Xenotropic Mouse Gammaretroviruses Isolated from Pre-Leukemic Tissues Include a Recombinant
Abstract
1. Introduction
2. Materials and Methods
2.1. Viruses and Sequencing
2.2. Phylogenetic Trees
2.3. Inbred Strain Distribution and Subspecies Origins of AKR6-Related ERVs
3. Results and Discussion
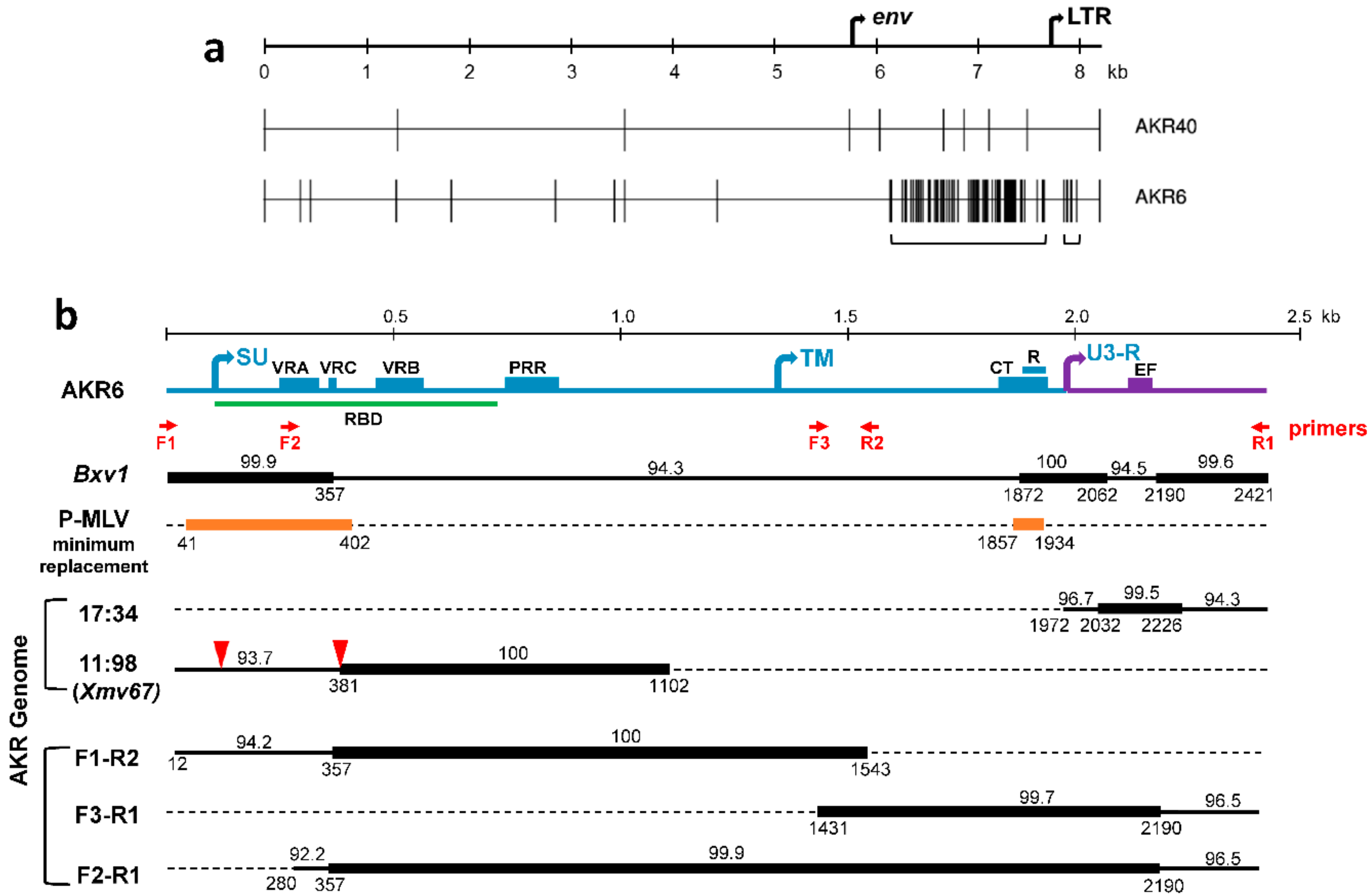
3.1. Sequence Analysis of Two X-MLVs
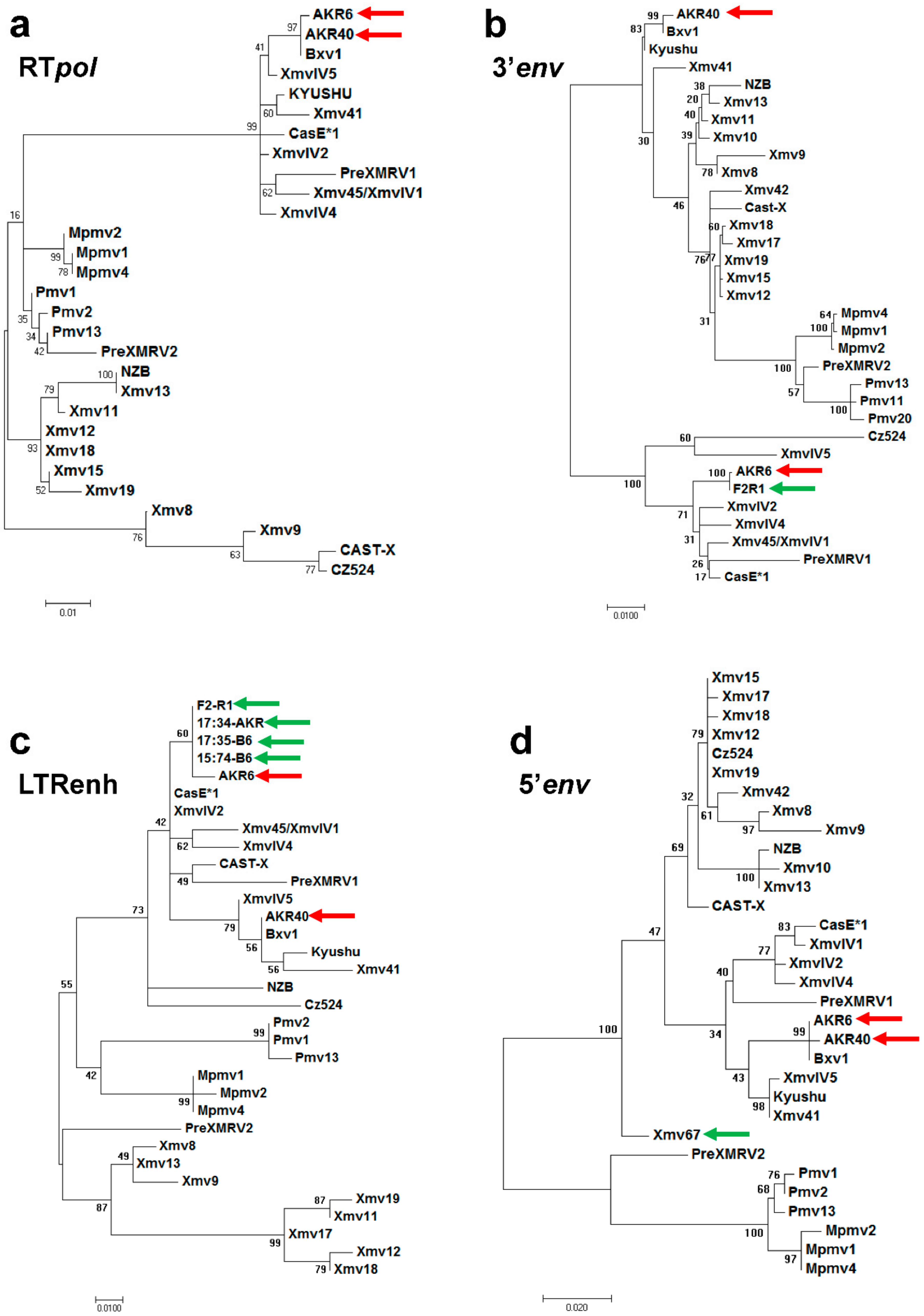
3.2. ERV Progenitors of the AKR6 Replacements
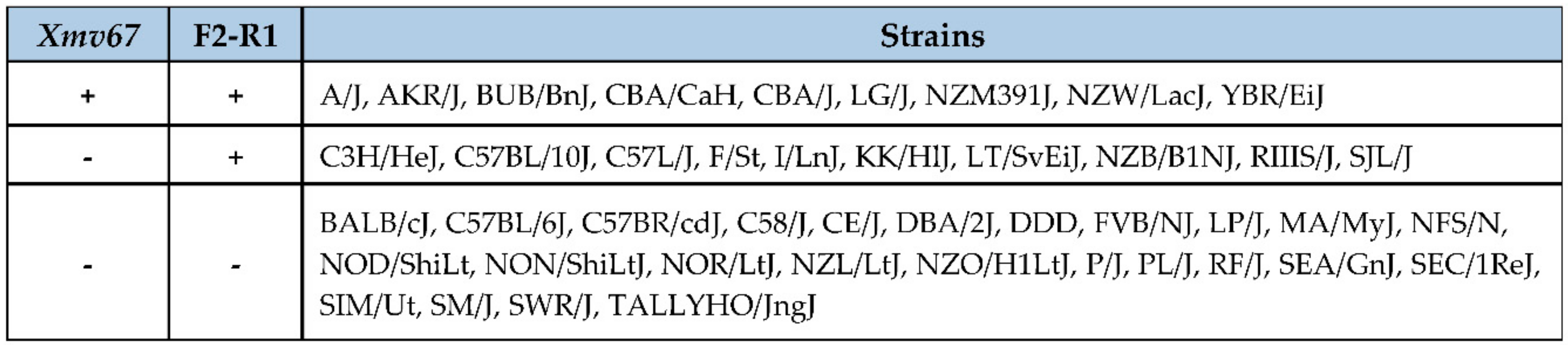
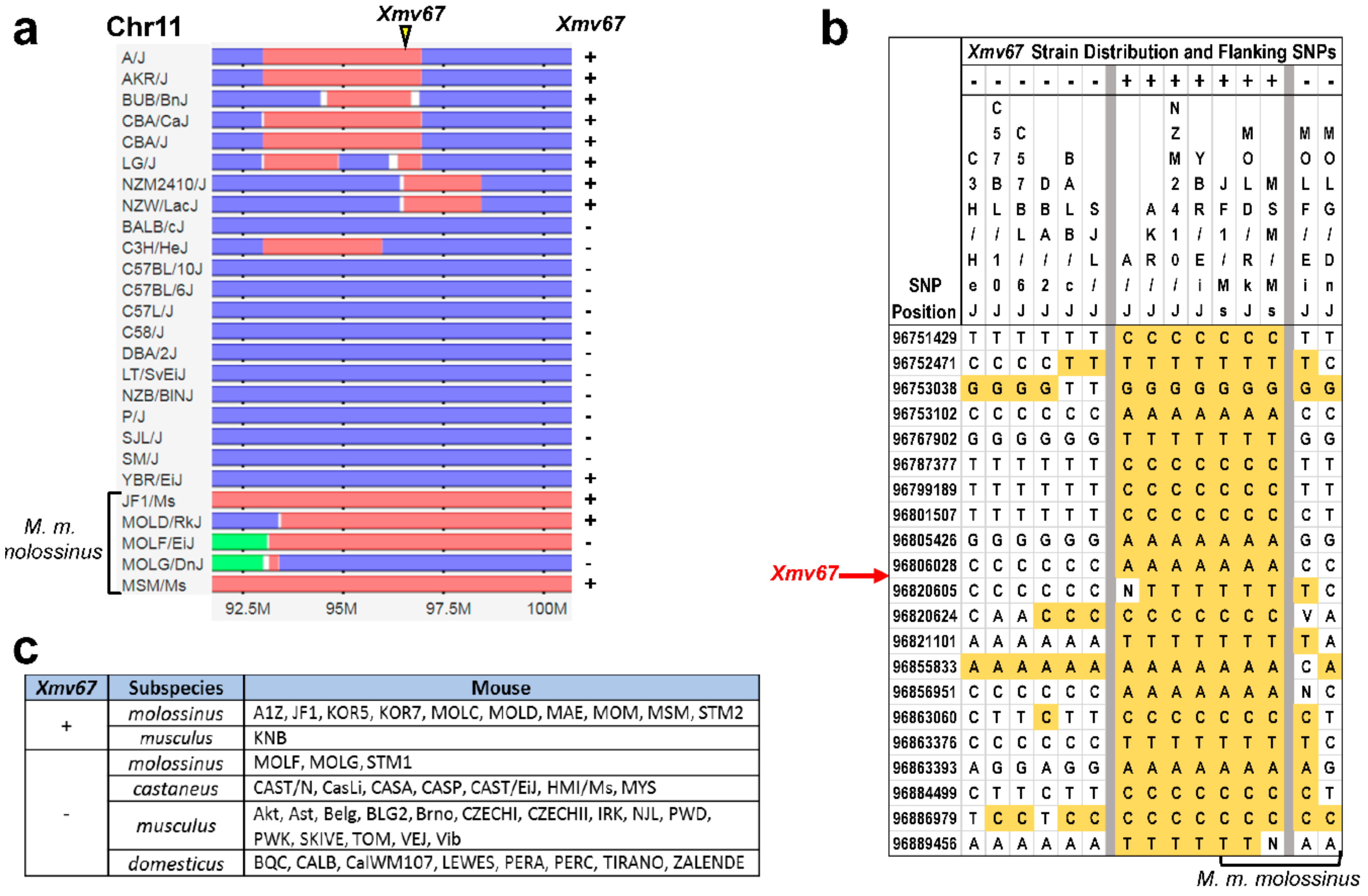
3.3. Acquisition of AKR6-Related ERVs in M. musculus Subspecies
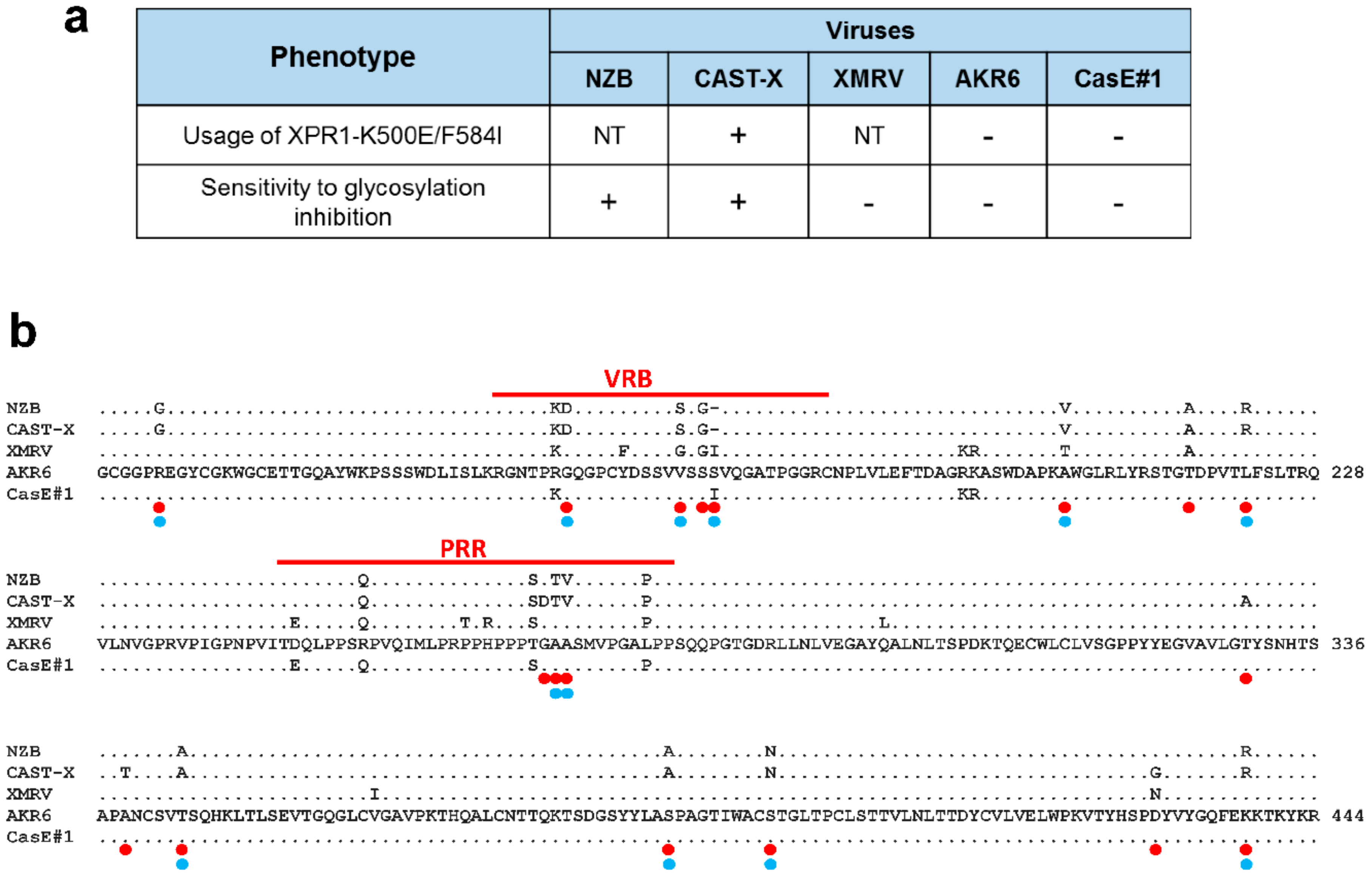
3.4. Env Replacement Mutations Linked to the Modified AKR6 Host Range
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kozak, C.A. Origins of the endogenous and infectious laboratory mouse gammaretroviruses. Viruses 2015, 7, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Oie, H.K.; Russell, E.K.; Dotson, J.H.; Rhoads, J.M.; Gazdar, A.F. Host range properties of murine xenotropic and ecotropic type-C viruses. J. Natl. Cancer Inst. 1976, 56, 423–426. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Liu, Q.; Wollenberg, K.; Martin, C.; Buckler-White, A.; Kozak, C.A. Evolution of functional and sequence variants of the mammalian XPR1 receptor for mouse xenotropic gammaretroviruses and the human-derived XMRV. J. Virol. 2010, 84, 11970–11980. [Google Scholar] [CrossRef] [PubMed]
- Albritton, L.M.; Tseng, L.; Scadden, D.; Cunningham, J.M. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell 1989, 57, 659–666. [Google Scholar] [CrossRef]
