Infectious Bronchitis Virus Nonstructural Protein 4 Alone Induces Membrane Pairing
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells, Viruses, and Plasmids
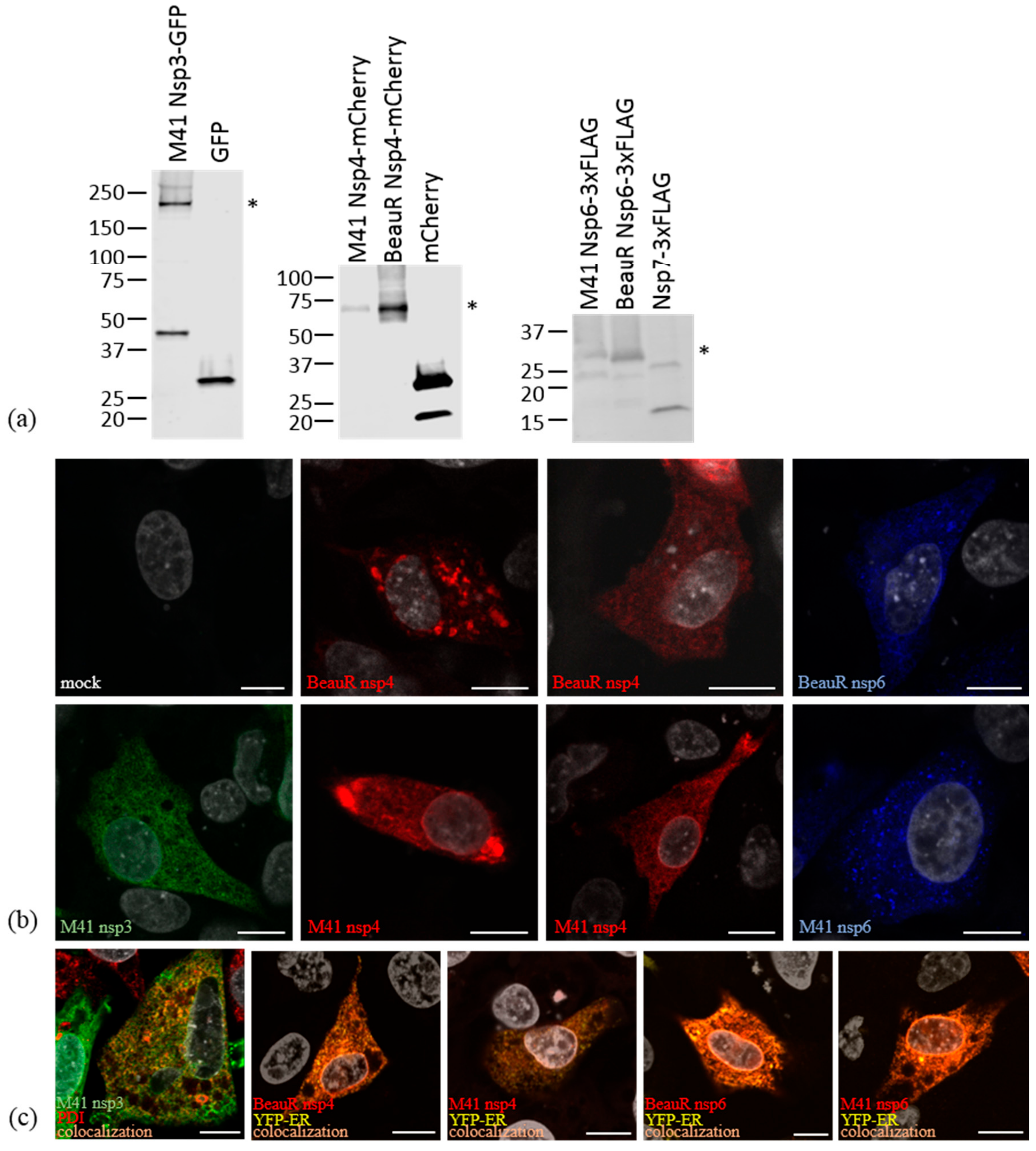
2.2. SDS-PAGE and Western Blotting
2.3. Immunofluorescence Labelling
2.4. Transmission Electron Microscopy
2.5. Electron Tomography
3. Results
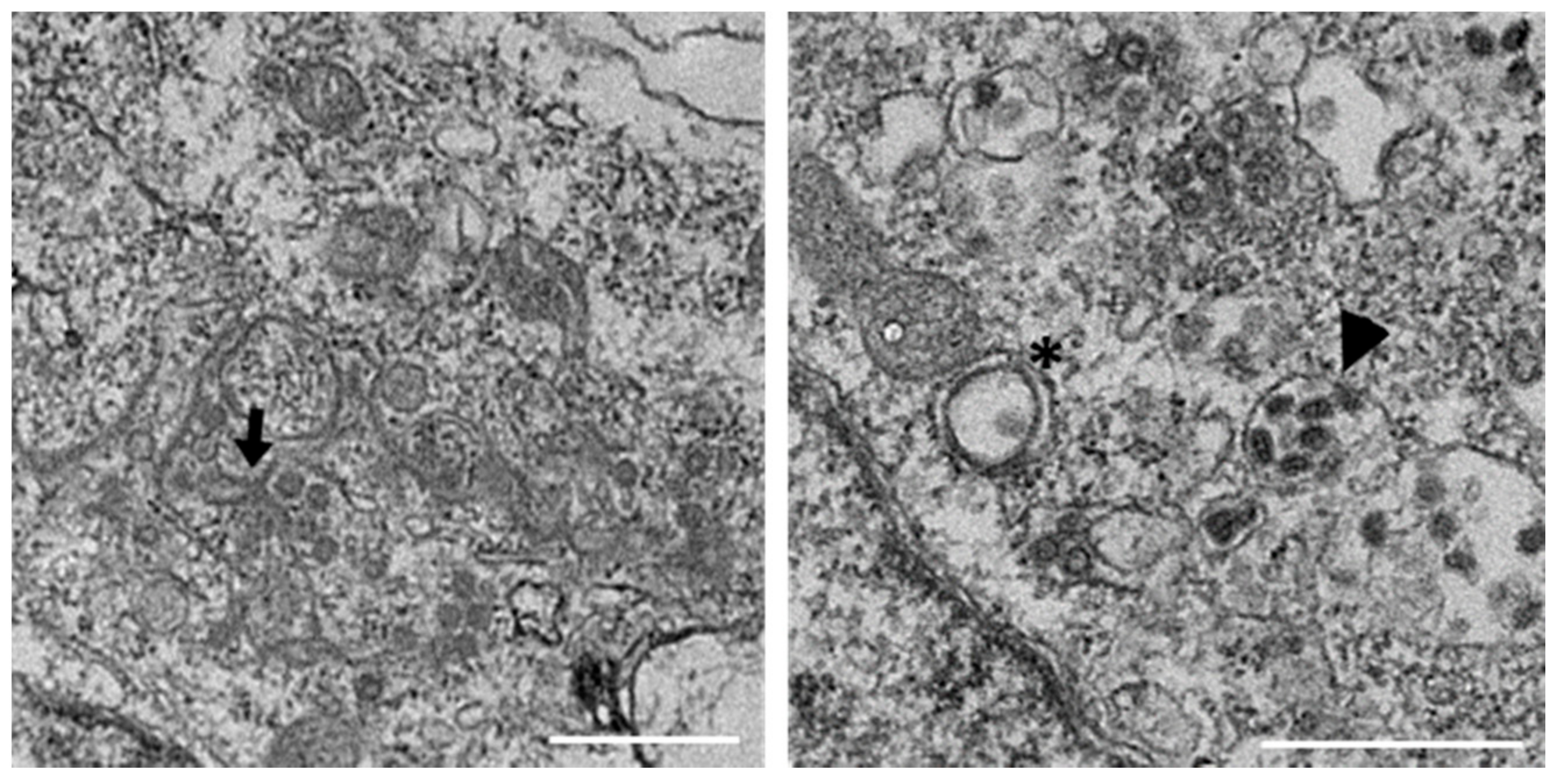
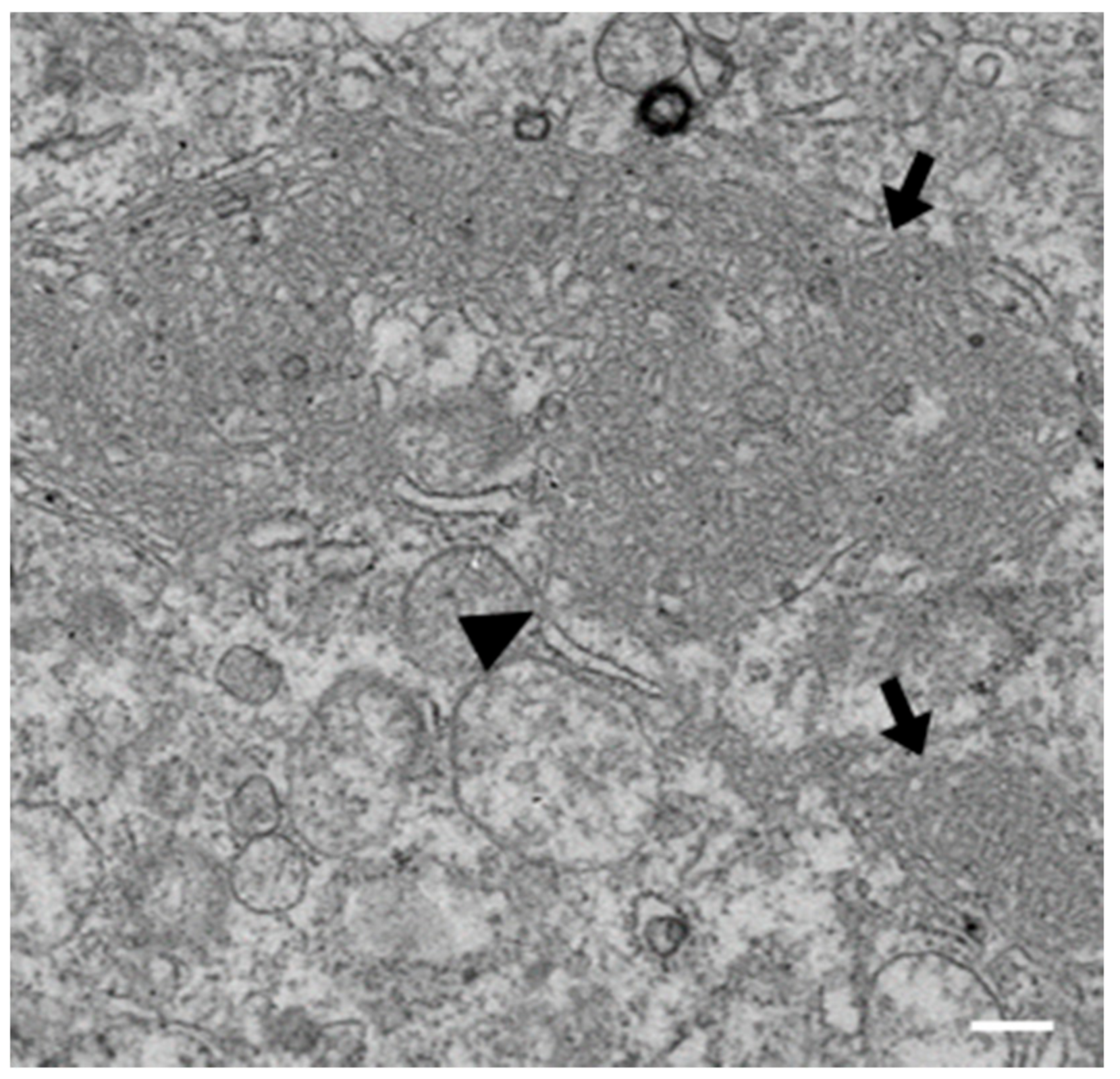
3.1. IBV Induces Typical Membrane Rearrangements in DF1 Cells
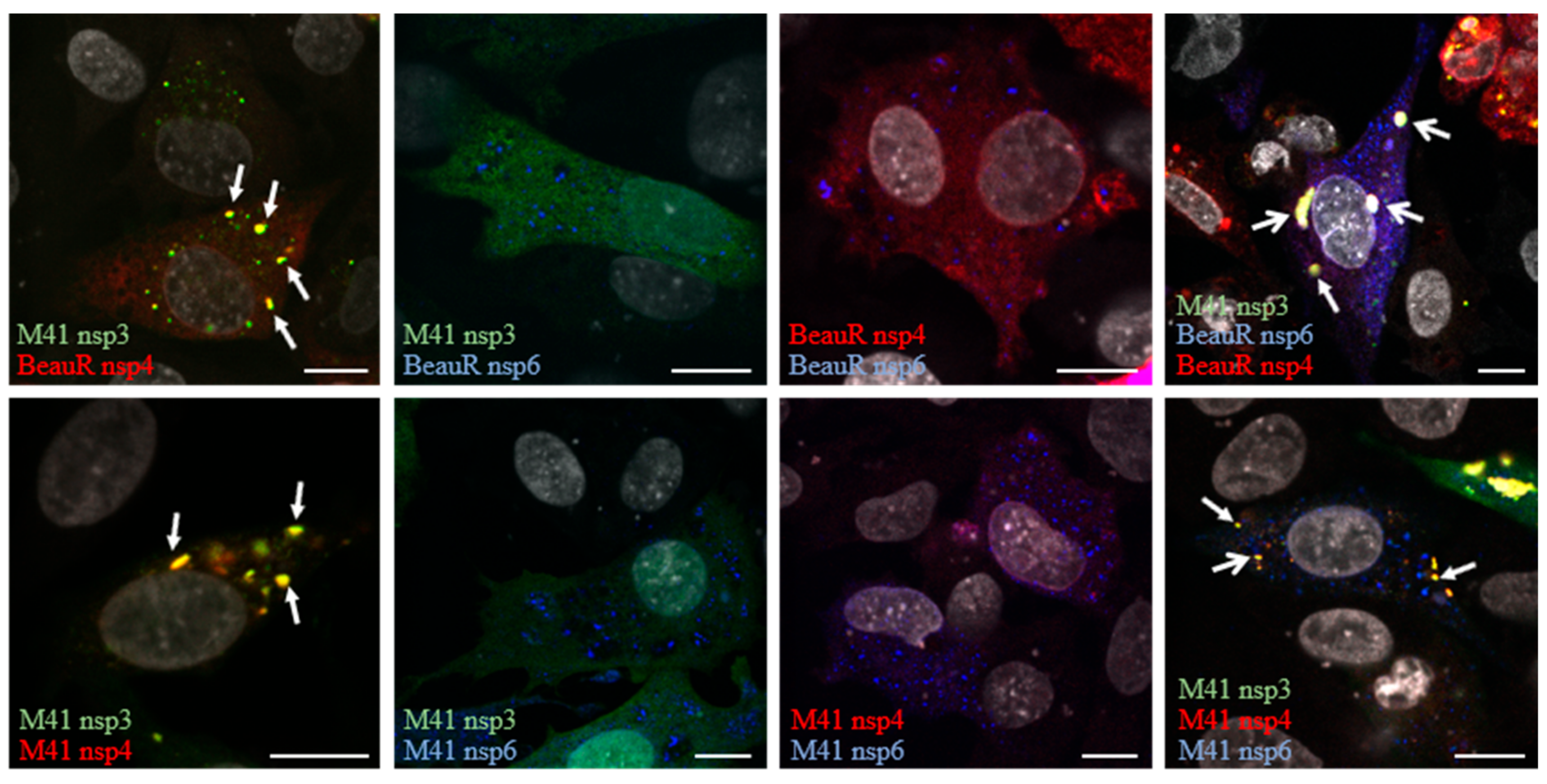
3.2. Non-Structural Proteins 3, 4, and 6 Relocalize upon Co-Expression
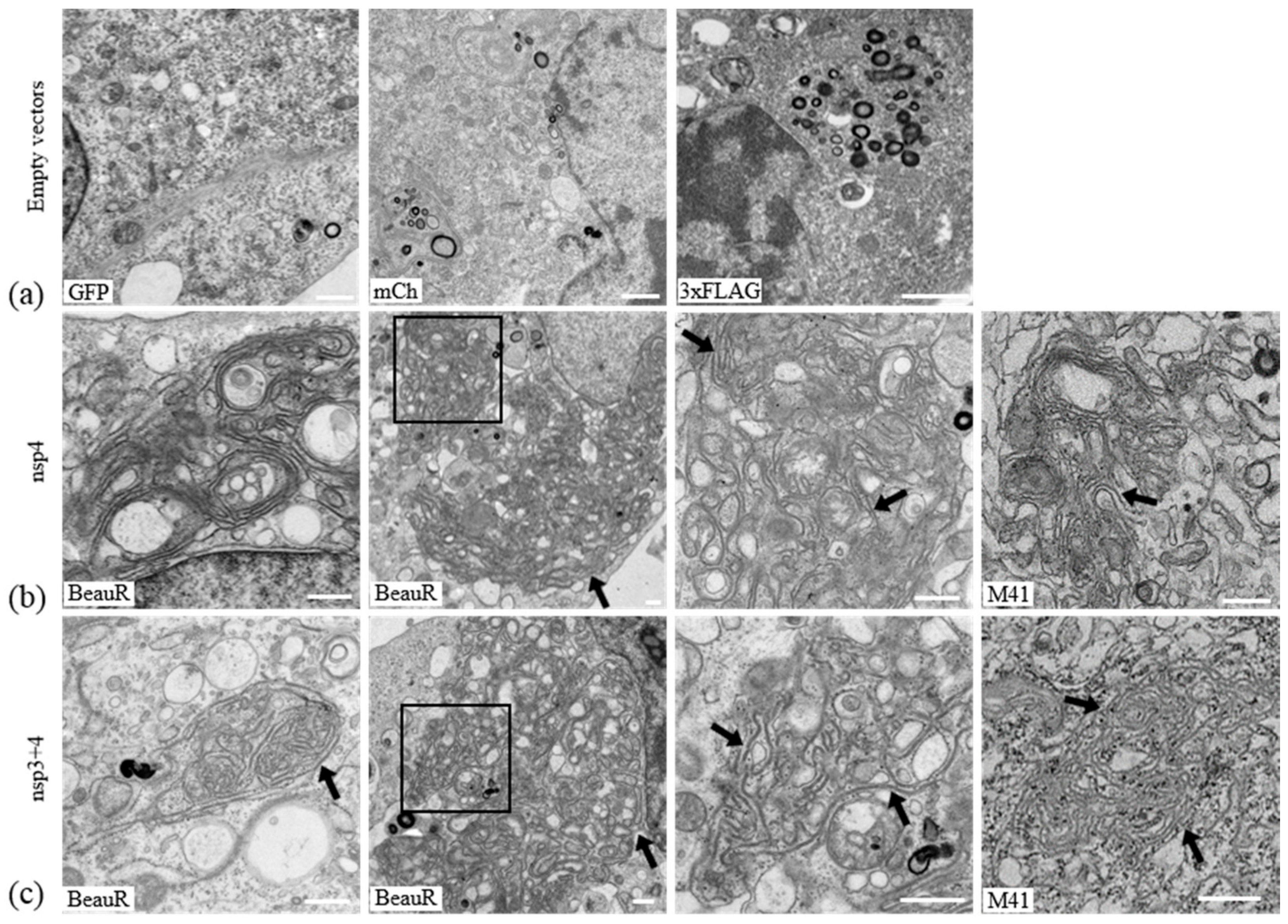
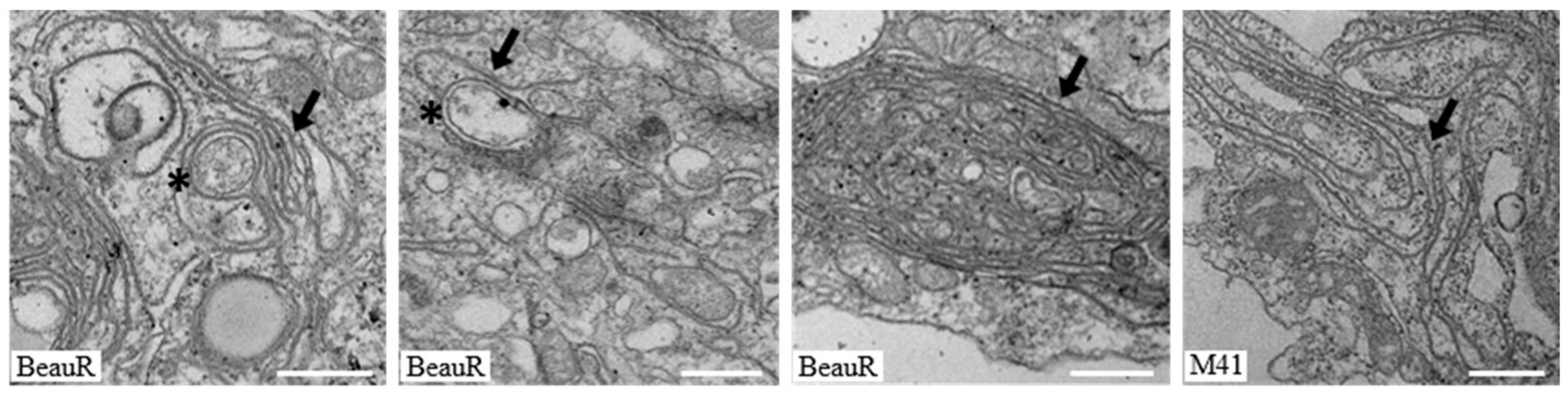
3.3. IBV nsp4 Alone Is Necessary and Sufficient to Induce Membrane Pairing
3.4. Nsp6 Induces the Formation of Piled, Disordered Membranes
3.5. Nsps 3, 4, and 6 Are Not Able to Recapitulate the IBV Replication Organelle
3.6. IBV nsp3 and 4 Do Not Induce DMVs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Miller, S.; Krijnse-Locker, J. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 2008, 6, 363. [Google Scholar] [CrossRef] [PubMed]
- Netherton, C.L.; Wileman, T. Virus factories, double membrane vesicles and viroplasm generated in animal cells. Curr. Opin. Virol. 2011, 1, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Boon, J.A.D.; Ahlquist, P. Organelle-Like Membrane Compartmentalization of Positive-Strand RNA Virus Replication Factories. Annu. Rev. Microbiol. 2010, 64, 241–256. [Google Scholar] [CrossRef] [PubMed]
- Neufeldt, C.J.; Joyce, M.A.; Van Buuren, N.; Levin, A.; Kirkegaard, K.; Gale, M., Jr.; Tyrrell, D.L.J.; Wozniak, R.W. The Hepatitis C Virus-Induced Membranous Web and Associated Nuclear Transport Machinery Limit Access of Pattern Recognition Receptors to Viral Replication Sites. PLoS Pathog. 2016, 12, e1005428. [Google Scholar] [CrossRef] [PubMed]
- Harak, C.; Lohmann, V. Ultrastructure of the replication sites of positive-strand RNA viruses. Virology 2015, 479–480, 418–433. [Google Scholar] [CrossRef] [PubMed]
- Paul, D.; Bartenschlager, R. Architecture and biogenesis of plus-strand RNA virus replication factories. World J. Virol. 2013, 2, 32–48. [Google Scholar] [CrossRef] [PubMed]
