Phage-Derived Peptidoglycan Degrading Enzymes: Challenges and Future Prospects for In Vivo Therapy
Abstract
1. The Antibiotic Resistance Crisis
2. Peptidoglycan-Degrading Enzymes—An Emerging Class of Antibacterial Agents
2.1. Biology and Diversity
2.2. Enzybiotics Lytic Activity and Efficacy
2.3. Recent Developments for Endolysins Targeting Gram-Negative Bacteria
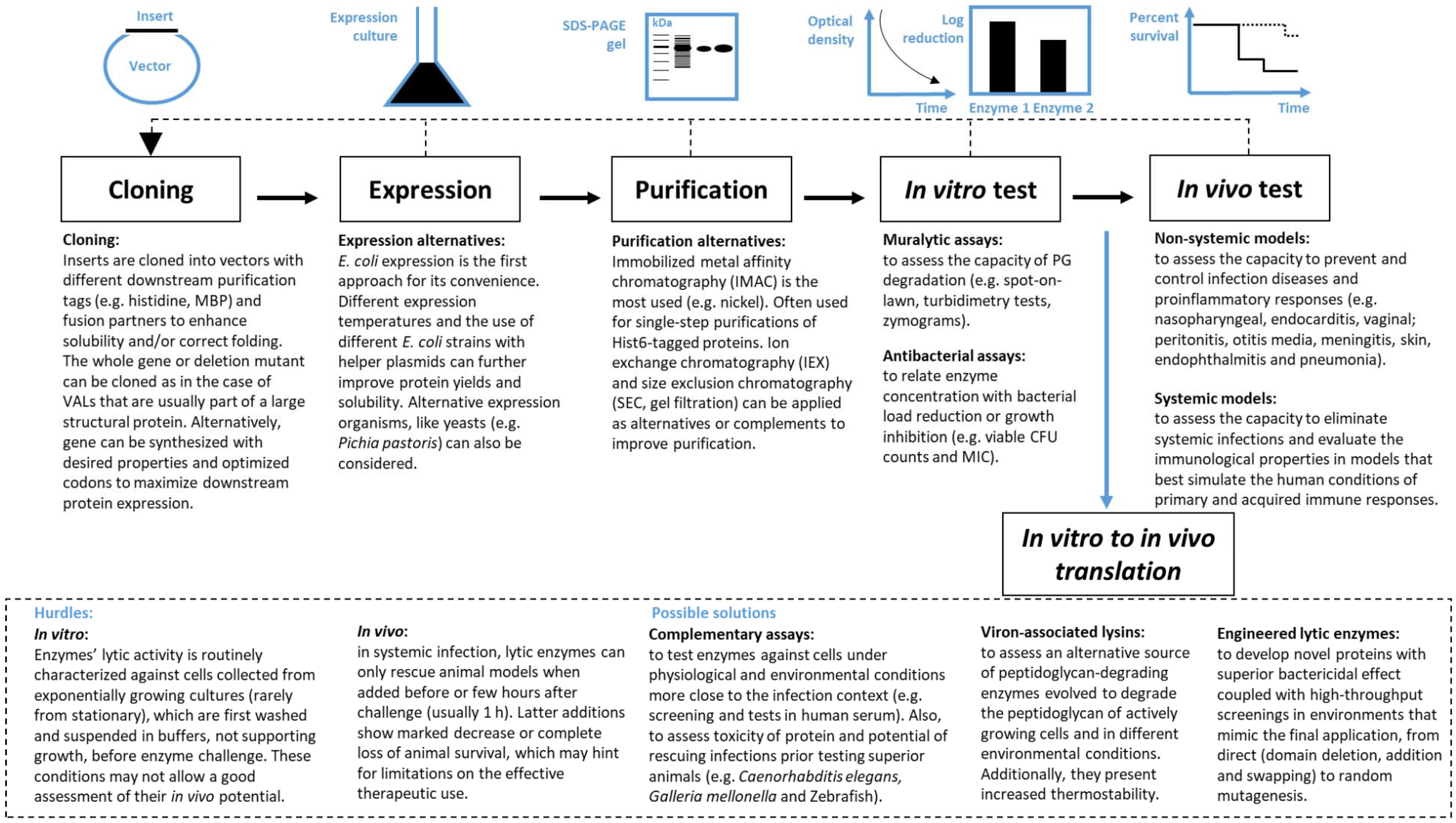
3. From Discovery to Therapeutic Application
3.1. In Vitro Performance Evaluation
3.2. In Vivo Performance Evaluation
4. In Vitro to in Vivo Translation
4.1. Hurdles
4.2. Possible Solutions
4.2.1. Complementary Screening Tests to Search for the Best Enzybiotic
4.2.2. Using VALs
4.2.3. Engineered Lytic Enzymes
5. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory tract infections in 195 countries: A systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect. Dis. 2017, 17, 1133–1161. [CrossRef]
- Heron, M. Deaths: Leading Causes for 2015. Natl. Vital Stat. Rap. 2017, 66, 1–76. [Google Scholar]
- Centers for Disease Control and Prevention (CDC). 2015. Available online: https://www.cdc.gov/drugresistance/pdf/national_action_plan_for_combating_antibotic-resistant_bacteria.pdf (accessed on 1 March 2017).
- World Health Organization (WHO). 2015. Available online: http://www.wpro.who.int/entity/drug_resistance/resources/global_action_plan_eng.pdf (accessed on 1 March 2017).
