Saracatinib Inhibits Middle East Respiratory Syndrome-Coronavirus Replication In Vitro
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
2.2. Chemical Compounds and Antibodies
2.3. Small-Molecule Compound Libraries
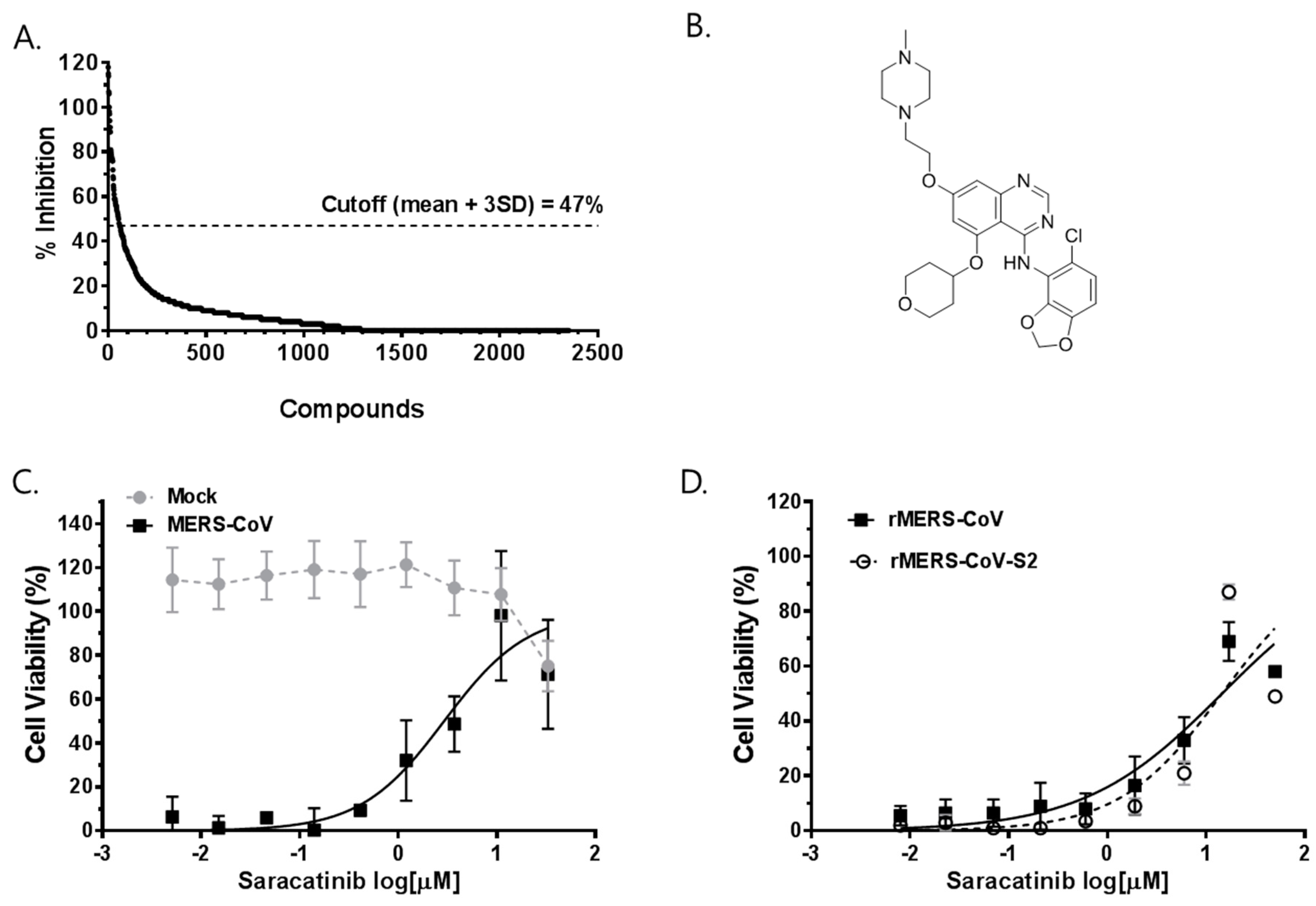
2.4. Primary Screening Assay
2.5. Time-Of-Addition Assay
2.6. Viral Plaque Assay
2.7. Quantitative Real-Time Reverse-Transcription PCR (RT-qPCR) Analysis
2.8. Western Blot Analysis
2.9. RNAi
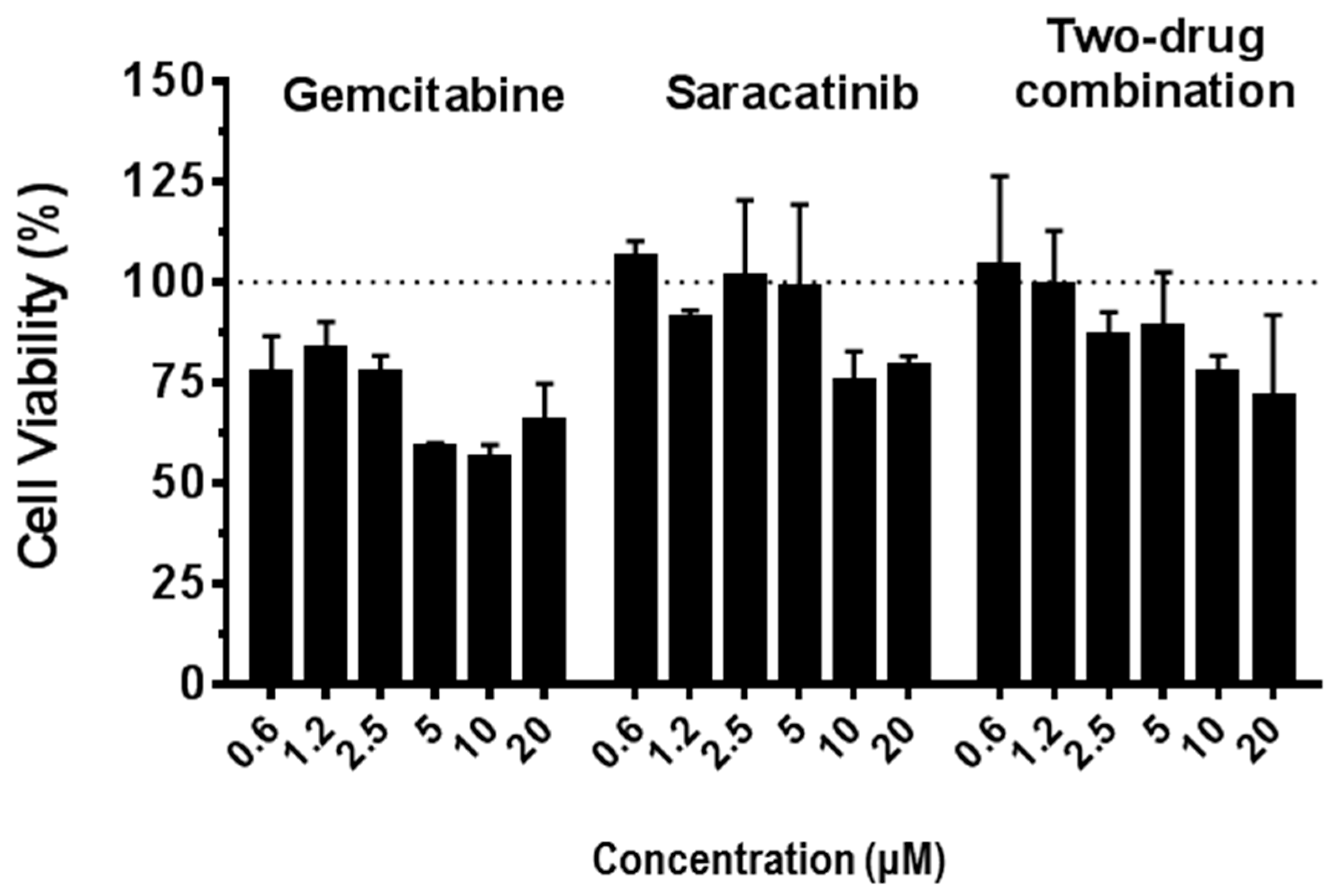
2.10. In Vitro Drug-Drug Combination Assay
3. Results
3.1. Identification of Saracatinib as an Anti-MERS-CoV Hit Compound
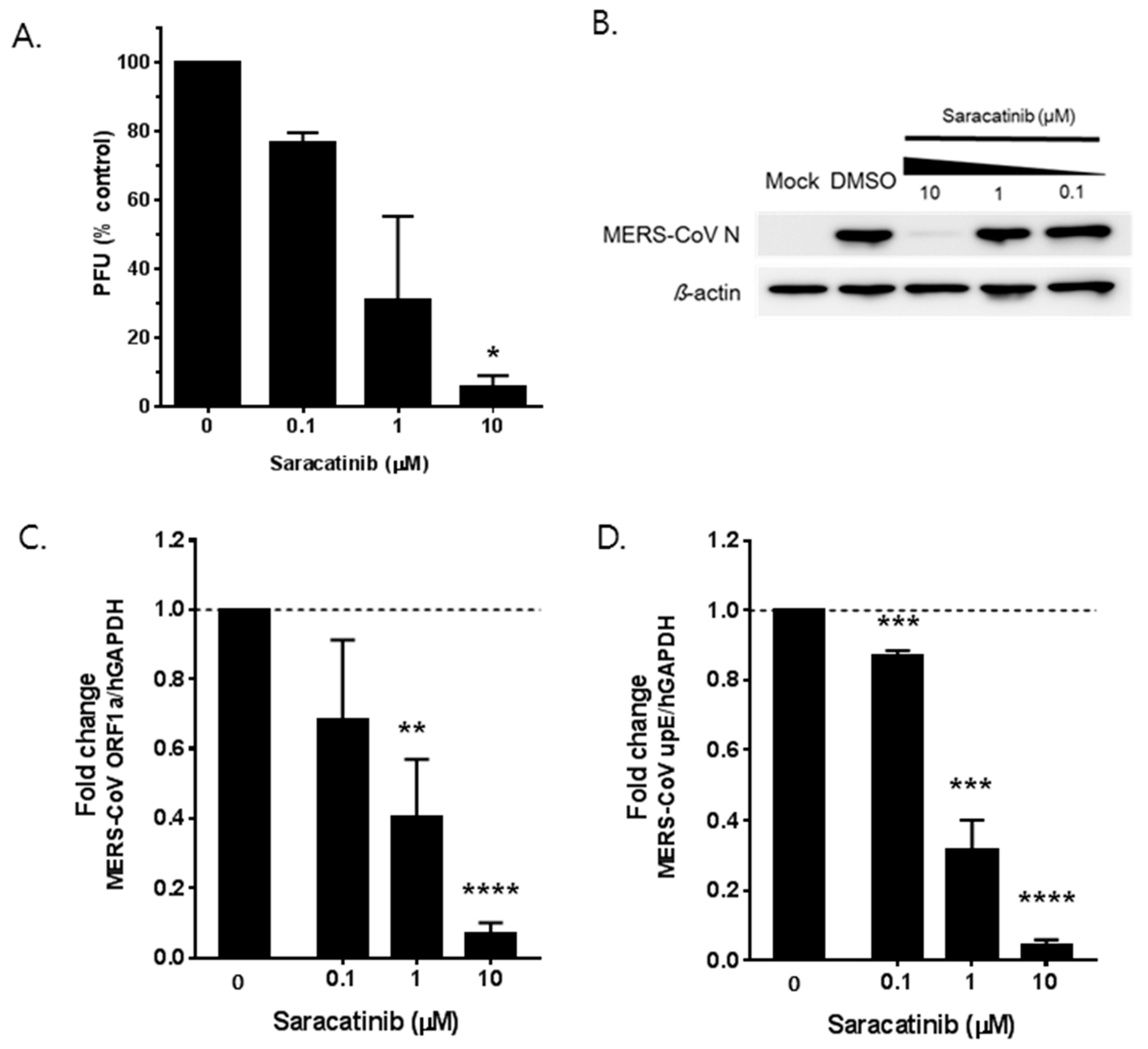
3.2. Saracatinib Inhibits MERS-CoV Replication In Vitro
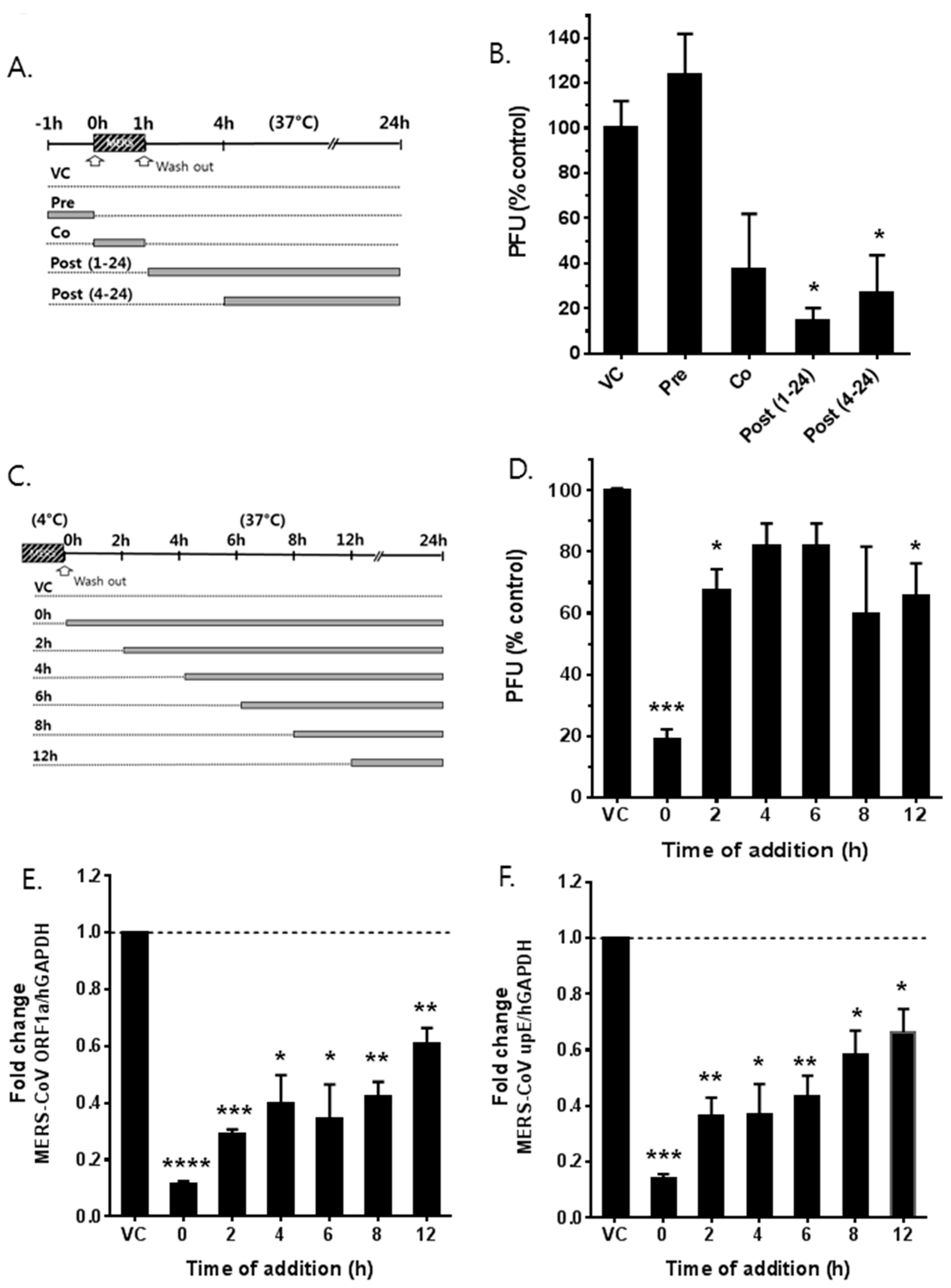
3.3. Saracatinib Inhibits the Early Stages of MERS-CoV Replication
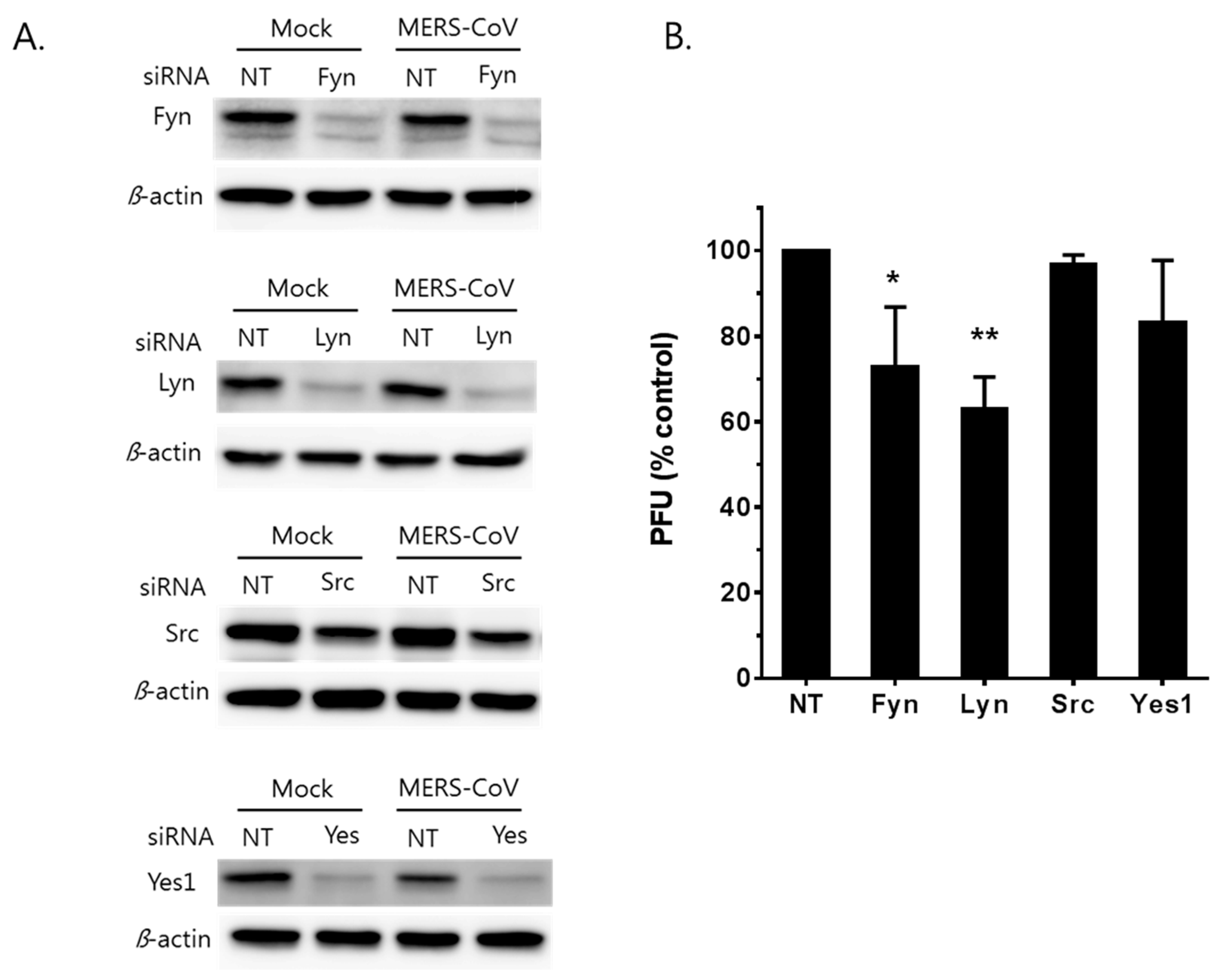
3.4. MERS-CoV Replication is Affected by Knockdown of Fyn and Lyn Kinases
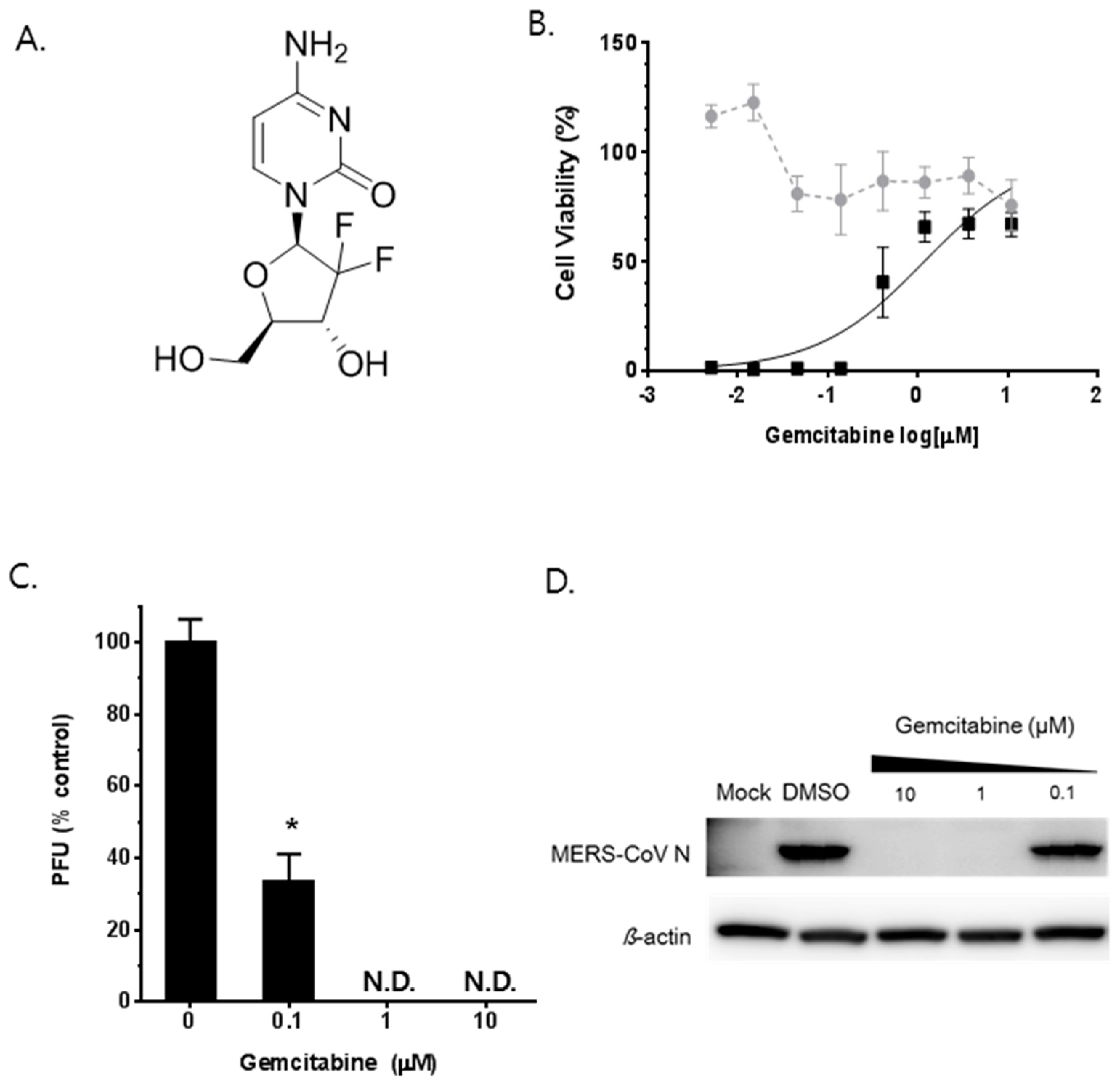
3.5. Synergistic Antiviral Effect of Saracatinib in Combination with Gemcitabine
4. Discussion
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- De Wit, E.; van Doremalen, N.; Falzarano, D.; Munster, V.J. SARS and MERS: Recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016, 14, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Zaki, A.M.; van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.; Fouchier, R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef] [PubMed]
