Insight into the Lytic Functions of the Lactococcal Prophage TP712
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. General Molecular Techniques
2.3. Plasmids Construction
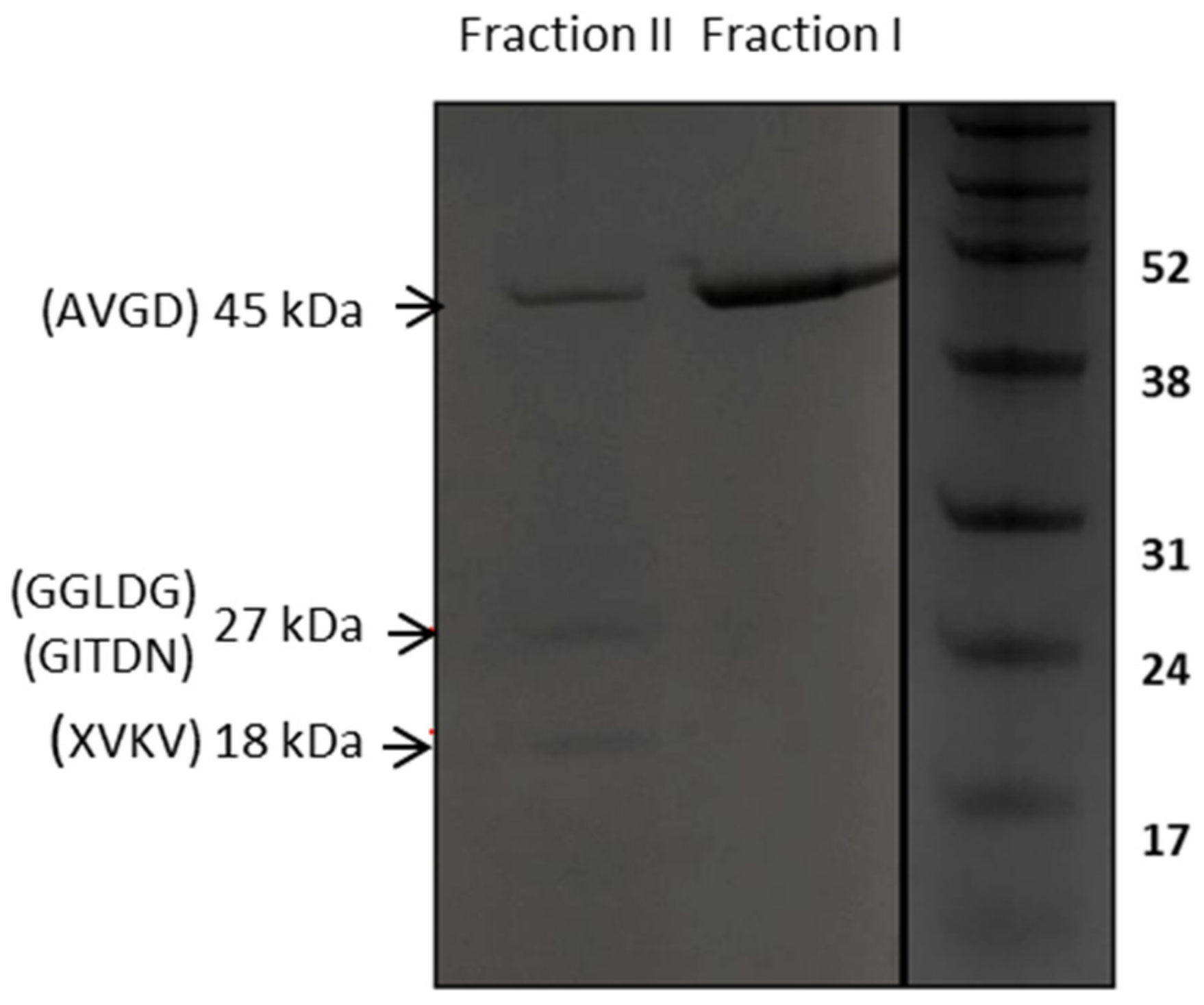
2.4. Affinity Purification of Recombinant LysTP712
2.5. N-Terminal Sequence Analysis
2.6. LysTP712 Activity Assays
2.7. Phage DNA Sequence Analysis
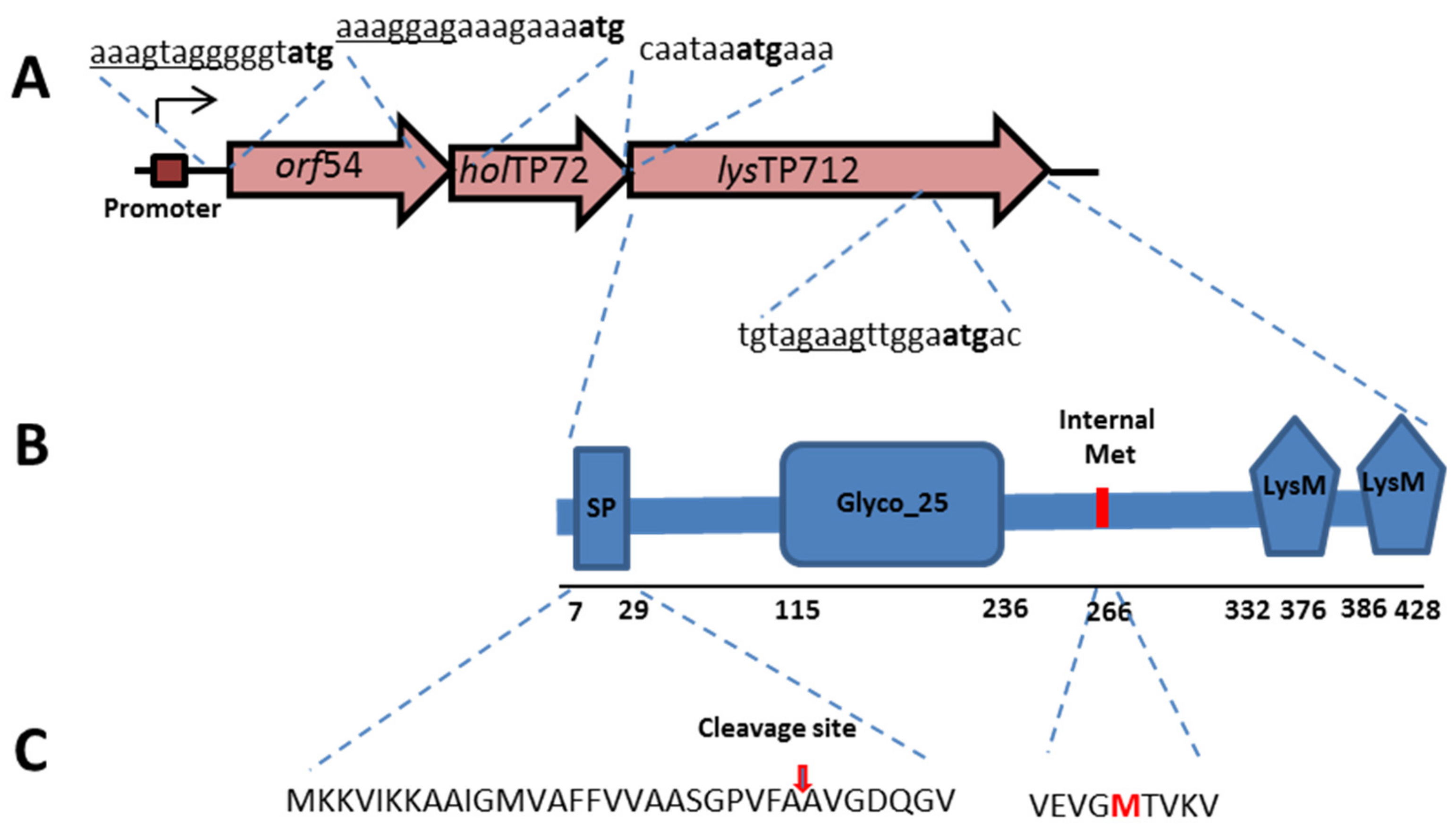
3. Results
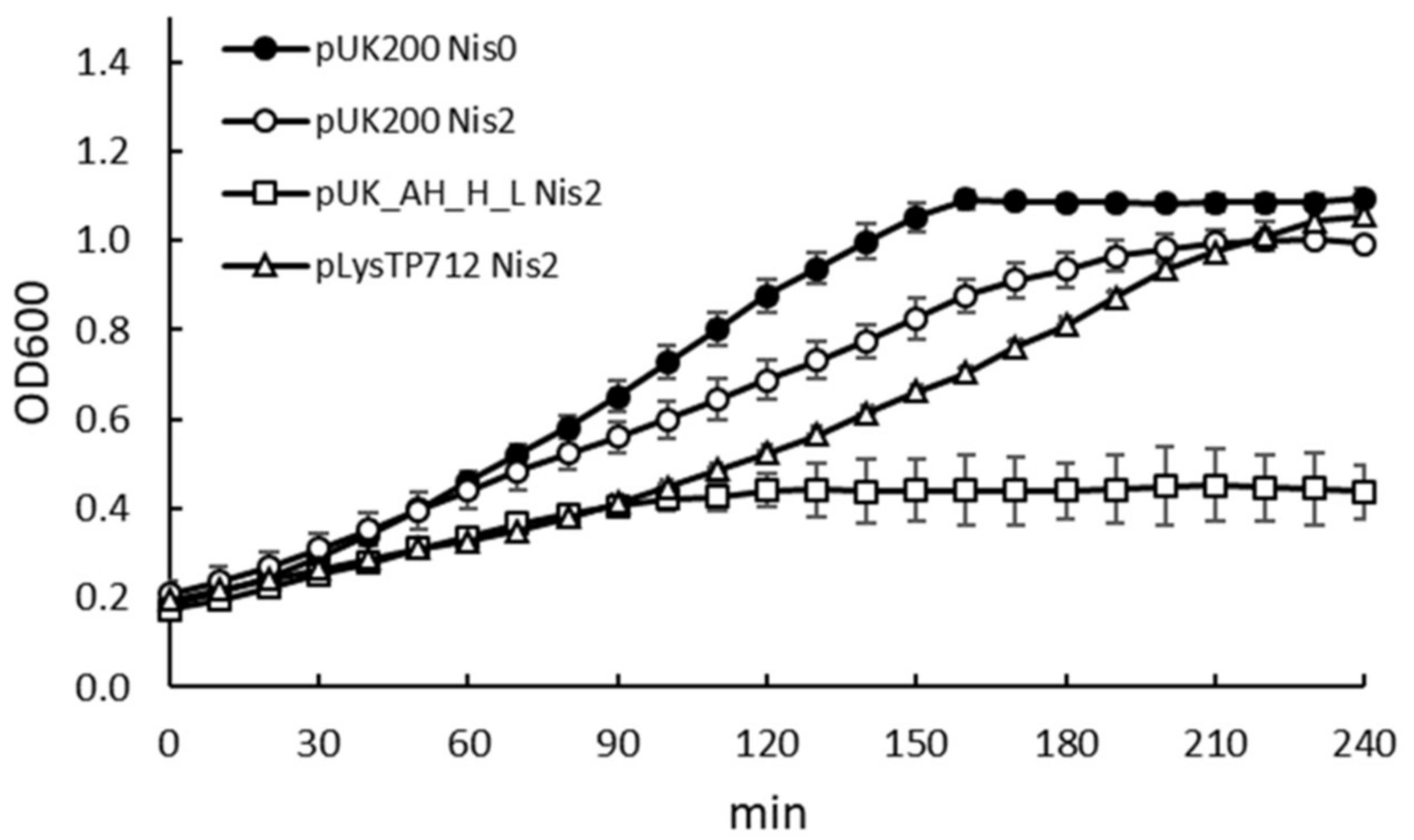
3.1. Expression of the TP712 Holin and Endolysin Genes Hampers Growth of L. lactis
3.2. LysTP712 Activity is Triggered by Membrane Depolarization
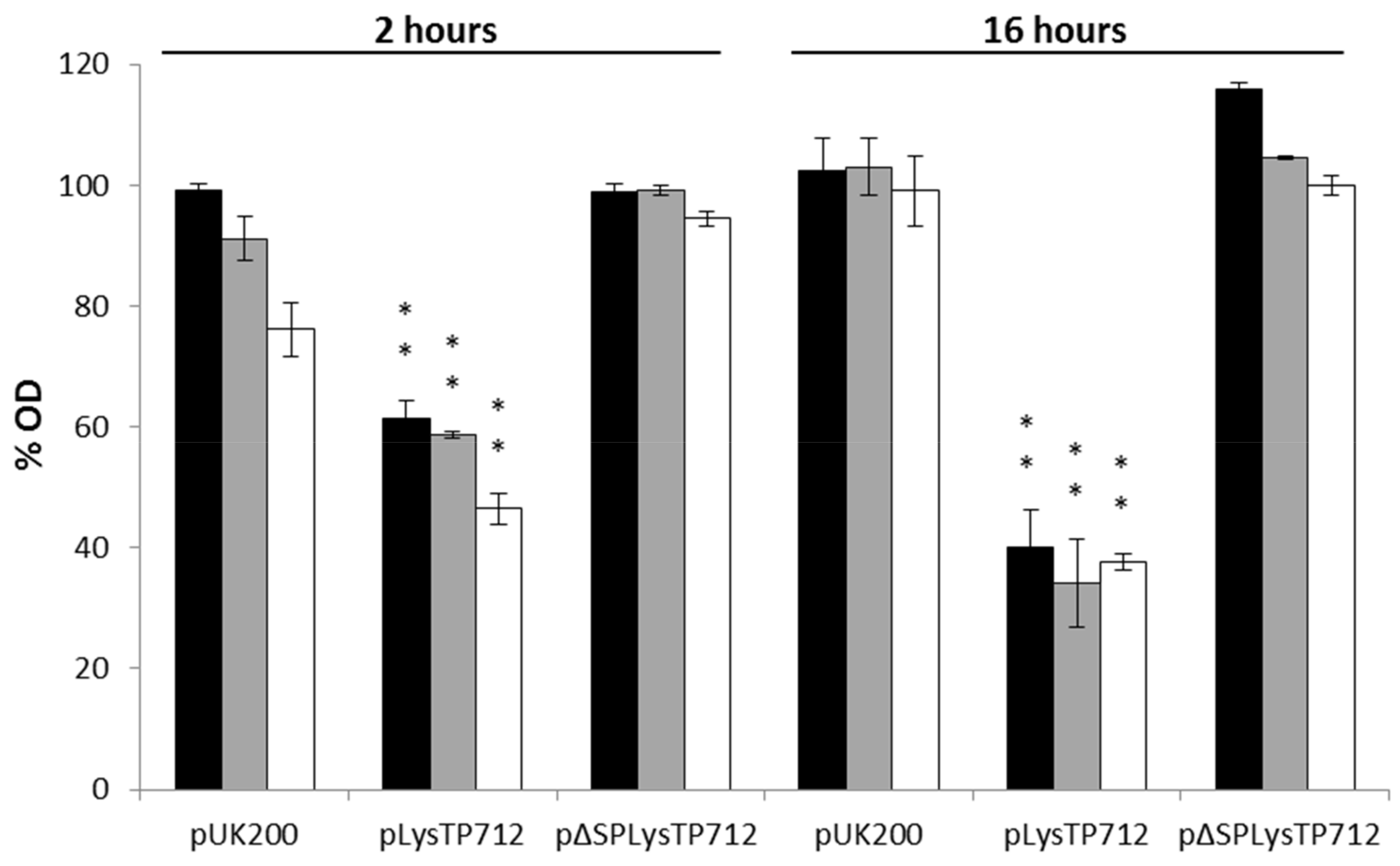
3.3. LysTP712 Is a Sec-Dependent Endolysin
3.4. LysTP712 may Require Additional Components for Activity
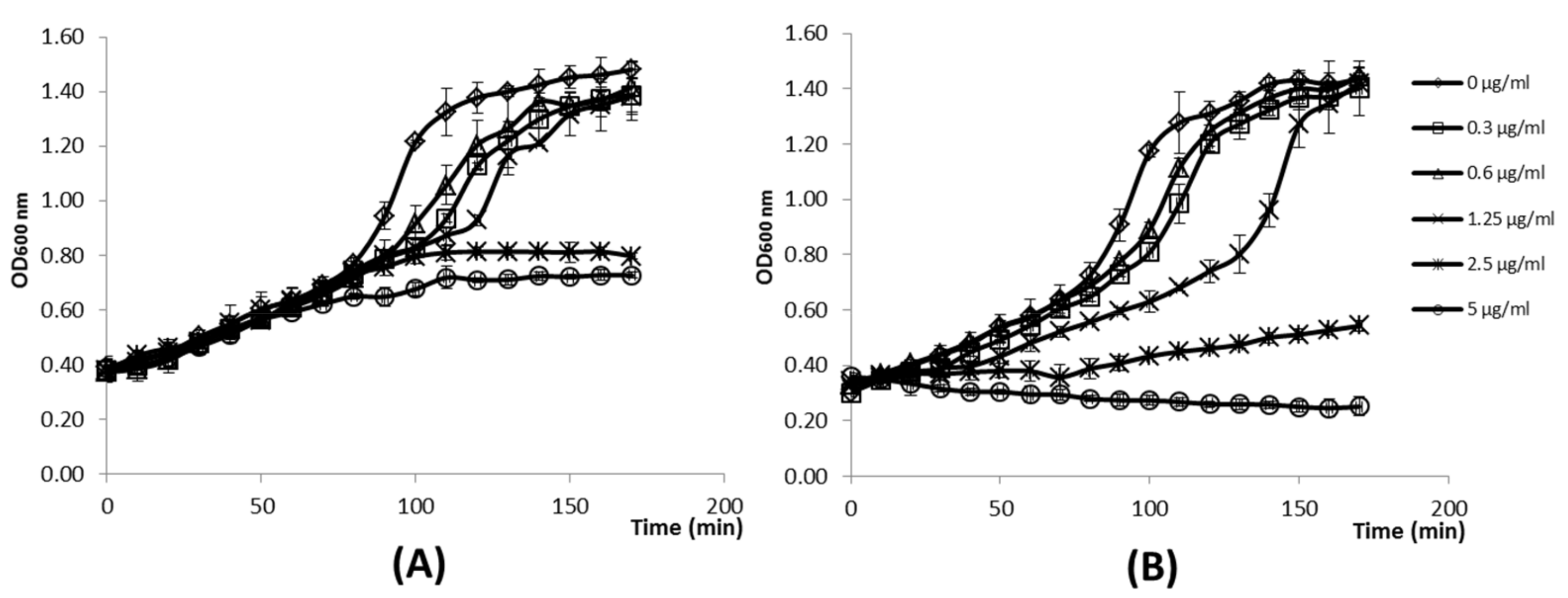
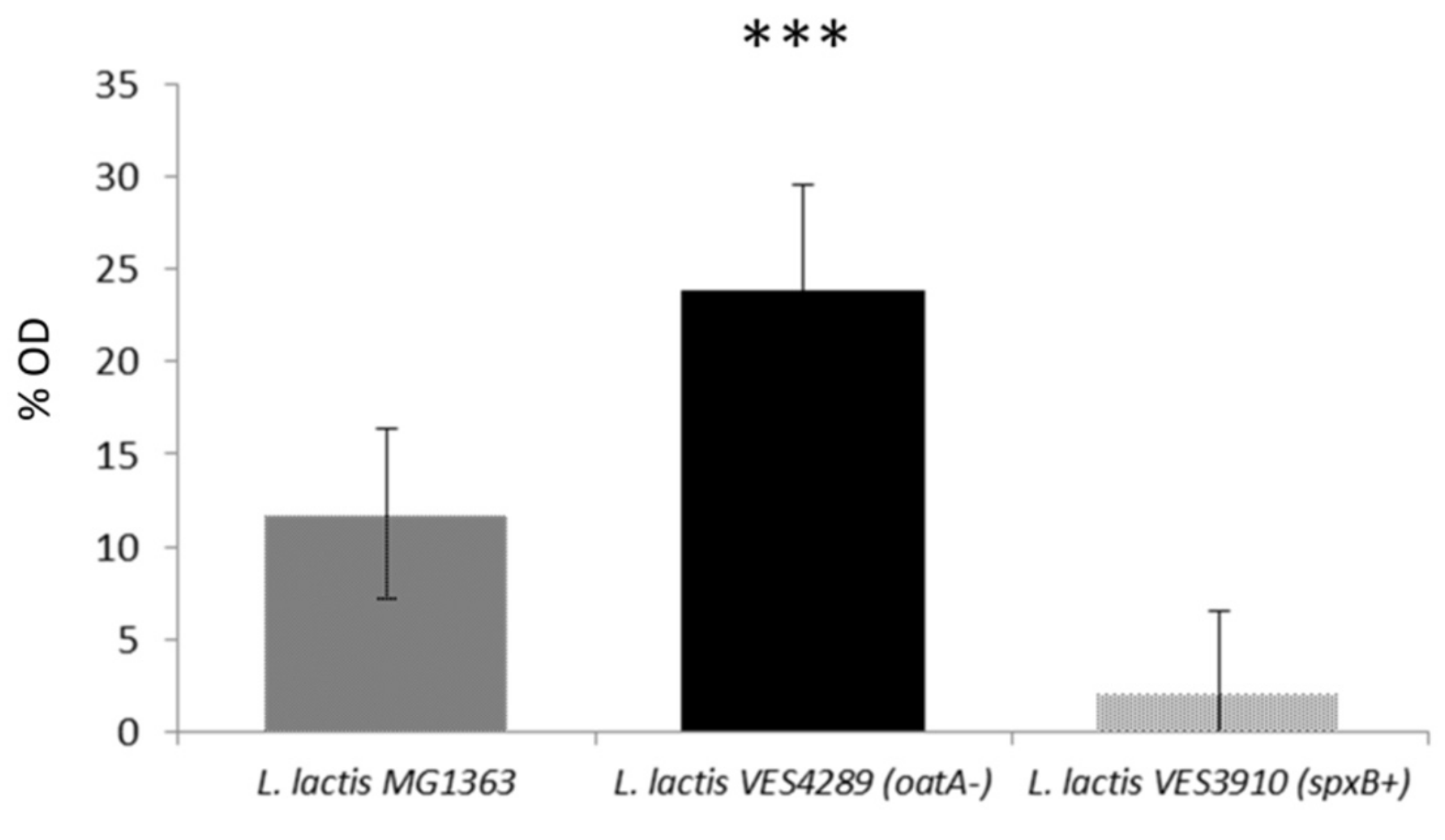
3.5. The Level of O-Acetylation of L. lactis Peptidoglycan Restricts the Inhibitory Activity of LysTP712
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Young, I.; Wang, I.; Roof, W.D. Phages will out: Strategies of host cell lysis. Trends Microbiol. 2000, 8, 120–128. [Google Scholar] [CrossRef]