- Bienz, K.; Egger, D.; Pfister, T.; Troxler, M. Structural and functional characterization of the poliovirus replication complex. J. Virol. 1992, 66, 2740–2747. [Google Scholar] [PubMed]
- Belov, G.A.; Nair, V.; Hansen, B.T.; Hoyt, F.H.; Fischer, E.R.; Ehrenfeld, E. Complex Dynamic Development of Poliovirus Membranous Replication Complexes. J. Virol. 2012, 86, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Gosert, R.; Egger, D.; Lohmann, V.; Bartenschlager, R.; Blum, H.E.; Bienz, K.; Moradpour, D. Identification of the Hepatitis C Virus RNA Replication Complex in Huh-7 Cells Harboring Subgenomic Replicons. J. Virol. 2003, 77, 5487–5492. [Google Scholar] [CrossRef] [PubMed]
- Ferraris, P.; Beaumont, E.; Uzbekov, R.; Brand, D.; Gaillard, J.; Blanchard, E.; Roingeard, P. Sequential biogenesis of host cell membrane rearrangements induced by hepatitis C virus infection. Cell. Mol. Life Sci. 2013, 70, 1297–1306. [Google Scholar] [CrossRef] [PubMed]
- Doerflinger, S.Y.; Cortese, M.; Romero-Brey, I.; Menne, Z.; Tubiana, T.; Schenk, C.; White, P.A.; Bartenschlager, R.; Bressanelli, S.; Hansman, G.S.; et al. Membrane alterations induced by nonstructural proteins of human norovirus. PLoS Pathog. 2017, 13, e1006705. [Google Scholar] [CrossRef] [PubMed]
- Ávila-Pérez, G.; Rejas, M.T.; Rodríguez, D. Ultrastructural characterization of membranous torovirus replication factories. Cell. Microbiol. 2016, 18, 1691–1708. [Google Scholar] [CrossRef] [PubMed]
- Kujala, P.; Ikäheimonen, A.; Ehsani, N.; Vihinen, H.; Auvinen, P.; Kääriäinen, L. Biogenesis of the Semliki Forest Virus RNA Replication Complex. J. Virol. 2001, 75, 3873–3884. [Google Scholar] [CrossRef] [PubMed]
- Welsch, S.; Miller, S.; Romero-Brey, I.; Merz, A.; Bleck, C.K.E.; Walther, P.; Fuller, S.D.; Antony, C.; Krijnse-Locker, J.; Bartenschlager, R. Composition and Three-Dimensional Architecture of the Dengue Virus Replication and Assembly Sites. Cell Host Microbe 2009, 5, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, L.K.; Hoenen, A.; Morgan, G.; Mackenzie, J.M. The Endoplasmic Reticulum Provides the Membrane Platform for Biogenesis of the Flavivirus Replication Complex. J. Virol. 2010, 84, 10438–10447. [Google Scholar] [CrossRef] [PubMed]