- Zumla, A.; Chan, J.F.; Azhar, E.I.; Hui, D.S.; Yuen, K.Y. Coronaviruses—Drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016, 15, 327–347. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.; Li, K.S.; To, K.K.; Cheng, V.C.; Chen, H.; Yuen, K.Y. Is the discovery of the novel human betacoronavirus 2c EMC/2012 (HCoV-EMC) the beginning of another SARS-like pandemic? J. Infect. 2012, 65, 477–489. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.; Lau, S.K.; Woo, P.C. The emerging novel Middle East respiratory syndrome coronavirus: The “knowns” and “unknowns”. J. Formos. Med. Assoc. 2013, 112, 372–381. [Google Scholar] [CrossRef] [PubMed]
- Azhar, E.I.; El-Kafrawy, S.A.; Farraj, S.A.; Hassan, A.M.; Al-Saeed, M.S.; Hashem, A.M.; Madani, T.A. Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med. 2014, 370, 2499–2505. [Google Scholar] [CrossRef] [PubMed]
- Drosten, C.; Meyer, B.; Muller, M.A.; Corman, V.M.; Al-Masri, M.; Hossain, R.; Madani, H.; Sieberg, A.; Bosch, B.J.; Lattwein, E.; et al. Transmission of MERS-coronavirus in household contacts. N. Engl. J. Med. 2014, 371, 828–835. [Google Scholar] [CrossRef] [PubMed]
- Oboho, I.K.; Tomczyk, S.M.; Al-Asmari, A.M.; Banjar, A.A.; Al-Mugti, H.; Aloraini, M.S.; Alkhaldi, K.Z.; Almohammadi, E.L.; Alraddadi, B.M.; Gerber, S.I.; et al. 2014 MERS-CoV outbreak in Jeddah—A link to health care facilities. N. Engl. J. Med. 2015, 372, 846–854. [Google Scholar] [CrossRef] [PubMed]
- Zumla, A.; Hui, D.S.; Perlman, S. Middle East respiratory syndrome. Lancet 2015, 386, 995–1007. [Google Scholar] [CrossRef]