- Tailor, C.S.; Nouri, A.; Lee, C.G.; Kozak, C.; Kabat, D. Cloning and characterization of a cell surface receptor for xenotropic and polytropic murine leukemia viruses. Proc. Natl. Acad. Sci. USA 1999, 96, 927–932. [Google Scholar] [CrossRef] [PubMed]
- Battini, J.L.; Rasko, J.E.; Miller, A.D. A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: Possible role in G protein-coupled signal transduction. Proc. Natl. Acad. Sci. USA 1999, 96, 1385–1390. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-L.; Guo, L.; Xu, S.; Holland, C.A.; Kitamura, T.; Hunter, K.; Cunningham, J.M. Receptors for polytropic and xenotropic mouse leukaemia viruses encoded by a single gene at Rmc1. Nat. Genet. 1999, 21, 216–219. [Google Scholar] [CrossRef] [PubMed]
- Baliji, S.; Liu, Q.; Kozak, C.A. Common inbred strains of the laboratory mouse that are susceptible to infection by mouse xenotropic gammaretroviruses and the human-derived retrovirus XMRV. J. Virol. 2010, 84, 12841–12849. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, N.A.; Copeland, N.G.; Taylor, B.A.; Lee, B.K. Organization, distribution, and stability of endogenous ecotropic murine leukemia virus DNA sequences in chromosomes of Mus musculus. J. Virol. 1982, 43, 26–36. [Google Scholar] [PubMed]
- Frankel, W.N.; Stoye, J.P.; Taylor, B.A.; Coffin, J.M. A linkage map of endogenous murine leukemia proviruses. Genetics 1990, 124, 221–236. [Google Scholar] [PubMed]
- Stoye, J.P.; Coffin, J.M. The four classes of endogenous murine leukemia virus: Structural relationships and potential for recombination. J. Virol. 1987, 61, 2659–2669. [Google Scholar] [PubMed]