- Laxminarayan, R.; Duse, A.; Wattal, C.; Zaidi, A.K.; Wertheim, H.F.; Sumpradit, N.; Vlieghe, E.; Hara, G.L.; Gould, I.M.; Goossens, H.; et al. Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 2013, 13, 1057–1098. [Google Scholar] [CrossRef]
- Magill, S.S.; Edwards, J.R.; Bamberg, W.; Beldavs, Z.G.; Dumyati, G.; Kainer, M.A.; Lynfield, R.; Maloney, M.; McAllister-Hollod, L.; Nadle, J.; et al. Multistate point-prevalence survey of health care-associated infections. N. Engl. J. Med. 2014, 370, 1198–1208. [Google Scholar] [CrossRef] [PubMed]
- Klevens, R.M.; Edwards, J.R.; Richards, C.L., Jr.; Horan, T.C.; Gaynes, R.P.; Pollock, D.A.; Cardo, D.M. Estimating health care-associated infections and deaths in US Hospitals, 2002. Public Health Rep. 2007, 122, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.C.; Schmelcher, M.; Rodriguez-Rubio, L.; Klumpp, J.; Pritchard, D.G.; Dong, S.; Donovan, D.M. Endolysins as antimicrobials. Adv. Virus Res. 2012, 83, 299–365. [Google Scholar] [PubMed]
- Schmelcher, M.; Donovan, D.M.; Loessner, M.J. Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 2012, 7, 1147–1171. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.; Loomis, L.; Fischetti, V.A. Prevention and elimination of upper respiratory colonization of mice by group a streptococci by using a bacteriophage lytic enzyme. Proc. Natl. Acad. Sci. USA 2001, 98, 4107–4112. [Google Scholar] [CrossRef] [PubMed]
- Latka, A.; Maciejewska, B.; Majkowska-Skrobek, G.; Briers, Y.; Drulis-Kawa, Z. Bacteriophage-encoded virion-associated enzymes to overcome the carbohydrate barriers during the infection process. Appl. Microbiol. Biotechnol. 2017, 101, 3103–3119. [Google Scholar] [CrossRef] [PubMed]
- Moak, M.; Molineux, I.J. Peptidoglycan hydrolytic activities associated with bacteriophage virions. Mol. Microbiol. 2004, 51, 1169–1183. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Rubio, L.; Quiles-Puchalt, N.; Martinez, B.; Rodriguez, A.; Penades, J.R.; Garcia, P. The peptidoglycan hydrolase of Staphylococcus aureus bacteriophage 11 plays a structural role in the viral particle. Appl. Environ. Microbiol. 2013, 79, 6187–6190. [Google Scholar] [CrossRef] [PubMed]
- Moak, M.; Molineux, I.J. Role of the GP16 lytic transglycosylase motif in bacteriophage T7 virions at the initiation of infection. Mol. Microbiol. 2000, 37, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Young, R. Phage lysis: Do we have the hole story yet? Curr. Opin. Microbiol. 2013, 16, 790–797. [Google Scholar] [CrossRef] [PubMed]
- Catalao, M.J.; Gil, F.; Moniz-Pereira, J.; Sao-Jose, C.; Pimentel, M. Diversity in bacterial lysis systems: Bacteriophages show the way. FEMS Microbiol. Rev. 2013, 37, 554–571. [Google Scholar] [CrossRef] [PubMed]
- Vollmer, W.; Blanot, D.; de Pedro, M.A. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 2008, 32, 149–167. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, H.; Melo, L.D.; Santos, S.B.; Nobrega, F.L.; Ferreira, E.C.; Cerca, N.; Azeredo, J.; Kluskens, L.D. Molecular aspects and comparative genomics of bacteriophage endolysins. J. Virol. 2013, 87, 4558–4570. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, H.; Azeredo, J.; Lavigne, R.; Kluskens, L.D. Bacteriophage endolysins as a response to emerging foodborne pathogens. Trends Food Sci. Technol. 2012, 28, 103–115. [Google Scholar] [CrossRef]
- Payne, K.M.; Hatfull, G.F. Mycobacteriophage endolysins: Diverse and modular enzymes with multiple catalytic activities. PLoS ONE 2012, 7, e34052. [Google Scholar] [CrossRef] [PubMed]
- Walmagh, M.; Briers, Y.; dos Santos, S.B.; Azeredo, J.; Lavigne, R. Characterization of modular bacteriophage endolysins from Myoviridae phages OBP, 201phi2-1 and PVP-SE1. PLoS ONE 2012, 7, e36991. [Google Scholar] [CrossRef] [PubMed]
- Briers, Y.; Volckaert, G.; Cornelissen, A.; Lagaert, S.; Michiels, C.W.; Hertveldt, K.; Lavigne, R. Muralytic activity and modular structure of the endolysins of Pseudomonas aeruginosa bacteriophages phiKZ and EL. Mol. Microbiol. 2007, 65, 1334–1344. [Google Scholar] [CrossRef] [PubMed]
- Loessner, M.J.; Kramer, K.; Ebel, F.; Scherer, S. C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol. Microbiol. 2002, 44, 335–349. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.; Flint, S.H.; Yu, P.L. Determination of the mode of action of enterolysin A, produced by Enterococcus faecalis B9510. J. Appl. Microbiol. 2013, 115, 484–494. [Google Scholar] [CrossRef] [PubMed]