- Chan, J.F.; Lau, S.K.; To, K.K.; Cheng, V.C.; Woo, P.C.; Yuen, K.Y. Middle East respiratory syndrome coronavirus: Another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 2015, 28, 465–522. [Google Scholar] [CrossRef] [PubMed]
- Al-Tawfiq, J.A.; Memish, Z.A. What are our pharmacotherapeutic options for MERS-CoV? Expert Rev. Clin. Pharmacol. 2014, 7, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Ashburn, T.T.; Thor, K.B. Drug repositioning: Identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004, 3, 673–683. [Google Scholar] [CrossRef] [PubMed]
- De Wilde, A.H.; Jochmans, D.; Posthuma, C.C.; Zevenhoven-Dobbe, J.C.; van Nieuwkoop, S.; Bestebroer, T.M.; van den Hoogen, B.G.; Neyts, J.; Snijder, E.J. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents Chemother. 2014, 58, 4875–4884. [Google Scholar] [CrossRef] [PubMed]
- Dyall, J.; Coleman, C.M.; Hart, B.J.; Venkataraman, T.; Holbrook, M.R.; Kindrachuk, J.; Johnson, R.F.; Olinger, G.G., Jr.; Jahrling, P.B.; Laidlaw, M.; et al. Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob. Agents Chemother. 2014, 58, 4885–4893. [Google Scholar] [CrossRef] [PubMed]
- Hart, B.J.; Dyall, J.; Postnikova, E.; Zhou, H.; Kindrachuk, J.; Johnson, R.F.; Olinger, G.G., Jr.; Frieman, M.B.; Holbrook, M.R.; Jahrling, P.B.; et al. Interferon-beta and mycophenolic acid are potent inhibitors of Middle East respiratory syndrome coronavirus in cell-based assays. J. Gen. Virol. 2014, 95, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Kindrachuk, J.; Ork, B.; Hart, B.J.; Mazur, S.; Holbrook, M.R.; Frieman, M.B.; Traynor, D.; Johnson, R.F.; Dyall, J.; Kuhn, J.H.; et al. Antiviral potential of ERK/MAPK and PI3K/AKT/mTOR signaling modulation for Middle East respiratory syndrome coronavirus infection as identified by temporal kinome analysis. Antimicrob. Agents Chemother. 