- Lung, M.L.; Hartley, J.W.; Rowe, W.P.; Hopkins, N.H. Large RNase T1-resistant oligonucleotides encoding p15E and the U3 region of the long terminal repeat distinguish two biological classes of mink cell focus-forming type C viruses of inbred mice. J. Virol. 1983, 45, 275–290. [Google Scholar] [PubMed]
- Chattopadhyay, S.K.; Cloyd, M.W.; Linemeyer, D.L.; Lander, M.R.; Rands, E.; Lowy, D.R. Cellular origin and role of mink cell focus-forming viruses in murine thymic lymphomas. Nature 1982, 295, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.S.; Rowe, W.P.; Martin, M.A. Cloning of endogenous murine leukemia virus-related sequences from chromosomal DNA of BALB/c and AKR/J mice: Identification of an env progenitor of AKR-247 mink cell focus-forming proviral DNA. J. Virol. 1982, 44, 625–636. [Google Scholar] [PubMed]
- Bamunusinghe, D.; Liu, Q.; Plishka, R.; Dolan, M.A.; Skorski, M.; Oler, A.J.; Yedavalli, V.R.K.; Buckler-White, A.; Hartley, J.W.; Kozak, C.A. Recombinant origins of pathogenic and nonpathogenic mouse gammaretroviruses with polytropic host range. J. Virol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Hartley, J.W.; Wolford, N.K.; Old, L.J.; Rowe, W.P. New class of murine leukemia-virus associated with development of spontaneous lymphomas. Proc. Natl. Acad. Sci. USA 1977, 74, 789–792. [Google Scholar] [CrossRef] [PubMed]
- Herr, W.; Gilbert, W. Somatically acquired recombinant murine leukemia proviruses in thymic leukemias of AKR/J mice. J. Virol. 1983, 46, 70–82. [Google Scholar] [PubMed]