- Thumm, G.; Gotz, F. Studies on prolysostaphin processing and characterization of the lysostaphin immunity factor (Lif) of Staphylococcus simulans biovar staphylolyticus. Mol. Microbiol. 1997, 23, 1251–1265. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Rubio, L.; Martinez, B.; Donovan, D.M.; Rodriguez, A.; Garcia, P. Bacteriophage virion-associated peptidoglycan hydrolases: Potential new enzybiotics. Crit. Rev. Microbiol. 2013, 39, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Nishima, W.; Kanamaru, S.; Arisaka, F.; Kitao, A. Screw motion regulates multiple functions of T4 phage protein gene product 5 during cell puncturing. J. Am. Chem. Soc. 2011, 133, 13571–13576. [Google Scholar] [CrossRef] [PubMed]
- Lavigne, R.; Noben, J.P.; Hertveldt, K.; Ceyssens, P.J.; Briers, Y.; Dumont, D.; Roucourt, B.; Krylov, V.N.; Mesyanzhinov, V.V.; Robben, J.; et al. The structural proteome of Pseudomonas aeruginosa bacteriophage phiKMV. Microbiology 2006, 152, 529–534. [Google Scholar] [CrossRef] [PubMed]
- Delbruck, M. The growth of bacteriophage and lysis of the host. J. Gen. Physiol. 1940, 23, 643–660. [Google Scholar] [CrossRef] [PubMed]
- Stockdale, S.R.; Mahony, J.; Courtin, P.; Chapot-Chartier, M.P.; van Pijkeren, J.P.; Britton, R.A.; Neve, H.; Heller, K.J.; Aideh, B.; Vogensen, F.K.; et al. The lactococcal phages Tuc2009 and TP901-1 incorporate two alternate forms of their tail fiber into their virions for infection specialization. J. Biol. Chem. 2013, 288, 5581–5590. [Google Scholar] [CrossRef] [PubMed]
- Cohen, D.N.; Sham, Y.Y.; Haugstad, G.D.; Xiang, Y.; Rossmann, M.G.; Anderson, D.L.; Popham, D.L. Shared catalysis in virus entry and bacterial cell wall depolymerization. J. Mol. Biol. 2009, 387, 607–618. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, L.; Martinez, B.; Zhou, Y.; Rodriguez, A.; Donovan, D.M.; Garcia, P. Lytic activity of the virion-associated peptidoglycan hydrolase HydH5 of Staphylococcus aureus bacteriophage vb_SauS-phiiPLA88. BMC Microbiol. 2011, 11, 138. [Google Scholar] [CrossRef] [PubMed]
- Takac, M.; Blasi, U. Phage P68 virion-associated protein 17 displays activity against clinical isolates of Staphylococcus aureus. Antimicrob. Agents Chemother. 2005, 49, 2934–2940. [Google Scholar] [CrossRef] [PubMed]
- Rashel, M.; Uchiyama, J.; Takemura, I.; Hoshiba, H.; Ujihara, T.; Takatsuji, H.; Honke, K.; Matsuzaki, S. Tail-associated structural protein gp61 of Staphylococcus aureus phage phi MR11 has bifunctional lytic activity. FEMS Microbiol. Lett. 2008, 284, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Caldentey, J.; Bamford, D.H. The lytic enzyme of the Pseudomonas phage phi 6. Purification and biochemical characterization. Biochim. Biophys. Acta 1992, 1159, 44–50. [Google Scholar] [CrossRef]
- Briers, Y.; Lavigne, R.; Plessers, P.; Hertveldt, K.; Hanssens, I.; Engelborghs, Y.; Volckaert, G. Stability analysis of the bacteriophage phiKMV lysin gp36C and its putative role during infection. Cell. Mol. Life Sci. 2006, 63, 1899–1905. [Google Scholar] [CrossRef] [PubMed]
- Briers, Y.; Miroshnikov, K.; Chertkov, O.; Nekrasov, A.; Mesyanzhinov, V.; Volckaert, G.; Lavigne, R. The structural peptidoglycan hydrolase gp181 of bacteriophage phiKZ. Biochem. Biophys. Res. Commun. 2008, 374, 747–751. [Google Scholar] [CrossRef] [PubMed]
- Schleifer, K.H.; Kandler, O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 1972, 36, 407–477. [Google Scholar] [PubMed]
- Rodriguez-Rubio, L.; Martinez, B.; Rodriguez, A.; Donovan, D.M.; Garcia, P. Enhanced staphylolytic activity of the Staphylococcus aureus bacteriophage vB_SauS-phiiPLA88 HydH5 virion-associated peptidoglycan hydrolase: Fusions, deletions, and synergy with lysh5. Appl. Environ. Microbiol. 2012, 78, 2241–2248. [Google Scholar] [CrossRef] [PubMed]
- Poonacha, N.; Nair, S.; Desai, S.; Tuppad, D.; Hiremath, D.; Mohan, T.; Vipra, A.; Sharma, U. Efficient killing of planktonic and biofilm-embedded coagulase-negative staphylococci by bactericidal protein P128. Antimicrob. Agents Chemother. 2017, 61, e00457-17. [Google Scholar] [CrossRef] [PubMed]
- Manoharadas, S.; Witte, A.; Blasi, U. Antimicrobial activity of a chimeric enzybiotic towards Staphylococcus aureus. J. Biotechnol. 2009, 139, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Saravanan, S.R.; Paul, V.D.; George, S.; Sundarrajan, S.; Kumar, N.; Hebbur, M.; Veena, A.; Maheshwari, U.; Appaiah, C.B.; Chidambaran, M.; et al. Properties and mutation studies of a bacteriophage-derived chimeric recombinant staphylolytic protein p128: Comparison to recombinant lysostaphin. Bacteriophage 2013, 3, e26564. [Google Scholar] [CrossRef] [PubMed]
- Proenca, D.; Leandro, C.; Garcia, M.; Pimentel, M.; Sao-Jose, C. Ec300: A phage-based, bacteriolysin-like protein with enhanced antibacterial activity against Enterococcus faecalis. Appl. Microbiol. Biotechnol. 2015, 99, 5137–5149. [Google Scholar] [CrossRef] [PubMed]
