A Virus in American Blackcurrant (Ribes americanum) with Distinct Genome Features Reshapes Classification in the Tymovirales
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Barba, M.; Czosnek, H.; Hadidi, A. Historical perspective, development and applications of next-generation sequencing in plant virology. Viruses 2014, 6, 106–136. [Google Scholar] [CrossRef] [PubMed]
- Di Bello, P.L.; Laney, A.G.; Druciarek, T.; Ho, T.; Gergerich, R.C.; Keller, K.E.; Martin, R.R.; Tzanetakis, I.E. A novel emaravirus is associated with redbud yellow ringspot disease. Virus Res. 2016, 222, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.; Tzanetakis, I.E. Developing a virus detection and discovery pipeline using next generation sequencing. Virology 2014, 471–473, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Massart, S.; Olmos, A.; Jijakli, H.; Candresse, T. Current impact and future directions of high throughput sequencing in plant virus diagnostics. Virus Res. 2014, 188, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Postman, J.; Hummer, K.; Stover, E.; Krueger, R.; Forsline, P.; Grauke, L.J.; Zee, F.; Ayala-Silva, T.; Irish, B. Fruit and nut genebanks in the US national plant germplasm system. HortScience 2006, 41, 1188–1194. [Google Scholar]
- Terry, L. Health-Promoting Properties of Fruit and Vegetables; CABI: Wallingford, UK, 2014; p. 432. [Google Scholar]
- Thekke-Veetil, T.; Ho, T.; Postman, J.D.; Tzanetakis, I.E. Molecular characterization of a new member of the genus Waikavirus. Phytopathology 2017, 107, 102. [Google Scholar]
- ICTV. Virus Taxonomy 2017 Release, Order Tymovirales. Available online: https://talk.ictvonline.org/taxonomy/ (accessed on 31 July 2018).
- Adams, M.J.; Antoniw, J.F.; Bar-Joseph, M.; Brunt, A.A.; Candresse, T.; Foster, G.D.; Martelli, G.P.; Milne, R.G.; Fauquet, C.M. The new plant virus family Flexiviridae and assessment of molecular criteria for species demarcation. Arch. Virol. 2004, 149, 1045–1060. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.; Martin, R.R.; Tzanetakis, I.E. Next-generation sequencing of elite berry germplasm and data analysis using a bioinformatics pipeline for virus detection and discovery. In Plant Pathology: Techniques and Protocols (Methods in Molecular Biology); Lacomme, C., Ed.; Springer: New York, NY, USA, 2015; pp. 301–313. [Google Scholar]