- Stoye, J.P.; Moroni, C.; Coffin, J.M. Virological events leading to spontaneous AKR thymomas. J. Virol. 1991, 65, 1273–1285. [Google Scholar] [PubMed]
- Cloyd, M.W.; Hartley, J.W.; Rowe, W.P. Lymphomagenicity of recombinant mink cell focus-inducing murine leukemia viruses. J. Exp. Med. 1980, 151, 542–552. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, S.; Pal, B.K.; Gardner, M.B. Characterization of a highly oncogenic murine leukemia virus from wild mice. Int. J. Cancer 1982, 29, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Pothlichet, J.; Mangeney, M.; Heidmann, T. Mobility and integration sites of a murine C57BL/6 melanoma endogenous retrovirus involved in tumor progression in vivo. Int. J. Cancer 2006, 119, 1869–1877. [Google Scholar] [CrossRef] [PubMed]
- Paprotka, T.; Delviks-Frankenberry, K.A.; Cingoz, O.; Martinez, A.; Kung, H.J.; Tepper, C.G.; Hu, W.S.; Fivash, M.J., Jr.; Coffin, J.M.; Pathak, V.K. Recombinant origin of the retrovirus XMRV. Science 2011, 333, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Bamunusinghe, D.; Naghashfar, Z.; Buckler-White, A.; Plishka, R.; Baliji, S.; Liu, Q.; Kassner, J.; Oler, A.; Hartley, J.; Kozak, C.A. Sequence diversity, intersubgroup relationships, and origins of the mouse leukemia gammaretroviruses of laboratory and wild mice. J. Virol. 2016, 90, 4186–4198. [Google Scholar] [CrossRef] [PubMed]
- Tomonaga, K.; Coffin, J.M. Structure and distribution of endogenous nonecotropic murine leukemia viruses in wild mice. J. Virol. 1998, 72, 8289–8300. [Google Scholar] [PubMed]