- Westaway, E.G.; Mackenzie, J.M.; Kenney, M.T.; Jones, M.K.; Khromykh, A.A. Ultrastructure of Kunjin virus-infected cells: Colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 1997, 71, 6650–6661. [Google Scholar] [PubMed]
- Cortese, M.; Goellner, S.; Acosta, E.G.; Neufeldt, C.J.; Oleksiuk, O.; Lampe, M.; Haselmann, U.; Funaya, C.; Schieber, N.; Ronchi, P.; et al. Ultrastructural Characterization of Zika Virus Replication Factories. Cell Rep. 2017, 18, 2113–2123. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, M.; Chen, J.; Janda, M.; Sullivan, M.; den Boon, J.; Ahlquist, P. A Positive-Strand RNA Virus Replication Complex Parallels Form and Function of Retrovirus Capsids. Mol. Cell 2002, 9, 505–514. [Google Scholar] [CrossRef]
- Snijder, E.J.; van der Meer, Y.; Zevenhoven-Dobbe, J.; Onderwater, J.J.M.; van der Meulen, J.; Koerten, H.K.; Mommaas, A.M. Ultrastructure and Origin of Membrane Vesicles Associated with the Severe Acute Respiratory Syndrome Coronavirus Replication Complex. J. Virol. 2006, 80, 5927–5940. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, C.S.; Tatti, K.M.; Ksiazek, T.G.; Rollin, P.E.; Comer, J.A.; Lee, W.W.; Rota, P.A.; Bankamp, B.; Bellini, W.J.; Zaki, S.R. Ultrastructural Characterization of SARS Coronavirus. Emerg. Infect. Dis. 2004, 10, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Gosert, R.; Kanjanahaluethai, A.; Egger, D.; Bienz, K.; Baker, S.C. RNA Replication of Mouse Hepatitis Virus Takes Place at Double-Membrane Vesicles. J. Virol. 2002, 76, 3697–3708. [Google Scholar] [CrossRef] [PubMed]
- Ulasli, M.; Verheije, M.H.; de Haan, C.A.; Reggiori, F. Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell. Microbiol. 2010, 12, 844–861. [Google Scholar] [CrossRef] [PubMed]
- De Wilde, A.H.; Raj, V.S.; Oudshoorn, D.; Bestebroer, T.M.; van Nieuwkoop, S.; Limpens, R.W.A.L.; Posthuma, C.C.; van der Meer, Y.; Bárcena, M.; Haagmans, B.L.; et al. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-α treatment. J. Gen. Virol. 2013, 94, 1749–1760. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Cong, Y.; Veenendaal, T.; Klumperman, J.; Shi, D.; Mari, M.; Reggiori, F. Ultrastructural Characterization of Membrane Rearrangements Induced by Porcine Epidemic Diarrhea Virus Infection. Viruses 2017, 9, 251. [Google Scholar] [CrossRef] [PubMed]