- Fernandes, S.; São-José, C. Enzymes and Mechanisms Employed by Tailed Bacteriophages to Breach the Bacterial Cell Barriers. Viruses 2018, 10, 396. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Struck, D.K.; Deaton, J.; Wang, I.N.; Young, R. A signal-arrest-release sequence mediates export and control of the phage P1 endolysin. Proc. Natl. Acad. Sci. USA 2004, 101, 6415–6420. [Google Scholar] [CrossRef] [PubMed]
- Sao-Jose, C.; Parreira, R.; Vieira, G.; Santos, M.A. The N-Terminal Region of the Oenococcus oeni Bacteriophage fOg44 Lysin Behaves as a Bona Fide Signal Peptide in Escherichia coli and as a cis-Inhibitory Element, Preventing Lytic Activity on Oenococcal Cells. J. Bacteriol. 2000, 182, 5823–5831. [Google Scholar] [CrossRef] [PubMed]
- Kakikawa, M.; Yokoi, K.; Kimoto, H.; Nakano, M.; Kawasaki, K.; Taketo, A.; Kodaira, K. Molecular analysis of the lysis protein Lys encoded by Lactobacillus plantarum phage ϕg1e. Gene 2002, 299, 227–234. [Google Scholar] [CrossRef]
- Frias, M.J.; Melo-Cristino, J.; Ramirez, M. Export of the pneumococcal phage SV1 lysin requires choline-containing teichoic acids and is holin-independent. Mol. Microbiol. 2013, 87, 430–445. [Google Scholar] [CrossRef]
- Catalão, M.J.; Gil, F.; Moniz-Pereira, J.; Pimentel, M. The mycobacteriophage Ms6 encodes a chaperone-like protein involved in the endolysin delivery to the peptidoglycan. Mol. Microbiol. 2010, 77, 672–686. [Google Scholar] [CrossRef] [PubMed]