2015, 59, 1088–1099. [Google Scholar] [CrossRef] [PubMed]
- Coleman, C.M.; Sisk, J.M.; Mingo, R.M.; Nelson, E.A.; White, J.M.; Frieman, M.B. Abelson Kinase Inhibitors Are Potent Inhibitors of Severe Acute Respiratory Syndrome Coronavirus and Middle East Respiratory Syndrome Coronavirus Fusion. J. Virol. 2016, 90, 8924–8933. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.W.; Kim, Y.J.; Park, S.H.; Yun, M.R.; Yang, J.S.; Kang, H.J.; Han, Y.W.; Lee, H.S.; Kim, H.M.; Kim, H.; et al. Variations in Spike Glycoprotein Gene of MERS-CoV, South Korea, 2015. Emerg. Infect. Dis. 2016, 22, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Shin, J.S.; Shie, J.J.; Ku, K.B.; Kim, C.; Go, Y.Y.; Huang, K.F.; Kim, M.; Liang, P.H. Identification and evaluation of potent Middle East respiratory syndrome coronavirus (MERS-CoV) 3CLPro inhibitors. Antivir. Res. 2017, 141, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.H.; Chung, T.D.; Oldenburg, K.R. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen. 1999, 4, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Go, Y.Y.; Kim, Y.S.; Cheon, S.; Nam, S.; Ku, K.B.; Kim, M.; Cho, N.H.; Park, H.; Alison Lee, P.Y.; Lin, Y.C.; et al. Evaluation and Clinical Validation of Two Field-Deployable Reverse Transcription-Insulated Isothermal PCR Assays for the Detection of the Middle East Respiratory Syndrome-Coronavirus. J. Mol. Diagn. 2017, 19, 817–827. [Google Scholar] [CrossRef] [PubMed]