- Maier, H.J.; Hawes, P.C.; Cottam, E.M.; Mantell, J.; Verkade, P.; Monaghan, P.; Wileman, T.; Britton, P. Infectious Bronchitis Virus Generates Spherules from Zippered Endoplasmic Reticulum Membranes. mBio 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Oudshoorn, D.; Rijs, K.; Limpens, R.W.A.L.; Groen, K.; Koster, A.J.; Snijder, E.J.; Kikkert, M.; Bárcena, M. Expression and Cleavage of Middle East Respiratory Syndrome Coronavirus nsp3-4 Polyprotein Induce the Formation of Double-Membrane Vesicles That Mimic Those Associated with Coronaviral RNA Replication. mBio 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Neuman, B.W. Bioinformatics and functional analyses of coronavirus nonstructural proteins involved in the formation of replicative organelles. Antiviral Res. 2016, 135, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Clementz, M.A.; Kanjanahaluethai, A.; O’Brien, T.E.; Baker, S.C. Mutation in murine coronavirus replication protein nsp4 alters assembly of double membrane vesicles. Virology 2008, 375, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Gadlage, M.J.; Sparks, J.S.; Beachboard, D.C.; Cox, R.G.; Doyle, J.D.; Stobart, C.C.; Denison, M.R. Murine Hepatitis Virus Nonstructural Protein 4 Regulates Virus-Induced Membrane Modifications and Replication Complex Function. J. Virol. 2010, 84, 280–290. [Google Scholar] [CrossRef] [PubMed]
- Beachboard, D.C.; Anderson-Daniels, J.M.; Denison, M.R. Mutations across Murine Hepatitis Virus nsp4 Alter Virus Fitness and Membrane Modifications. J. Virol. 2015, 89, 2080–2089. [Google Scholar] [CrossRef] [PubMed]
- Knoops, K.; Kikkert, M.; Worm, S.H.; Zevenhoven-Dobbe, J.C.; van der Meer, Y.; Koster, A.J.; Mommaas, A.M.; Snijder, E.J. SARS-Coronavirus Replication Is Supported by a Reticulovesicular Network of Modified Endoplasmic Reticulum. PLoS Biol. 2008, 6, e226. [Google Scholar] [CrossRef] [PubMed]