- Wheeler, D.L.; Church, D.M.; Federhen, S.; Lash, A.E.; Madden, T.L.; Pontius, J.U.; Schuler, G.D.; Schriml, L.M.; Sequeira, E.; Tatusova, T.A.; et al. Database resources of the National Center for Biotechnology. Nucleic Acids Res. 2003, 31, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Gromiha, M.M.; Raghava, G.P.S. Prediction of RNA binding sites in a protein using SVM and PSSM profile. Proteins 2008, 71, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999, 292, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Kosugi, S.; Hasebe, M.; Tomita, M.; Yanagawa, H. Systematic identification of yeast cell cycle-dependent nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. USA 2009, 106, 10171–10176. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Dolja, V.V. Evolution and taxonomy of positive-strand RNA viruses: Implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 1993, 28, 375–430. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Mock, R. Characterization of a flowering cherry strain of cherry necrotic rusty mottle virus. Arch. Virol. 2008, 153, 973–978. [Google Scholar] [CrossRef] [PubMed]
- Sabanadzovic, S.; Abou Ghanem-Sabanadzovic, N.; Tzanetakis, I.E. Blackberry virus E: An unusual flexivirus. Arch. Virol. 2011, 156, 1665–1669. [Google Scholar] [CrossRef] [PubMed]
- Villamor, D.V.; Druffel, K.L.; Eastwell, K.C. Complete nucleotide sequence of a virus associated with rusty mottle disease of sweet cherry (Prunus avium). Arch. Virol. 2013, 158, 1805–1810. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.P.; Kirkpatrick, B.C.; Smart, C.D.; Uyemoto, J.K. cDNA cloning and molecular characterization of cherry green ring mottle virus. J. Gen. Virol. 1998, 79, 2275–2281. [Google Scholar] [CrossRef] [PubMed]
- Verchot-Lubicz, J.; Ye, C.M.; Bamunusinghe, D. Molecular biology of potexviruses: Recent advances. J. Gen. Virol. 2007, 88, 1643–1655. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.M.; Mahtani, P.H.; Lee, K.C.; Yu, H.H.; Tan, Y.; Neo, K.K.; Chan, Y.; Wu, M.; Chng, C.G. Cymbidium mosaic potexvirus RNA: Complete nucleotide sequence and phylogenetic analysis. Arch. Virol. 1997, 142, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Martelli, G.P.; Adams, M.J.; Kreuze, J.F.; Dolja, V.V. Family Flexiviridae: A case study in virion and genome plasticity. Annu. Rev. Phytopathol. 2007, 45, 73–100. [Google Scholar] [CrossRef] [PubMed]
- Morozov, S.Y.; Solovyev, A.G. Triple gene block: Modular design of a multifunctional machine for plant virus movement. J. Gen. Virol. 2003, 84, 1351–1366. [Google Scholar] [CrossRef] [PubMed]
