nanotubes. Viruses 2019, 11, 901. https://doi.org/10.3390/v11100901
Furnon W, Fender P, Confort M-P, Desloire S, Nangola S, Kitidee K, Leroux C, Ratinier M, Arnaud F, Lecollinet S, et al. Remodeling of the Actin Network Associated with the Non-Structural Protein 1 (NS1) of West Nile Virus and Formation of NS1-Containing Tunneling Nanotubes. Viruses. 2019; 11(10):901. https://doi.org/10.3390/v11100901
Chicago/Turabian StyleFurnon, Wilhelm, Pascal Fender, Marie-Pierre Confort, Sophie Desloire, Sawitree Nangola, Kuntida Kitidee, Caroline Leroux, Maxime Ratinier, Frédérick Arnaud, Sylvie Lecollinet, and et al. 2019. "Remodeling of the Actin Network Associated with the Non-Structural Protein 1 (NS1) of West Nile Virus and Formation of NS1-Containing Tunneling Nanotubes" Viruses 11, no. 10: 901. https://doi.org/10.3390/v11100901
APA StyleFurnon, W., Fender, P., Confort, M.-P., Desloire, S., Nangola, S., Kitidee, K., Leroux, C., Ratinier, M., Arnaud, F., Lecollinet, S., Boulanger, P., & Hong, S.-S. (2019). Remodeling of the Actin Network Associated with the Non-Structural Protein 1 (NS1) of West Nile Virus and Formation of NS1-Containing Tunneling Nanotubes. Viruses, 11(10), 901. https://doi.org/10.3390/v11100901