- Posthuma, C.C.; Pedersen, K.W.; Lu, Z.; Joosten, R.G.; Roos, N.; Zevenhoven-Dobbe, J.C.; Snijder, E.J. Formation of the Arterivirus Replication/Transcription Complex: A Key Role for Nonstructural Protein 3 in the Remodeling of Intracellular Membranes. J. Virol. 2008, 82, 4480–4491. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, K.W.; van der Meer, Y.; Roos, N.; Snijder, E.J. Open Reading Frame 1a-Encoded Subunits of the Arterivirus Replicase Induce Endoplasmic Reticulum-Derived Double-Membrane Vesicles Which Carry the Viral Replication Complex. J. Virol. 1999, 73, 2016–2026. [Google Scholar] [PubMed]
- Snijder, E.J.; van Tol, H.; Roos, N.; Pedersen, K.W. Non-structural proteins 2 and 3 interact to modify host cell membranes during the formation of the arterivirus replication complex. J. Gen. Virol. 2001, 82, 985–994. [Google Scholar] [CrossRef] [PubMed]
- Gorbalenya, A.E.; Enjuanes, L.; Ziebuhr, J.; Snijder, E.J. Nidovirales: Evolving the largest RNA virus genome. Virus Res. 2006, 117, 17–37. [Google Scholar] [CrossRef] [PubMed]
- Hagemeijer, M.C.; Monastyrska, I.; Griffith, J.; van der Sluijs, P.; Voortman, J.; van Bergen en Henegouwen, P.M.; Vonk, A.M.; Rottier, P.J.M.; Reggiori, F.; de Haan, C.A.M. Membrane rearrangements mediated by coronavirus nonstructural proteins 3 and 4. Virology 2014, 458–459, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Hagemeijer, M.C.; Ulasli, M.; Vonk, A.M.; Reggiori, F.; Rottier, P.J.M.; de Haan, C.A.M. Mobility and Interactions of Coronavirus Nonstructural Protein 4. J. Virol. 2011, 85, 4572–4577. [Google Scholar] [CrossRef] [PubMed]
- Angelini, M.M.; Akhlaghpour, M.; Neuman, B.W.; Buchmeier, M.J. Severe Acute Respiratory Syndrome Coronavirus Nonstructural Proteins 3, 4, and 6 Induce Double-Membrane Vesicles. mBio 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Lundin, A.; Dijkman, R.; Bergström, T.; Kann, N.; Adamiak, B.; Hannoun, C.; Kindler, E.; Jónsdóttir, H.R.; Muth, D.; Kint, J.; et al. Targeting Membrane-Bound Viral RNA Synthesis Reveals Potent Inhibition of Diverse Coronaviruses Including the Middle East Respiratory Syndrome Virus. PLoS Pathog. 2014, 10, e1004166. [Google Scholar] [CrossRef] [PubMed]