- Rupasov, V.V.; Morozov, S.Y.; Kanyuka, K.V.; Zavriev, S.K. Partial nucleotide sequence of potato virus M RNA shows similarities to potexviruses in gene arrangement and the encoded amino acid sequences. J. Gen. Virol. 1989, 70, 1861–1869. [Google Scholar] [CrossRef] [PubMed]
- Kozak, M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 1986, 44, 283–292. [Google Scholar] [CrossRef]
- Sánchez-Puig, J.M.; Blasco, R. AUG context and mRNA translation in vaccinia virus. Span. J. Agric. Res. 2008, 6, 73–80. [Google Scholar] [CrossRef]
- Dolja, V.V.; Boyko, V.P.; Agranovsky, A.A.; Koonin, E.V. Phylogeny of capsid proteins of rod-shaped and filamentous RNA plant viruses: Two families with distinct patterns of sequence and probably structure conservation. Virology 1991, 184, 79–86. [Google Scholar] [CrossRef]
- Choi, Y.G.; Rao, A.L.N. Molecular studies on bromovirus capsid protein: VII. Selective packaging of BMV RNA4 by specific N-terminal arginine residues. Virology 2000, 275, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Meguid, S.; Yamane, S.T.; Fukuyama, K.; Rossmann, M.G. The location of calcium ions in southern bean mosaic virus. Virology 1981, 114, 81–85. [Google Scholar] [CrossRef]
- Durham, A.C.; Hendry, D.A. Cation binding by tobacco mosaic virus. Virology 1977, 77, 510–519. [Google Scholar] [CrossRef]
- Gajardo, R.; Vende, P.; Poncet, D.; Cohen, J. Two proline residues are essential in the calcium-binding activity of rotavirus VP7 outer capsid protein. J. Virol. 1997, 71, 2211–2216. [Google Scholar] [PubMed]
- Marais, A.; Faure, C.; Mustafayev, E.; Candresse, T. Characterization of new isolates of apricot vein clearing-associated virus and of a new Prunus-Infecting virus: Evidence for recombination as a driving force in Betaflexiviridae evolution. PLoS ONE 2015, 10, e0129469. [Google Scholar] [CrossRef] [PubMed]
- Alabi, O.J.; Al Rwahnih, M.; Mekuria, T.A.; Naidu, R.A. Genetic diversity of grapevine virus A in Washington and California vineyards. Phytopathology 2014, 104, 548–560. [Google Scholar] [CrossRef] [PubMed]
- Singh, L.; Hallan, V.; Martin, D.P.; Ram, R.; Zaidi, A.A. Genomic sequence analysis of four new chrysanthemum virus B isolates: Evidence of RNA recombination. Arch. Virol. 2012, 157, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Villamor, D.E.; Eastwell, K.C. Viruses associated with rusty mottle and twisted leaf diseases of sweet cherry are distinct species. Phytopathology 2013, 103, 1287–1295. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.Y.; Joa, J.H.; Choi, K.S.; Do, K.S.; Lim, H.C.; Chung, B.M. Genetic diversity of a natural population of apple stem pitting virus isolated from apple in Korea. Plant Pathol. J. 2014, 30, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Zanardo, L.G.; Silva, F.N.; Lima, A.T.M.; Milanesi, D.F.; Castilho-Urquiza, G.P.; Almeida, A.M.R. Molecular variability of cowpea mild mottle virus infecting soybean in Brazil. Arch. Virol. 2014, 159, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Salminen, M. Detecting recombination in viral sequences. In The Phylogenetic Handbook: A Practical Approach to DNA and Protein Phylogeny; Salemi, M., Vandamme, A.M., Eds.; University Press: Cambridge, UK, 2003; pp. 348–377. [Google Scholar]

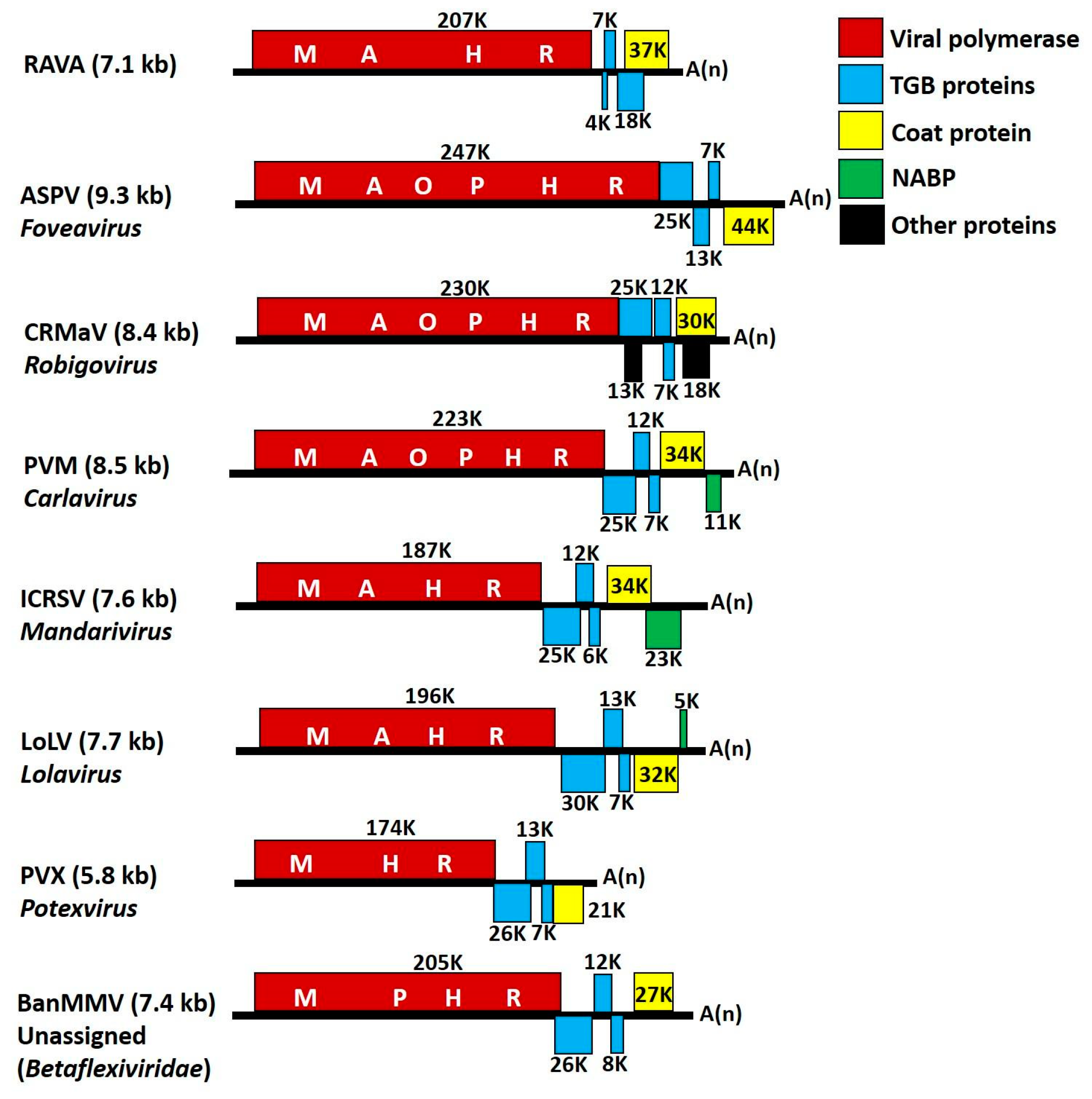

| Virus Acronyms | Virus Names | Genus (Family) | RdRp/MTR |
|---|---|---|---|
| ACLSV | Apple chlorotic leaf spot virus | Trichovirus (β) | NP040551.1 |
| CMLV | Cherry mottle leaf virus | Trichovirus (β) | AOY07780.1 |
| GVA | Grapevine virus A | Vitivirus (β) | AFV73358.1 |
| GVB | Grapevine virus B | Vitivirus (β) | AIL90366.1 |
| CNRMV | Cherry necrotic rusty mottle virus | Robigovirus (β) | NP059937.1 |
| AOPRV | African oil palm ringspot virus | Robigovirus (β) | YP002776347.1 |
| CtChV-1 | Carrot Ch virus 1 | Chordovirus (β) | AHA85534.1 |
| CtChV-2 | Carrot Ch virus 2 | Chordovirus (β) | AHA85531.1 |
| DiVA | Diuris virus A | Divavirus (β) | YP006905850.1 |
| DiVB | Diuris virus B | Divavirus (β) | AFV57240.1 |
| AVCaV | Apricot vein clearing associated virus | Prunevirus (β) | AKN09002.1 |
| CPrV | Caucasus prunus virus | Prunevirus (β) | AKN08994.1 |
| PVT | Potato virus T | Tepovirus (β) | AFU55321.1 |
| PrVT | Prunus virus T | Tepovirus (β) | YP009051684.1 |
| ASGV | Apple stem grooving virus | Capillovirus (β) | APT42870.1 |
| CVA | Cherry virus A | Capillovirus (β) | AMH87272.1 |
| GarCLV | Garlic common latent virus | Carlavirus (β) | AGG13282.1 |
| PVM | Potato virus M | Carlavirus (β) | AHL30493.1 |
| ASPV | Apple stem pitting virus | Foveavirus (β) | NP604464.1 |
| ApLV | Apricot latent virus | Foveavirus (β) | YP004089619.1 |
| CLBV | Citrus leaf blotch virus | Citrivirus (β) | NP624333.1 |
| BanMMV | Banana mild mosaic virus | Unassigned (β) | NP112029.1 |
| SCSMaV | Sugarcane striate mosaic-associated virus | Unassigned (β) | NP624313.1 |
| BVX | Banana virus X | Unassigned (β) | AAW50958.1 |
| BotV-F | Botrytis virus F | Mycoflexivirus (γ) | NP068549.1 |
| ShVX | Shallot virus X | Allexivirus (α) | NP620648.1 |