- Chou, T.C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 2006, 58, 621–681. [Google Scholar] [CrossRef] [PubMed]
- Go, Y.Y. Evaluation of Antiviral Activity of Saracatinib against Virus Attachment; Korea Research Institute of Chemical Technology: Daejeon, Korea, 2018. [Google Scholar]
- Green, T.P.; Fennell, M.; Whittaker, R.; Curwen, J.; Jacobs, V.; Allen, J.; Logie, A.; Hargreaves, J.; Hickinson, D.M.; Wilkinson, R.W.; et al. Preclinical anticancer activity of the potent, oral Src inhibitor AZD0530. Mol. Oncol. 2009, 3, 248–261. [Google Scholar] [CrossRef] [PubMed]
- Hennequin, L.F.; Allen, J.; Breed, J.; Curwen, J.; Fennell, M.; Green, T.P.; Lambert-van der Brempt, C.; Morgentin, R.; Norman, R.A.; Olivier, A.; et al. N-(5-chloro-1,3-benzodioxol-4-yl)-7-[2-(4-methylpiperazin-1-yl)ethoxy]-5-(tetrahydro-2H-pyran-4-yloxy)quinazolin-4-amine, a novel, highly selective, orally available, dual-specific c-Src/Abl kinase inhibitor. J. Med. Chem. 2006, 49, 6465–6488. [Google Scholar] [CrossRef] [PubMed]
- Plunkett, W.; Huang, P.; Gandhi, V. Preclinical characteristics of gemcitabine. Anticancer Drugs 1995, 6 (Suppl. 6), 7–13. [Google Scholar] [CrossRef] [PubMed]
- Plunkett, W.; Huang, P.; Xu, Y.Z.; Heinemann, V.; Grunewald, R.; Gandhi, V. Gemcitabine: Metabolism, mechanisms of action, and self-potentiation. Semin. Oncol. 1995, 22, 3–10. [Google Scholar] [PubMed]