- Maier, H.J.; Neuman, B.W.; Bickerton, E.; Keep, S.M.; Alrashedi, H.; Hall, R.; Britton, P. Extensive coronavirus-induced membrane rearrangements are not a determinant of pathogenicity. Sci. Rep. 2016, 6, 27126. [Google Scholar] [CrossRef] [PubMed]
- Casais, R.; Thiel, V.; Siddell, S.G.; Cavanagh, D.; Britton, P. Reverse Genetics System for the Avian Coronavirus Infectious Bronchitis Virus. J. Virol. 2001, 75, 12359–12369. [Google Scholar] [CrossRef] [PubMed]
- Darbyshire, J.H.; Rowell, J.G.; Cook, J.K.A.; Peters, R.W. Taxonomic studies on strains of avian infectious bronchitis virus using neutralisation tests in tracheal organ cultures. Arch. Virol. 1979, 61, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Kremer, J.R.; Mastronarde, D.N.; McIntosh, J.R. Computer Visualization of Three-Dimensional Image Data Using IMOD. J. Struct. Biol. 1996, 116, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.G.; Shen, S.; Tay, F.P.L.; Liu, D.X. Selection of and recombination between minor variants lead to the adaptation of an avian coronavirus to primate cells. Biochem. Biophys. Res. Commun. 2005, 336, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Promkuntod, N.; Wickramasinghe, I.N.A.; de Vrieze, G.; Gröne, A.; Verheije, M.H. Contributions of the S2 spike ectodomain to attachment and host range of infectious bronchitis virus. Virus Res. 2013, 177, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Almazán, F.; González, J.M.; Pénzes, Z.; Izeta, A.; Calvo, E.; Plana-Durán, J.; Enjuanes, L. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 2000, 97, 5516–5521. [Google Scholar] [CrossRef] [PubMed]
- Yount, B.; Curtis, K.M.; Baric, R.S. Strategy for Systematic Assembly of Large RNA and DNA Genomes: Transmissible Gastroenteritis Virus Model. J. Virol. 2000, 74, 10600–10611. [Google Scholar] [CrossRef] [PubMed]
- Yount, B.; Denison, M.R.; Weiss, S.R.; Baric, R.S. Systematic Assembly of a Full-Length Infectious cDNA of Mouse Hepatitis Virus Strain A59. J. Virol. 2002, 76, 11065–11078. [Google Scholar] [CrossRef] [PubMed]
- Cottam, E.M.; Maier, H.J.; Manifava, M.; Vaux, L.C.; Chandra-Schoenfelder, P.; Gerner, W.; Britton, P.; Ktistakis, N.T.; Wileman, T. Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy 2011, 7, 1335–1347. [Google Scholar] [CrossRef] [PubMed]