- Catalao, M.J.; Gil, F.; Moniz-Pereira, J.; Pimentel, M. The Endolysin-Binding Domain Encompasses the N-Terminal Region of the Mycobacteriophage Ms6 Gp1 Chaperone. J. Bacteriol. 2011, 193, 5002–5006. [Google Scholar] [CrossRef]
- Fernandes, S.; São-José, C. More than a hole: The holin lethal function may be required to fully sensitize bacteria to the lytic action of canonical endolysins. Mol. Microbiol. 2016, 102, 92–106. [Google Scholar] [CrossRef]
- Nascimento, J.G.; Guerreiro-Pereira, M.C.; Costa, S.F.; Sao-Jose, C.; Santos, M.A. Nisin-Triggered Activity of Lys44, the Secreted Endolysin from Oenococcus oeni Phage fOg44. J. Bacteriol. 2008, 190, 457–461. [Google Scholar] [CrossRef]
- García, P.; Martínez, B.; Rodríguez, L.; Rodríguez, A. Synergy between the phage endolysin LysH5 and nisin to kill Staphylococcus aureus in pasteurized milk. Int. J. Food Microbiol. 2010, 141, 151–155. [Google Scholar] [CrossRef] [PubMed]
- Broendum, S.S.; Buckle, A.M.; McGowan, S. Catalytic diversity and cell wall binding repeats in the phage-encoded endolysins. Mol. Microbiol. 2018, 110, 879–896. [Google Scholar] [CrossRef] [PubMed]
- Schmelcher, M.; Donovan, D.M.; Loessner, M.J. Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 2012, 7, 1147–1171. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, H.; Melo, L.D.R.; Santos, S.B.; Nobrega, F.L.; Ferreira, E.C.; Cerca, N.; Azeredo, J.; Kluskens, L.D. Molecular Aspects and Comparative Genomics of Bacteriophage Endolysins. J. Virol. 2013, 87, 4558–4570. [Google Scholar] [CrossRef] [PubMed]
- Mesnage, S.; Dellarole, M.; Baxter, N.J.; Rouget, J.B.; Dimitrov, J.D.; Wang, N.; Fujimoto, Y.; Hounslow, A.M.; Lacroix-Desmazes, S.; Fukase, K.; et al. Molecular basis for bacterial peptidoglycan recognition by LysM domains. Nat. Commun. 2014, 5, 4269. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.; Schuch, R.; Chahales, P.; Zhu, S.; Fischetti, V.A. PlyC: A multimeric bacteriophage lysin. Proc. Natl. Acad. Sci. USA 2006, 103, 10765–10770. [Google Scholar] [CrossRef] [PubMed]
- Proença, D.; Velours, C.; Leandro, C.; García, M.; Pimentel, M.; São-José, C. A two-component, multimeric endolysin encoded by a single gene. Mol. Microbiol. 2015, 95, 739–753. [Google Scholar] [CrossRef]
- Dunne, M.; Leicht, S.; Krichel, B.; Mertens, H.D.T.; Thompson, A.; Krijgsveld, J.; Svergun, D.I.; Gomez-Torres, N.; Garde, S.; Uetrecht, C.; et al. Crystal Structure of the CTP1L Endolysin Reveals How Its Activity Is Regulated by a Secondary Translation Product. J. Biol. Chem. 2016, 291, 4882–4893. [Google Scholar] [CrossRef]