| ICRSV | Indian citrus ringspot virus | Mandarivirus (α) | AAK97522.1 |
| PVX | Potato virus X | Potexvirus (α) | BAE07083.1 |
| LoLV | Lolium latent virus | Lolavirus (α) | YP001718499.1 |
| BVE | Blackberry virus E | Unassigned (α) | AEI17897.1 |
| SaDFV1 | Soybean associated deltaflexivirus 1 | Deltaflexivirus (Λ) | ALM62223.1 |
| FgDFV1 | Fusarium deltaflexivirus 1 | Deltaflexivirus (Λ) | YP009268710.1 |
| SsDFV1 | Sclerotinia deltaflexivirus 1 | Deltaflexivirus (Λ) | AMD16208.1 |
| GFkV | Grapevine fleck virus | Maculavirus (T) | NP542612.1 |
| MRFV | Maize rayado fino virus | Marafivirus (T) | NP115454.1 |
| TYMV | Turnip yellow mosaic virus | Tymovirus (T) | AMH40140.1 |
| p2 |
| MNFKSYLLKKIKSVGIGLASSLIIYIASFVFNVLYRRSF |
| p3 |
| MCYIDVAFDLVCLFICVLILVALLKLTYCNSSAFCVALALTIYSLFLNFNLLVLLYDLSR |
| p4 |
| MYGYNNGIRKTSDRFSKGSVSKDKYGQRYNCGTDRLPFLVMADVSKLKIDFENATENMLFQIVSLLLHFCVLQNIGQ RKAKRGKIKKKKAAYNEYRRNKDGASSSYQGGGGLARTRDSQENERQVDAARDKRAEFYS |
| DSSSTEGDGDGSGQTRNERHFV |
| CP |
| MESEKLVIVSAKVPFRRTSMAKDTTAARTDFLSSLWRMSLNSKLISKMRQRTCYSRLCHSCCTSAYCKILARGRQREERLRRRKLPIMSTGEIRMEPLPVTREGEAWLELEIARKMKGKLTLQETNGRNSILTVAQPKEMV |
| MDLDRPEMRDILFNLDFTKRLIDQDVFVCSYLVKKAKRVGVEVCTDFHCYFVDTDMTVSALLDAIEIASFFGCINSAVFEICATGSCLCKVGLRELIIEVEKRTIEIPLKCGYHGIKHLTEVEDRQWKVLCANPLIKLEEIEE IYIFWNSLGLKNHERHVKALLDVNGLKES TLRILGAI |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thekke-Veetil, T.; Ho, T.; Postman, J.D.; Martin, R.R.; Tzanetakis, I.E. A Virus in American Blackcurrant (Ribes americanum) with Distinct Genome Features Reshapes Classification in the Tymovirales. Viruses 2018, 10, 406. https://doi.org/10.3390/v10080406
Thekke-Veetil T, Ho T, Postman JD, Martin RR, Tzanetakis IE. A Virus in American Blackcurrant (Ribes americanum) with Distinct Genome Features Reshapes Classification in the Tymovirales. Viruses. 2018; 10(8):406. https://doi.org/10.3390/v10080406
Chicago/Turabian StyleThekke-Veetil, Thanuja, Thien Ho, Joseph D. Postman, Robert R. Martin, and Ioannis E. Tzanetakis. 2018. "A Virus in American Blackcurrant (Ribes americanum) with Distinct Genome Features Reshapes Classification in the Tymovirales" Viruses 10, no. 8: 406. https://doi.org/10.3390/v10080406
APA StyleThekke-Veetil, T., Ho, T., Postman, J. D., Martin, R. R., & Tzanetakis, I. E. (2018). A Virus in American Blackcurrant (Ribes americanum) with Distinct Genome Features Reshapes Classification in the Tymovirales. Viruses, 10(8), 406. https://doi.org/10.3390/v10080406