- Cloyd, M.W.; Hartley, J.W.; Rowe, W.P. Cell-surface antigens associated with recombinant mink cell focus-inducing murine leukemia viruses. J. Exp. Med. 1979, 149, 702–712. [Google Scholar] [CrossRef] [PubMed]
- Keele, B.F.; Giorgi, E.E.; Salazar-Gonzalez, J.F.; Decker, J.M.; Pham, K.T.; Salazar, M.G.; Sun, C.; Grayson, T.; Wang, S.; Li, H.; et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. USA 2008, 105, 7552–7557. [Google Scholar] [CrossRef] [PubMed]
- Van Hoeven, N.S.; Miller, A.D. Use of different but overlapping determinants in a retrovirus receptor accounts for non-reciprocal interference between xenotropic and polytropic murine leukemia viruses. Retrovirology 2005, 2, 76. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Jern, P.; Stoye, J.P.; Coffin, J.M. Role of APOBEC3 in genetic diversity among endogenous murine leukemia viruses. PLoS Genet. 2007, 3, e183. [Google Scholar] [CrossRef] [PubMed]
- Lamont, C.; Culp, P.; Talbott, R.L.; Phillips, T.R.; Trauger, R.J.; Frankel, W.N.; Wilson, M.C.; Coffin, J.M.; Elder, J.H. Characterization of endogenous and recombinant proviral elements of a highly tumorigenic AKR cell line. J. Virol. 1991, 65, 4619–4628. [Google Scholar] [PubMed]
- Kent, W.J. BLAT—The BLAST-like alignment tool. Genome Res. 2002, 12, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Zerbino, D.R.; Achuthan, P.; Akanni, W.; Amode, M.R.; Barrell, D.; Bhai, J.; Billis, K.; Cummins, C.; Gall, A.; Giron, C.G.; et al. Ensembl 2018. Nucleic Acids Res. 2018, 46, D754–D761. [Google Scholar] [CrossRef] [PubMed]