- Baliji, S.; Cammer, S.A.; Sobral, B.; Baker, S.C. Detection of Nonstructural Protein 6 in Murine Coronavirus-Infected Cells and Analysis of the Transmembrane Topology by Using Bioinformatics and Molecular Approaches. J. Virol. 2009, 83, 6957–6962. [Google Scholar] [CrossRef] [PubMed]
- Kanjanahaluethai, A.; Chen, Z.; Jukneliene, D.; Baker, S.C. Membrane topology of murine coronavirus replicase nonstructural protein 3. Virology 2007, 361, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Oostra, M.; Hagemeijer, M.C.; van Gent, M.; Bekker, C.P.J.; te Lintelo, E.G.; Rottier, P.J.M.; de Haan, C.A.M. Topology and Membrane Anchoring of the Coronavirus Replication Complex: Not All Hydrophobic Domains of nsp3 and nsp6 Are Membrane Spanning. J. Virol. 2008, 82, 12392–12405. [Google Scholar] [CrossRef] [PubMed]
- Oostra, M.; te Lintelo, E.G.; Deijs, M.; Verheije, M.H.; Rottier, P.J.M.; de Haan, C.A.M. Localization and Membrane Topology of Coronavirus Nonstructural Protein 4: Involvement of the Early Secretory Pathway in Replication. J. Virol. 2007, 81, 12323–12336. [Google Scholar] [CrossRef] [PubMed]
- Maier, H.J.; Cottam, E.M.; Stevenson-Leggett, P.; Wilkinson, J.A.; Harte, C.J.; Wileman, T.; Britton, P. Visualizing the autophagy pathway in avian cells and its application to studying infectious bronchitis virus. Autophagy 2013, 9, 496–509. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.A.; Peng, X.; Gao, Y.; Li, Z.; Lu, X.; Chen, Y.; Ishaq, M.; Liu, D.; DeDiego, M.L.; Enjuanes, L.; et al. Genome-Wide Analysis of Protein-Protein Interactions and Involvement of Viral Proteins in SARS-CoV Replication. PLoS ONE 2008, 3, e3299. [Google Scholar] [CrossRef] [PubMed]
- Von Brunn, A.; Teepe, C.; Simpson, J.C.; Pepperkok, R.; Friedel, C.C.; Zimmer, R.; Roberts, R.; Baric, R.; Haas, J. Analysis of Intraviral Protein-Protein Interactions of the SARS Coronavirus ORFeome. PLoS ONE 2007, 2, e459. [Google Scholar] [CrossRef] [PubMed]
- Van der Hoeven, B.; Oudshoorn, D.; Koster, A.J.; Snijder, E.J.; Kikkert, M.; Bárcena, M. Biogenesis and architecture of arterivirus replication organelles. Virus Res. 2016, 220, 70–90. [Google Scholar] [CrossRef] [PubMed]