- Freimer, E.H.; Krause, R.M.; Mc, C.M. Studies of l forms and protoplasts of group A streptococci. I. Isolation, growth, and bacteriologic characteristics. J. Exp. Med. 1959, 110, 853–874. [Google Scholar] [CrossRef] [PubMed]
- Rashel, M.; Uchiyama, J.; Ujihara, T.; Uehara, Y.; Kuramoto, S.; Sugihara, S.; Yagyu, K.; Muraoka, A.; Sugai, M.; Hiramatsu, K.; et al. Efficient elimination of multidrug-resistant Staphylococcus aureus by cloned lysin derived from bacteriophage phi MR11. J. Infect. Dis. 2007, 196, 1237–1247. [Google Scholar] [CrossRef] [PubMed]
- O'Flaherty, S.; Coffey, A.; Meaney, W.; Fitzgerald, G.F.; Ross, R.P. The recombinant phage lysin lysk has a broad spectrum of lytic activity against clinically relevant staphylococci, including methicillin-resistant Staphylococcus aureus. J. Bacteriol. 2005, 187, 7161–7164. [Google Scholar] [CrossRef] [PubMed]
- Yoong, P.; Schuch, R.; Nelson, D.; Fischetti, V.A. Identification of a broadly active phage lytic enzyme with lethal activity against antibiotic-resistant Enterococcus faecalis and Enterococcus faecium. J. Bacteriol. 2004, 186, 4808–4812. [Google Scholar] [CrossRef] [PubMed]
- Jado, I.; Lopez, R.; Garcia, E.; Fenoll, A.; Casal, J.; Garcia, P. Phage lytic enzymes as therapy for antibiotic-resistant Streptococcus pneumoniae infection in a murine sepsis model. J. Antimicrob. Chemother. 2003, 52, 967–973. [Google Scholar] [CrossRef] [PubMed]
- Djurkovic, S.; Loeffler, J.M.; Fischetti, V.A. Synergistic killing of Streptococcus pneumoniae with the bacteriophage lytic enzyme Cpl-1 and penicillin or gentamicin depends on the level of penicillin resistance. Antimicrob. Agents Chemother. 2005, 49, 1225–1228. [Google Scholar] [CrossRef] [PubMed]
- Gerstmans, H.; Criel, B.; Briers, Y. Synthetic biology of modular endolysins. Biotechnol. Adv. 2018, 36, 624–640. [Google Scholar] [CrossRef] [PubMed]
- Sao-Jose, C. Engineering of phage-derived lytic enzymes: Improving their potential as antimicrobials. Antibiotics 2018, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- Diaz, E.; Lopez, R.; Garcia, J.L. Chimeric phage-bacterial enzymes: A clue to the modular evolution of genes. Proc. Natl. Acad. Sci. USA 1990, 87, 8125–8129. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, S.; Proenca, D.; Cantante, C.; Silva, F.A.; Leandro, C.; Lourenco, S.; Milheirico, C.; de Lencastre, H.; Cavaco-Silva, P.; Pimentel, M.; et al. Novel chimerical endolysins with broad antimicrobial activity against methicillin-resistant Staphylococcus aureus. Microb. Drug Resist. 2012, 18, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Daniel, A.; Euler, C.; Collin, M.; Chahales, P.; Gorelick, K.J.; Fischetti, V.A. Synergism between a novel chimeric lysin and oxacillin protects against infection by methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2010, 54, 1603–1612. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Fischetti, V.A. Mutagenesis of a bacteriophage lytic enzyme plyGBS significantly increases its antibacterial activity against group B streptococci. Appl. Microbiol. Biotechnol. 2007, 74, 1284–1291. [Google Scholar] [CrossRef] [PubMed]
- Horgan, M.; O'Flynn, G.; Garry, J.; Cooney, J.; Coffey, A.; Fitzgerald, G.F.; Ross, R.P.; McAuliffe, O. Phage lysin lysk can be truncated to its CHAP domain and retain lytic activity against live antibiotic-resistant staphylococci. Appl. Environ. Microbiol. 2009, 75, 872–874. [Google Scholar] [CrossRef] [PubMed]
- Low, L.Y.; Yang, C.; Perego, M.; Osterman, A.; Liddington, R.C. Structure and lytic activity of a Bacillus anthracis prophage endolysin. J. Biol. Chem. 2005, 280, 35433–35439. [Google Scholar] [CrossRef] [PubMed]
- Low, L.Y.; Yang, C.; Perego, M.; Osterman, A.; Liddington, R. Role of net charge on catalytic domain and influence of cell wall binding domain on bactericidal activity, specificity, and host range of phage lysins. J. Biol. Chem. 2011, 286, 34391–34403. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.J.; Garefalaki, V.; Spoerl, R.; Narbad, A.; Meijers, R. Structure-based modification of a Clostridium difficile-targeting endolysin affects activity and host range. J. Bacteriol. 2011, 193, 5477–5486. [Google Scholar] [CrossRef] [PubMed]