- Bamunusinghe, D.; Liu, Q.; Lu, X.; Oler, A.; Kozak, C.A. Endogenous gammaretrovirus acquisition in Mus musculus subspecies carrying functional variants of the XPR1 virus receptor. J. Virol. 2013, 87, 9845–9855. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.R.; de Villena, F.P.; McMillan, L. Comparative analysis and visualization of multiple collinear genomes. BMC Bioinform. 2012, 13 (Suppl. 3), S13. [Google Scholar]
- Cloyd, M.W.; Thompson, M.M.; Hartley, J.W. Host range of mink cell focus-inducing viruses. Virology 1985, 140, 239–248. [Google Scholar] [CrossRef]
- Kozak, C.; Rowe, W.P. Genetic mapping of xenotropic leukemia virus-inducing loci in two mouse strains. Science 1978, 199, 1448–1449. [Google Scholar] [CrossRef] [PubMed]
- Kozak, C.A.; Rowe, W.P. Genetic mapping of xenotropic murine leukemia virus-inducing loci in five mouse strains. J. Exp. Med. 1980, 152, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Golemis, E.A.; Speck, N.A.; Hopkins, N. Alignment of U3 region sequences of mammalian type C viruses: Identification of highly conserved motifs and implications for enhancer design. J. Virol. 1990, 64, 534–542. [Google Scholar] [PubMed]
- Morrison, H.L.; Soni, B.; Lenz, J. Long terminal repeat enhancer core sequences in proviruses adjacent to c-myc in T-cell lymphomas induced by a murine retrovirus. J. Virol. 1995, 69, 446–455. [Google Scholar] [PubMed]
- Spies, C.P.; Ritter, G.D., Jr.; Mulligan, M.J.; Compans, R.W. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein alters the conformation of the external domain. J. Virol. 1994, 68, 585–591. [Google Scholar] [PubMed]