- Sakai, Y.; Kawachi, K.; Terada, Y.; Omori, H.; Matsuura, Y.; Kamitani, W. Two-amino acids change in the nsp4 of SARS coronavirus abolishes viral replication. Virology 2017, 510, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Kallio, K.; Hellström, K.; Balistreri, G.; Spuul, P.; Jokitalo, E.; Ahola, T. Template RNA Length Determines the Size of Replication Complex Spherules for Semliki Forest Virus. J. Virol. 2013, 87, 9125–9134. [Google Scholar] [CrossRef] [PubMed]
- Hellström, K.; Kallio, K.; Utt, A.; Quirin, T.; Jokitalo, E.; Merits, A.; Ahola, T. Partially Uncleaved Alphavirus Replicase Forms Spherule Structures in the Presence and Absence of RNA Template. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Kopek, B.G.; Settles, E.W.; Friesen, P.D.; Ahlquist, P. Nodavirus-Induced Membrane Rearrangement in Replication Complex Assembly Requires Replicase Protein A, RNA Templates, and Polymerase Activity. J. Virol. 2010, 84, 12492–12503. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Doyle, N.; Neuman, B.W.; Simpson, J.; Hawes, P.C.; Mantell, J.; Verkade, P.; Alrashedi, H.; Maier, H.J. Infectious Bronchitis Virus Nonstructural Protein 4 Alone Induces Membrane Pairing. Viruses 2018, 10, 477. https://doi.org/10.3390/v10090477
Doyle N, Neuman BW, Simpson J, Hawes PC, Mantell J, Verkade P, Alrashedi H, Maier HJ. Infectious Bronchitis Virus Nonstructural Protein 4 Alone Induces Membrane Pairing. Viruses. 2018; 10(9):477. https://doi.org/10.3390/v10090477
Chicago/Turabian StyleDoyle, Nicole, Benjamin W. Neuman, Jennifer Simpson, Philippa C. Hawes, Judith Mantell, Paul Verkade, Hasan Alrashedi, and Helena J. Maier. 2018. "Infectious Bronchitis Virus Nonstructural Protein 4 Alone Induces Membrane Pairing" Viruses 10, no. 9: 477. https://doi.org/10.3390/v10090477
APA StyleDoyle, N., Neuman, B. W., Simpson, J., Hawes, P. C., Mantell, J., Verkade, P., Alrashedi, H., & Maier, H. J. (2018). Infectious Bronchitis Virus Nonstructural Protein 4 Alone Induces Membrane Pairing. Viruses, 10(9), 477. https://doi.org/10.3390/v10090477