- Kang, H.; Kim, C.; Kim, D.E.; Song, J.H.; Choi, M.; Choi, K.; Kang, M.; Lee, K.; Kim, H.S.; Shin, J.S.; et al. Synergistic antiviral activity of gemcitabine and ribavirin against enteroviruses. Antivir. Res. 2015, 124, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Manicassamy, S. Sotrastaurin, a protein kinase C inhibitor for the prevention of transplant rejection and treatment of psoriasis. Curr. Opin. Investig. Drugs 2009, 10, 1225–1235. [Google Scholar] [PubMed]
- Thomas, S.M.; Brugge, J.S. Cellular functions regulated by Src family kinases. Annu. Rev. Cell Dev. Biol. 1997, 13, 513–609. [Google Scholar] [CrossRef] [PubMed]
- Ricono, J.M.; Huang, M.; Barnes, L.A.; Lau, S.K.; Weis, S.M.; Schlaepfer, D.D.; Hanks, S.K.; Cheresh, D.A. Specific cross-talk between epidermal growth factor receptor and integrin αvβ5 promotes carcinoma cell invasion and metastasis. Cancer Res. 2009, 69, 1383–1391. [Google Scholar] [CrossRef] [PubMed]
- Colicelli, J. ABL tyrosine kinases: Evolution of function, regulation, and specificity. Sci. Signal 2010, 3, re6. [Google Scholar] [CrossRef] [PubMed]
- Johnsen, I.B.; Nguyen, T.T.; Bergstroem, B.; Fitzgerald, K.A.; Anthonsen, M.W. The tyrosine kinase c-Src enhances RIG-I (retinoic acid-inducible gene I)-elicited antiviral signaling. J. Biol. Chem. 2009, 284, 19122–19131. [Google Scholar] [CrossRef] [PubMed]
- Johnsen, I.B.; Nguyen, T.T.; Ringdal, M.; Tryggestad, A.M.; Bakke, O.; Lien, E.; Espevik, T.; Anthonsen, M.W. Toll-like receptor 3 associates with c-Src tyrosine kinase on endosomes to initiate antiviral signaling. EMBO J. 2006, 25, 3335–3346. [Google Scholar] [CrossRef] [PubMed]