- Yang, H.; Bell, T.A.; Churchill, G.A.; Pardo-Manuel de Villena, F. On the subspecific origin of the laboratory mouse. Nat. Genet. 2007, 39, 1100–1107. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, J.R.; Didion, J.P.; Buus, R.J.; Bell, T.A.; Welsh, C.E.; Bonhomme, F.; Yu, A.H.; Nachman, M.W.; Pialek, J.; et al. Subspecific origin and haplotype diversity in the laboratory mouse. Nat. Genet. 2011, 43, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Miller, D.G.; Miller, A.D. Tunicamycin treatment of CHO cells abrogates multiple blocks to retrovirus infection, one of which is due to a secreted inhibitor. J. Virol. 1992, 66, 78–84. [Google Scholar] [PubMed]
- Yan, Y.; Liu, Q.; Kozak, C.A. Six host range variants of the xenotropic/polytropic gammaretroviruses define determinants for entry in the XPR1 cell surface receptor. Retrovirology 2009, 6, 87. [Google Scholar] [CrossRef] [PubMed]
- Battini, J.L.; Heard, J.M.; Danos, O. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J. Virol. 1992, 66, 1468–1475. [Google Scholar] [PubMed]
- Argaw, T.; Wilson, C.A. Detailed mapping of determinants within the porcine endogenous retrovirus envelope surface unit identifies critical residues for human cell infection within the proline-rich region. J. Virol. 2012, 86, 9096–9104. [Google Scholar] [CrossRef] [PubMed]
- Bahrami, S.; Duch, M.; Pedersen, F.S. Change of tropism of SL3-2 murine leukemia virus, using random mutational libraries. J. Virol. 2004, 78, 9343–9351. [Google Scholar] [CrossRef] [PubMed]