- Ventura, M.; Zomer, A.; Canchaya, C.; O’Connell-Motherway, M.; Kuipers, O.; Turroni, F.; Ribbera, A.; Foroni, E.; Buist, G.; Wegmann, U.; et al. Comparative Analyses of Prophage-Like Elements Present in Two Lactococcus lactis Strains. Appl. Environ. Microbiol. 2007, 73, 7771–7780. [Google Scholar] [CrossRef]
- Wegmann, U.; Overweg, K.; Jeanson, S.; Gasson, M.; Shearman, C. Molecular characterization and structural instability of the industrially important composite metabolic plasmid pLP712. Microbiology 2012, 158, 2936–2945. [Google Scholar] [CrossRef]
- Roces, C.; Wegmann, U.; Campelo, A.B.; García, P.; Rodriguez, A.; Martinez, B. Lack of the host membrane protease FtsH hinders release of the Lactococcus lactis bacteriophage TP712. J. Gen. Virol. 2013, 94, 2814–2818. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Roces, C.; Campelo, A.B.; Escobedo, S.; Wegmann, U.; García, P.; Rodríguez, A.; Martínez, B. Reduced Binding of the Endolysin LysTP712 to Lactococcus lactis ΔftsH Contributes to Phage Resistance. Front. Microbiol. 2016, 7, 138. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Gasson, M.J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 1983, 154, 1–9. [Google Scholar] [PubMed]
- Kuipers, O.P.; de Ruyter, P.G.G.A.; Kleerebezem, M.; de Vos, W.M. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 1998, 64, 15–21. [Google Scholar] [CrossRef]