- Hahn, A.S.; Kaufmann, J.K.; Wies, E.; Naschberger, E.; Panteleev-Ivlev, J.; Schmidt, K.; Holzer, A.; Schmidt, M.; Chen, J.; Konig, S.; et al. The ephrin receptor tyrosine kinase A2 is a cellular receptor for Kaposi’s sarcoma-associated herpesvirus. Nat. Med. 2012, 18, 961–966. [Google Scholar] [CrossRef] [PubMed]
- Eierhoff, T.; Hrincius, E.R.; Rescher, U.; Ludwig, S.; Ehrhardt, C. The epidermal growth factor receptor (EGFR) promotes uptake of influenza A viruses (IAV) into host cells. PLoS Pathog. 2010, 6, e1001099. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Sharma, N.R.; Ly, H.; Parslow, T.G.; Liang, Y. Receptor tyrosine kinase inhibitors that block replication of influenza a and other viruses. Antimicrob. Agents Chemother. 2011, 55, 5553–5559. [Google Scholar] [CrossRef] [PubMed]
- Brindley, M.A.; Hunt, C.L.; Kondratowicz, A.S.; Bowman, J.; Sinn, P.L.; McCray, P.B., Jr.; Quinn, K.; Weller, M.L.; Chiorini, J.A.; Maury, W. Tyrosine kinase receptor Axl enhances entry of Zaire ebolavirus without direct interactions with the viral glycoprotein. Virology 2011, 415, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Coyne, C.B.; Bergelson, J.M. Virus-induced Abl and Fyn kinase signals permit coxsackievirus entry through epithelial tight junctions. Cell 2006, 124, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Pfannkuche, A.; Buther, K.; Karthe, J.; Poenisch, M.; Bartenschlager, R.; Trilling, M.; Hengel, H.; Willbold, D.; Haussinger, D.; Bode, J.G. c-Src is required for complex formation between the hepatitis C virus-encoded proteins NS5A and NS5B: A prerequisite for replication. Hepatology 2011, 53, 1127–1136. [Google Scholar] [CrossRef] [PubMed]