- Schmelcher, M.; Tchang, V.S.; Loessner, M.J. Domain shuffling and module engineering of Listeria phage endolysins for enhanced lytic activity and binding affinity. Microb. Biotechnol. 2011, 4, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Fischetti, V.A. Bacteriophage endolysins: A novel anti-infective to control Gram-positive pathogens. Int. J. Med. Microbiol. 2010, 300, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Morita, M.; Tanji, Y.; Orito, Y.; Mizoguchi, K.; Soejima, A.; Unno, H. Functional analysis of antibacterial activity of bacillus amyloliquefaciens phage endolysin against Gram-negative bacteria. FEBS Lett. 2001, 500, 56–59. [Google Scholar] [CrossRef]
- During, K.; Porsch, P.; Mahn, A.; Brinkmann, O.; Gieffers, W. The non-enzymatic microbicidal activity of lysozymes. FEBS Lett. 1999, 449, 93–100. [Google Scholar] [CrossRef]
- Oliveira, H.; Vilas Boas, D.; Mesnage, S.; Kluskens, L.D.; Lavigne, R.; Sillankorva, S.; Secundo, F.; Azeredo, J. Structural and enzymatic characterization of ABgp46, a novel phage endolysin with broad anti-gram-negative bacterial activity. Front. Microbiol. 2016, 7, 208. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, H.; Thiagarajan, V.; Walmagh, M.; Sillankorva, S.; Lavigne, R.; Neves-Petersen, M.T.; Kluskens, L.D.; Azeredo, J. A thermostable Salmonella phage endolysin, Lys68, with broad bactericidal properties against gram-negative pathogens in presence of weak acids. PLoS ONE 2014, 9, e108376. [Google Scholar] [CrossRef] [PubMed]
- Briers, Y.; Walmagh, M.; Lavigne, R. Use of bacteriophage endolysin EL188 and outer membrane permeabilizers against Pseudomonas aeruginosa. J. Appl. Microbiol. 2011, 110, 778–785. [Google Scholar] [CrossRef] [PubMed]
- Briers, Y.; Walmagh, M.; Van Puyenbroeck, V.; Cornelissen, A.; Cenens, W.; Aertsen, A.; Oliveira, H.; Azeredo, J.; Verween, G.; Pirnay, J.P.; et al. Engineered endolysin-based “Artilysins” to combat multidrug-resistant gram-negative pathogens. MBio 2014, 5, e01379-14. [Google Scholar] [CrossRef] [PubMed]
- Briers, Y.; Walmagh, M.; Grymonprez, B.; Biebl, M.; Pirnay, J.P.; Defraine, V.; Michiels, J.; Cenens, W.; Aertsen, A.; Miller, S.; et al. Art-175 is a highly efficient antibacterial against multidrug-resistant strains and persisters of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2014, 58, 3774–3784. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Becker, S.C.; Dong, S.; Baker, J.R.; Foster-Frey, J.; Pritchard, D.G.; Donovan, D.M. LysK CHAP endopeptidase domain is required for lysis of live staphylococcal cells. FEMS Microbiol. Lett. 2009, 294, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Lavigne, R.; Briers, Y.; Hertveldt, K.; Robben, J.; Volckaert, G. Identification and characterization of a highly thermostable bacteriophage lysozyme. Cell. Mol. Life Sci. 2004, 61, 2753–2759. [Google Scholar] [CrossRef] [PubMed]
- Briers, Y.; Lavigne, R.; Volckaert, G.; Hertveldt, K. A standardized approach for accurate quantification of murein hydrolase activity in high-throughput assays. J. Biochem. Biophys. Methods 2007, 70, 531–533. [Google Scholar] [CrossRef] [PubMed]
- Loeffler, J.M.; Djurkovic, S.; Fischetti, V.A. Phage lytic enzyme Cpl-1 as a novel antimicrobial for pneumococcal bacteremia. Infect. Immun. 2003, 71, 6199–6204. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, A.; Leite, M.; Kluskens, L.D.; Santos, S.B.; Melo, L.D.; Azeredo, J. The first Paenibacillus larvae bacteriophage endolysin (PlyPl23) with high potential to control american foulbrood. PLoS ONE 2015, 10, e0132095. [Google Scholar]
- Shen, Y.; Koller, T.; Kreikemeyer, B.; Nelson, D.C. Rapid degradation of Streptococcus pyogenes biofilms by PlyC, a bacteriophage-encoded endolysin. J. Antimicrob. Chemother. 2013, 68, 1818–1824. [Google Scholar] [CrossRef] [PubMed]
- Ceri, H.; Olson, M.E.; Stremick, C.; Read, R.R.; Morck, D.; Buret, A. The Calgary Biofilm Device: New technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 1999, 37, 1771–1776. [Google Scholar] [PubMed]
- Gilmer, D.B.; Schmitz, J.E.; Thandar, M.; Euler, C.W.; Fischetti, V.A. The Phage lysin PlySs2 decolonizes streptococcus suis from murine intranasal mucosa. PLoS ONE 2017, 12, e0169180. [Google Scholar] [CrossRef] [PubMed]
- Díez-Martínez, R.; De Paz, H.D.; Garcia-Fernandez, E.; Bustamante, N.; Euler, C.W.; Fischetti, V.A.; Menendez, M.; Garcia, P. A novel chimeric phage lysin with high in vitro and in vivo bactericidal activity against Streptococcus pneumoniae. J. Antimicrob. Chemother. 2015, 70, 1763–1773. [Google Scholar] [CrossRef] [PubMed]
- Abaev, I.; Foster-Frey, J.; Korobova, O.; Shishkova, N.; Kiseleva, N.; Kopylov, P.; Pryamchuk, S.; Schmelcher, M.; Becker, S.C.; Donovan, D.M. Staphylococcal Phage 2638A endolysin is lytic for Staphylococcus aureus and harbors an inter-lytic-domain secondary translational start site. Appl. Microbiol. Biotechnol. 2013, 97, 3449–3456. [Google Scholar] [CrossRef] [PubMed]