- Veiga, P.; Bulbarela-Sampieri, C.; Furlan, S.; Maisons, A.; Chapot-Chartier, M.P.; Erkelenz, M.; Mervelet, P.; Noirot, P.; Frees, D.; Kuipers, O.P.; et al. SpxB Regulates O-Acetylation-dependent Resistance of Lactococcus lactis Peptidoglycan to Hydrolysis. J. Biol. Chem. 2007, 282, 19342–19354. [Google Scholar] [CrossRef]
- Wegmann, U.; Klein, J.R.; Drumm, I.; Kuipers, O.P.; Henrich, B. Introduction of peptidase genes from Lactobacillus delbrueckii subsp. lactis into Lactococcus lactis and controlled expression. Appl. Environ. Microbiol. 1999, 65, 4729–4733. [Google Scholar] [PubMed]
- Laemmli, U.K.; Mölbert, E.; Showe, M.; Kellenberger, E. Form-determining function of the genes required for the assembly of the head of bacteriophage T4. J. Mol. Biol. 1970, 49, 99–113. [Google Scholar] [CrossRef]
- Lepeuple, A.S.; van Gemert, E.; Chapot-Chartier, M.P. Analysis of the bacteriolytic enzymes of the autolytic lactococcus lactis subsp. cremoris strain AM2 by renaturing polyacrylamide gel electrophoresis: Identification of a prophage-encoded enzyme. Appl. Environ. Microbiol. 1998, 64, 4142–4148. [Google Scholar]
- Expasy Bioinformatics Resource Portal. Available online: http://web.expasy.org/protparam (accessed on 12 June 2019).
- Center of Biological sequence and analysis (CBS). Available online: http://www.cbs.dtu.dk/services/SignalP (accessed on 3 February 2019).