- Sherr, C.J.; Lieber, M.M.; Todaro, G.J. Mixed splenocyte cultures and graft versus host reactions selectively induce an S-tropic murine type C virus. Cell 1974, 1, 55–58. [Google Scholar] [CrossRef]
- Rosenke, K.; Lavignon, M.; Malik, F.; Kolokithas, A.; Hendrick, D.; Virtaneva, K.; Peterson, K.; Evans, L.H. Profound amplification of pathogenic murine polytropic retrovirus release from coinfected cells. J. Virol. 2012, 86, 7241–7248. [Google Scholar] [CrossRef] [PubMed]
- Barnett, A.L.; Davey, R.A.; Cunningham, J.M. Modular organization of the Friend murine leukemia virus envelope protein underlies the mechanism of infection. Proc. Natl. Acad. Sci. USA 2001, 98, 4113–4118. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bamunusinghe, D.; Skorski, M.; Buckler-White, A.; Kozak, C.A. Xenotropic Mouse Gammaretroviruses Isolated from Pre-Leukemic Tissues Include a Recombinant. Viruses 2018, 10, 418. https://doi.org/10.3390/v10080418
Bamunusinghe D, Skorski M, Buckler-White A, Kozak CA. Xenotropic Mouse Gammaretroviruses Isolated from Pre-Leukemic Tissues Include a Recombinant. Viruses. 2018; 10(8):418. https://doi.org/10.3390/v10080418
Chicago/Turabian StyleBamunusinghe, Devinka, Matthew Skorski, Alicia Buckler-White, and Christine A. Kozak. 2018. "Xenotropic Mouse Gammaretroviruses Isolated from Pre-Leukemic Tissues Include a Recombinant" Viruses 10, no. 8: 418. https://doi.org/10.3390/v10080418
APA StyleBamunusinghe, D., Skorski, M., Buckler-White, A., & Kozak, C. A. (2018). Xenotropic Mouse Gammaretroviruses Isolated from Pre-Leukemic Tissues Include a Recombinant. Viruses, 10(8), 418. https://doi.org/10.3390/v10080418