- Becker, S.C.; Swift, S.; Korobova, O.; Schischkova, N.; Kopylov, P.; Donovan, D.M.; Abaev, I. Lytic activity of the staphylolytic Twort phage endolysin CHAP domain is enhanced by the SH3b cell wall binding domain. FEMS Microbiol. Lett. 2015, 362. [Google Scholar] [CrossRef] [PubMed]
- Loeffler, J.M.; Nelson, D.; Fischetti, V.A. Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science 2001, 294, 2170–2172. [Google Scholar] [CrossRef] [PubMed]
- Corsini, B.; Diez-Martinez, R.; Aguinagalde, L.; Gonzalez-Camacho, F.; Garcia-Fernandez, E.; Letrado, P.; Garcia, P.; Yuste, J. Chemotherapy with phage lysins reduces pneumococcal colonization of the respiratory tract. Antimicrob. Agents Chemother. 2018. [Google Scholar] [CrossRef] [PubMed]
- Doehn, J.M.; Fischer, K.; Reppe, K.; Gutbier, B.; Tschernig, T.; Hocke, A.C.; Fischetti, V.A.; Loffler, J.; Suttorp, N.; Hippenstiel, S.; et al. Delivery of the endolysin Cpl-1 by inhalation rescues mice with fatal pneumococcal pneumonia. J. Antimicrob. Chemother. 2013, 68, 2111–2117. [Google Scholar] [CrossRef] [PubMed]
- Witzenrath, M.; Schmeck, B.; Doehn, J.M.; Tschernig, T.; Zahlten, J.; Loeffler, J.M.; Zemlin, M.; Muller, H.; Gutbier, B.; Schutte, H.; et al. Systemic use of the endolysin Cpl-1 rescues mice with fatal pneumococcal pneumonia. Crit. Care Med. 2009, 37, 642–649. [Google Scholar] [CrossRef] [PubMed]
- McCullers, J.A.; Karlstrom, A.; Iverson, A.R.; Loeffler, J.M.; Fischetti, V.A. Novel strategy to prevent otitis media caused by colonizing Streptococcus pneumoniae. PLoS Pathog. 2007, 3, e28. [Google Scholar] [CrossRef] [PubMed]
- Grandgirard, D.; Loeffler, J.M.; Fischetti, V.A.; Leib, S.L. Phage lytic enzyme Cpl-1 for antibacterial therapy in experimental pneumococcal meningitis. J. Infect. Dis. 2008, 197, 1519–1522. [Google Scholar] [CrossRef] [PubMed]
- Entenza, J.M.; Loeffler, J.M.; Grandgirard, D.; Fischetti, V.A.; Moreillon, P. Therapeutic effects of bacteriophage Cpl-1 lysin against Streptococcus pneumoniae endocarditis in rats. Antimicrob. Agents Chemother. 2005, 49, 4789–4792. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Nelson, D.; Zhu, S.; Fischetti, V.A. Removal of group B streptococci colonizing the vagina and oropharynx of mice with a bacteriophage lytic enzyme. Antimicrob. Agents Chemother. 2005, 49, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Vouillamoz, J.; Entenza, J.M.; Giddey, M.; Fischetti, V.A.; Moreillon, P.; Resch, G. Bactericidal synergism between daptomycin and the phage lysin Cpl-1 in a mouse model of pneumococcal bacteraemia. Int. J. Antimicrob. Agents 2013, 42, 416–421. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Li, D.; Wang, H.; Ma, Z.; Lu, C.; Dai, J. Prophage lysin Ply30 protects mice from Streptococcus suis and Streptococcus equi subsp. Zooepidemicus infections. Appl. Environ. Microbiol. 2015, 81, 7377–7384. [Google Scholar] [CrossRef] [PubMed]
- Lood, R.; Raz, A.; Molina, H.; Euler, C.W.; Fischetti, V.A. A highly active and negatively charged Streptococcus pyogenes lysin with a rare D-alanyl-L-alanine endopeptidase activity protects mice against streptococcal bacteremia. Antimicrob. Agents Chemother. 2014, 58, 3073–3084. [Google Scholar] [CrossRef] [PubMed]
- Oechslin, F.; Daraspe, J.; Giddey, M.; Moreillon, P.; Resch, G. In vitro characterization of PlySK1249, a novel phage lysin, and assessment of its antibacterial activity in a mouse model of streptococcus agalactiae bacteremia. Antimicrob. Agents Chemother. 2013, 57, 6276–6283. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Linden, S.B.; Wang, J.; Yu, J.; Nelson, D.C.; Wei, H. A chimeolysin with extended-spectrum streptococcal host range found by an induced lysis-based rapid screening method. Sci. Rep. 2015, 5, 17257. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.K.; Donovan, D.M.; Kumar, A. Intravitreal injection of the chimeric phage endolysin ply187 protects mice from Staphylococcus aureus endophthalmitis. Antimicrob. Agents Chemother. 2014, 58, 4621–4629. [Google Scholar] [CrossRef] [PubMed]
- Becker, S.C.; Roach, D.R.; Chauhan, V.S.; Shen, Y.; Foster-Frey, J.; Powell, A.M.; Bauchan, G.; Lease, R.A.; Mohammadi, H.; Harty, W.J.; et al. Triple-acting lytic enzyme treatment of drug-resistant and intracellular Staphylococcus aureus. Sci. Rep. 2016, 6, 25063. [Google Scholar] [CrossRef] [PubMed]
- Fenton, M.; Casey, P.G.; Hill, C.; Gahan, C.G.; Ross, R.P.; McAuliffe, O.; O'Mahony, J.; Maher, F.; Coffey, A. The truncated phage lysin CHAP(k) eliminates Staphylococcus aureus in the nares of mice. Bioeng. Bugs 2010, 1, 404–407. [Google Scholar] [CrossRef] [PubMed]