- Basic Local Alignment Search Tool (BLAST). Available online: http://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 7 August 2019).
- European Bioinformatics Institute (EMBL-EBI). Available online: https://pfam.xfam.org (accessed on 2 November 2018).
- Softberry BPROM. Available online: http://www.softberry.com/berry.phtml?topic=bprom (accessed on 23 April 2019).
- Wiedemann, I.; Breukink, E.; van Kraaij, C.; Kuipers, O.P.; Bierbaum, G.; de Kruijff, B.; Sahl, H.G. Specific Binding of Nisin to the Peptidoglycan Precursor Lipid II Combines Pore Formation and Inhibition of Cell Wall Biosynthesis for Potent Antibiotic Activity. J. Biol. Chem. 2001, 276, 1772–1779. [Google Scholar] [CrossRef]
- Breukink, E. Use of the Cell Wall Precursor Lipid II by a Pore-Forming Peptide Antibiotic. Science 1999, 286, 2361–2364. [Google Scholar] [CrossRef]
- Neef, J.; Koedijk, D.G.A.M.; Bosma, T.; van Dijl, J.M.; Buist, G. Efficient production of secreted staphylococcal antigens in a non-lysing and proteolytically reduced Lactococcus lactis strain. Appl. Microbiol. Biotechnol. 2014, 98, 10131–10141. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Carbohydrate-Active enZYmes Database (CAZY). Available online: www.cazy.org (accessed on 20 May 2019).