- Paul, V.D.; Rajagopalan, S.S.; Sundarrajan, S.; George, S.E.; Asrani, J.Y.; Pillai, R.; Chikkamadaiah, R.; Durgaiah, M.; Sriram, B.; Padmanabhan, S. A novel bacteriophage tail-associated muralytic enzyme (TAME) from phage k and its development into a potent antistaphylococcal protein. BMC Microbiol. 2011, 11, 226. [Google Scholar] [CrossRef] [PubMed]
- Xia, F.; Li, X.; Wang, B.; Gong, P.; Xiao, F.; Yang, M.; Zhang, L.; Song, J.; Hu, L.; Cheng, M.; et al. Combination therapy of LysGH15 and apigenin as a new strategy for treating pneumonia caused by Staphylococcus aureus. Appl. Environ. Microbiol. 2015, 82, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Pastagia, M.; Euler, C.; Chahales, P.; Fuentes-Duculan, J.; Krueger, J.G.; Fischetti, V.A. A novel chimeric lysin shows superiority to mupirocin for skin decolonization of methicillin-resistant and -sensitive Staphylococcus aureus strains. Antimicrob. Agents Chemother. 2011, 55, 738–744. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Kong, L.; Liu, Y.; Fu, Q.; Cui, Z.; Wang, J.; Ma, J.; Wang, H.; Yan, Y.; Sun, J. A cell-penetrating peptide fused phage lysin kills intracellular MRSA in keratinocytes and treatment for skin infections of mice. Appl. Environ. Microbiol. 2018, 84. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhang, Y.; Yu, J.; Huang, Y.; Zhang, X.E.; Wei, H. Novel chimeric lysin with high-level antimicrobial activity against methicillin-resistant Staphylococcus aureus in vitro and in vivo. Antimicrob. Agents Chemother. 2014, 58, 536–542. [Google Scholar] [CrossRef] [PubMed]
- Chopra, S.; Harjai, K.; Chhibber, S. Potential of combination therapy of endolysin MR-10 and minocycline in treating MRSA induced systemic and localized burn wound infections in mice. Int. J. Med. Microbiol. 2016, 306, 707–716. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Xu, W.; Lei, L.; Huang, J.; Feng, X.; Sun, C.; Du, C.; Zuo, J.; Li, Y.; Du, T.; et al. Lysgh15, a novel bacteriophage lysin, protects a murine bacteremia model efficiently against lethal methicillin-resistant Staphylococcus aureus infection. J. Clin. Microbiol. 2011, 49, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Schuch, R.; Lee, H.M.; Schneider, B.C.; Sauve, K.L.; Law, C.; Khan, B.K.; Rotolo, J.A.; Horiuchi, Y.; Couto, D.E.; Raz, A.; et al. Combination therapy with lysin CF-301 and antibiotic is superior to antibiotic alone for treating methicillin-resistant Staphylococcus aureus-induced murine bacteremia. J. Infect. Dis. 2014, 209, 1469–1478. [Google Scholar] [CrossRef] [PubMed]
- Gilmer, D.B.; Schmitz, J.E.; Euler, C.W.; Fischetti, V.A. Novel bacteriophage lysin with broad lytic activity protects against mixed infection by Streptococcus pyogenes and methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2013, 57, 2743–2750. [Google Scholar] [CrossRef] [PubMed]
- Jun, S.Y.; Jung, G.M.; Yoon, S.J.; Oh, M.D.; Choi, Y.J.; Lee, W.J.; Kong, J.C.; Seol, J.G.; Kang, S.H. Antibacterial properties of a pre-formulated recombinant phage endolysin, SAL-1. Int. J. Antimicrob. Agents 2013, 41, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Schmelcher, M.; Shen, Y.; Nelson, D.C.; Eugster, M.R.; Eichenseher, F.; Hanke, D.C.; Loessner, M.J.; Dong, S.; Pritchard, D.G.; Lee, J.C.; et al. Evolutionarily distinct bacteriophage endolysins featuring conserved peptidoglycan cleavage sites protect mice from MRSA infection. J. Antimicrob. Chemother. 2015, 70, 1453–1465. [Google Scholar] [CrossRef] [PubMed]
- Yoong, P.; Schuch, R.; Nelson, D.; Fischetti, V.A. Plyph, a bacteriolytic enzyme with a broad pH range of activity and lytic action against Bacillus anthracis. J. Bacteriol. 2006, 188, 2711–2714. [Google Scholar] [CrossRef] [PubMed]
- Schuch, R.; Nelson, D.; Fischetti, V.A. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature 2002, 418, 884–889. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.; Zhang, Y.; Li, X.; Liang, J.; Hu, L.; Gong, P.; Zhang, L.; Cai, R.; Zhang, H.; Ge, J.; et al. Endolysin Lysef-P10 shows potential as an alternative treatment strategy for multidrug-resistant Enterococcus faecalis infections. Sci. Rep. 2017, 7, 10164. [Google Scholar] [CrossRef] [PubMed]
- Briers, Y.; Lavigne, R. Breaking barriers: Expansion of the use of endolysins as novel antibacterials against Gram-negative bacteria. Future Microbiol 2015, 10, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Lood, R.; Winer, B.Y.; Pelzek, A.J.; Diez-Martinez, R.; Thandar, M.; Euler, C.W.; Schuch, R.; Fischetti, V.A. Novel phage lysin capable of killing the multidrug-resistant gram-negative bacterium Acinetobacter baumannii in a mouse bacteremia model. Antimicrob. Agents Chemother. 2015, 59, 1983–1991. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.Y.; You, R.I.; Lai, M.J.; Lin, N.T.; Chen, L.K.; Chang, K.C. Highly potent antimicrobial modified peptides derived from the Acinetobacter baumannii phage endolysin LysAB2. Sci. Rep. 2017, 7, 11477. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhang, H.; Wang, J.; Yu, J.; Wei, H. A novel chimeric lysin with robust antibacterial activity against planktonic and biofilm methicillin-resistant Staphylococcus aureus. Sci. Rep. 2017, 7, 40182. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.Z.; Schmelcher, M.; Harty, W.J.; Foster-Frey, J.; Donovan, D.M. Chimeric Ply187 endolysin kills Staphylococcus aureus more effectively than the parental enzyme. FEMS Microbiol. Lett. 2013, 342, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, D.; Li, X.; Hu, L.; Cheng, M.; Xia, F.; Gong, P.; Wang, B.; Ge, J.; Zhang, H.; et al. Lysgh15 kills Staphylococcus aureus without being affected by the humoral immune response or inducing inflammation. Sci. Rep. 2016, 6, 29344. [Google Scholar] [CrossRef] [PubMed]
- Typas, A.; Banzhaf, M.; Gross, C.A.; Vollmer, W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 2011, 10, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, S.; Sao-Jose, C. More than a hole: The holin lethal function may be required to fully sensitize bacteria to the lytic action of canonical endolysins. Mol. Microbiol. 2016, 102, 92–106. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.J.; Lin, N.T.; Hu, A.; Soo, P.C.; Chen, L.K.; Chen, L.H.; Chang, K.C. Antibacterial activity of Acinetobacter baumannii phage varphiAB2 endolysin (LysAB2) against both gram-positive and gram-negative bacteria. Appl. Microbiol. Biotechnol. 2011, 90, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, H.; Pinto, G.; Oliveira, A.; Oliveira, C.; Faustino, M.A.; Briers, Y.; Domingues, L.; Azeredo, J. Characterization and genome sequencing of a Citrobacter freundii phage Cfp1 harboring a lysin active against multidrug-resistant isolates. Appl. Microbiol. Biotechnol. 2016, 100, 10543–10553. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, H.; Azeredo, J. (University of Minho, Braga, Portugal) Characterization of a A. baumannii phage endolysin. Unpublished work. 2017. [Google Scholar]
- Yang, H.; Wang, M.; Yu, J.; Wei, H. Antibacterial activity of a novel peptide-modified lysin against Acinetobacter baumannii and Pseudomonas aeruginosa. Front. Microbiol. 2015, 6, 1471. [Google Scholar] [CrossRef] [PubMed]
- Glavis-Bloom, J.; Muhammed, M.; Mylonakis, E. Of model hosts and man: Using Caenorhabditis elegans, Drosophila melanogaster and Galleria mellonella as model hosts for infectious disease research. Adv. Exp. Med. Biol. 2012, 710, 11–17. [Google Scholar] [PubMed]
- Tsai, C.J.; Loh, J.M.; Proft, T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 2016, 7, 214–229. [Google Scholar] [CrossRef] [PubMed]
- Heo, Y.J.; Lee, Y.R.; Jung, H.H.; Lee, J.; Ko, G.; Cho, Y.H. Antibacterial efficacy of phages against Pseudomonas aeruginosa infections in mice and Drosophila melanogaster. Antimicrob. Agents Chemother. 2009, 53, 2469–2474. [Google Scholar] [CrossRef] [PubMed]
- Diez-Martinez, R.; de Paz, H.D.; Bustamante, N.; Garcia, E.; Menendez, M.; Garcia, P. Improving the lethal effect of cpl-7, a pneumococcal phage lysozyme with broad bactericidal activity, by inverting the net charge of its cell wall-binding module. Antimicrob. Agents Chemother. 2013, 57, 5355–5365. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Barros, M.; Vennemann, T.; Gallagher, D.T.; Yin, Y.; Linden, S.B.; Heselpoth, R.D.; Spencer, D.J.; Donovan, D.M.; Moult, J.; et al. A bacteriophage endolysin that eliminates intracellular streptococci. Elife 2016, 5, e13152. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oliveira, H.; São-José, C.; Azeredo, J. Phage-Derived Peptidoglycan Degrading Enzymes: Challenges and Future Prospects for In Vivo Therapy. Viruses 2018, 10, 292. https://doi.org/10.3390/v10060292
Oliveira H, São-José C, Azeredo J. Phage-Derived Peptidoglycan Degrading Enzymes: Challenges and Future Prospects for In Vivo Therapy. Viruses. 2018; 10(6):292. https://doi.org/10.3390/v10060292
Chicago/Turabian StyleOliveira, Hugo, Carlos São-José, and Joana Azeredo. 2018. "Phage-Derived Peptidoglycan Degrading Enzymes: Challenges and Future Prospects for In Vivo Therapy" Viruses 10, no. 6: 292. https://doi.org/10.3390/v10060292
APA StyleOliveira, H., São-José, C., & Azeredo, J. (2018). Phage-Derived Peptidoglycan Degrading Enzymes: Challenges and Future Prospects for In Vivo Therapy. Viruses, 10(6), 292. https://doi.org/10.3390/v10060292