- Fischetti, V.A. Bacteriophage lytic enzymes: Novel anti-infectives. Trends Microbiol. 2005, 13, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Smit, G.; Smit, B.; Engels, W. Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol. Rev. 2005, 29, 591–610. [Google Scholar] [CrossRef] [PubMed]
- De Ruyter, P.G.; Kuipers, O.P.; Meijer, W.C.; de Vos, W.M. Food-grade controlled lysis of Lactococcus lactis for accelerated cheese ripening. Nat. Biotechnol. 1997, 15, 976–979. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Steen, A.; van Schalkwijk, S.; Buist, G.; Twigt, M.; Szeliga, M.; Meijer, W.; Kuipers, O.P.; Kok, J.; Hugenholtz, J. Lytr, a phage-derived amidase is most effective in induced lysis of Lactococcus lactis compared with other lactococcal amidases and glucosaminidases. Int. Dairy J. 2007, 17, 926–936. [Google Scholar] [CrossRef]
- Van Tilburg, A.Y.; Cao, H.; van der Meulen, S.B.; Solopova, A.; Kuipers, O.P. Metabolic engineering and synthetic biology employing Lactococcus lactis and Bacillus subtilis cell factories. Curr. Opin. Biotechnol. 2019, 59, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Labrie, S.; Vukov, N.; Loessner, M.J.; Moineau, S. Distribution and composition of the lysis cassette of Lactococcus lactis phages and functional analysis of bacteriophage ul36 holin. FEMS Microbiol. Lett. 2004, 233, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Abaev, I.; Foster-Frey, J.; Korobova, O.; Shishkova, N.; Kiseleva, N.; Kopylov, P.; Pryamchuk, S.; Schmelcher, M.; Becker, S.C.; Donovan, D.M. Staphylococcal phage 2638A endolysin is lytic for Staphylococcus aureus and harbors an inter-lytic-domain secondary translational start site. Appl. Microbiol. Biotechnol. 2013, 97, 3449–3456. [Google Scholar] [CrossRef]
- Martínez, B.; Zomer, A.L.; Rodríguez, A.; Kok, J.; Kuipers, O.P. Cell envelope stress induced by the bacteriocin Lcn972 is sensed by the lactococcal two-component system CesSR. Mol. Microbiol. 2007, 64, 473–486. [Google Scholar] [CrossRef]
- Chapot-Chartier, M.P.; Kulakauskas, S. Cell wall structure and function in lactic acid bacteria. Microb. Cell Fact. 2014, 13. [Google Scholar] [CrossRef]
- Pfeffer, J.M.; Strating, H.; Weadge, J.T.; Clarke, A.J. Peptidoglycan O Acetylation and Autolysin Profile of Enterococcus faecalis in the Viable but Nonculturable State. J. Bacteriol. 2006, 188, 902–908. [Google Scholar] [CrossRef] [PubMed]
- Bera, A.; Herbert, S.; Jakob, A.; Vollmer, W.; Götz, F. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol. Microbiol. 2005, 55, 778–787. [Google Scholar] [CrossRef] [PubMed]
- Hebert, L.; Courtin, P.; Torelli, R.; Sanguinetti, M.; Chapot-Chartier, M.P.; Auffray, Y.; Benachour, A. Enterococcus faecalis Constitutes an Unusual Bacterial Model in Lysozyme Resistance. Infect. Immun. 2007, 75, 5390–5398. [Google Scholar] [CrossRef] [PubMed]

| Strain | Description | Reference |
|---|---|---|
| E. coli DH10B | E. coli cloning host | Invitrogen |
| E. coli BL21 (pLys) | Gene expression host | Novagen |
| L. lactis MG1363 | Plasmid-free derivative of NCDO712 | [24] |
| L. lactis NZ9000 | MG1363 pepN::nisRK. Host for nisin inducible gene expression | [25] |
| L. lactis UKLc10 TP712 | TP712 lysogen and host for nisin inducible expression | [21] |
| L. lactis VES3910 | MG1363 carrying pVES3910 (spxB+) | [26] |
| L. lactis VES4289 | MG1363 lacking a functional oatA (oatA-) | [26] |
| Plasmid | Description * | Reference |
|---|---|---|
| pUK200 | Nisin inducible L. lactis expression vector. CmR. | [27] |
| pET21a | IPTG inducible E. coli expression vector. AmpR. | EMD Biosciences |
| pUK_AH_H_L | holTP712-lysTP712 under the nisin inducible promoter in pUK200. | This work |
| pLysTP712 | lysTP712-His6 under the nisin inducible promoter in pUK200. | This work |
| pΔSPLysTP712 | leaderless LysTP712 under the nisin inducible promoter in pUK200. | This work |
| pETLysTP712 | pET21a::ΔSPlysTP712-His6. | This work |
| pETLysTP712CBD | pET21a::lysTP712CBD-His6. | This work |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Escobedo, S.; Campelo, A.B.; Wegmann, U.; García, P.; Rodríguez, A.; Martínez, B. Insight into the Lytic Functions of the Lactococcal Prophage TP712. Viruses 2019, 11, 881. https://doi.org/10.3390/v11100881
Escobedo S, Campelo AB, Wegmann U, García P, Rodríguez A, Martínez B. Insight into the Lytic Functions of the Lactococcal Prophage TP712. Viruses. 2019; 11(10):881. https://doi.org/10.3390/v11100881
Chicago/Turabian StyleEscobedo, Susana, Ana Belén Campelo, Udo Wegmann, Pilar García, Ana Rodríguez, and Beatriz Martínez. 2019. "Insight into the Lytic Functions of the Lactococcal Prophage TP712" Viruses 11, no. 10: 881. https://doi.org/10.3390/v11100881
APA StyleEscobedo, S., Campelo, A. B., Wegmann, U., García, P., Rodríguez, A., & Martínez, B. (2019). Insight into the Lytic Functions of the Lactococcal Prophage TP712. Viruses, 11(10), 881. https://doi.org/10.3390/v11100881