- Supekova, L.; Supek, F.; Lee, J.; Chen, S.; Gray, N.; Pezacki, J.P.; Schlapbach, A.; Schultz, P.G. Identification of human kinases involved in hepatitis C virus replication by small interference RNA library screening. J. Biol. Chem. 2008, 283, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, A.J.; Medigeshi, G.R.; Meyers, H.L.; DeFilippis, V.; Fruh, K.; Briese, T.; Lipkin, W.I.; Nelson, J.A. The Src family kinase c-Yes is required for maturation of West Nile virus particles. J. Virol. 2005, 79, 11943–11951. [Google Scholar] [CrossRef] [PubMed]
- Tintori, C.; Laurenzana, I.; La Rocca, F.; Falchi, F.; Carraro, F.; Ruiz, A.; Este, J.A.; Kissova, M.; Crespan, E.; Maga, G.; et al. Identification of Hck inhibitors as hits for the development of antileukemia and anti-HIV agents. ChemMedChem 2013, 8, 1353–1360. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, S.D.; Sakac, D.; Neschadim, A.; Branch, D.R. c-SRC protein tyrosine kinase regulates early HIV-1 infection post-entry. AIDS 2016, 30, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Musumeci, F.; Schenone, S.; Brullo, C.; Desogus, A.; Botta, L.; Tintori, C. Hck inhibitors as potential therapeutic agents in cancer and HIV infection. Curr. Med. Chem. 2015, 22, 1540–1564. [Google Scholar] [CrossRef] [PubMed]
- Coiras, M.; Ambrosioni, J.; Cervantes, F.; Miro, J.M.; Alcami, J. Tyrosine kinase inhibitors: Potential use and safety considerations in HIV-1 infection. Expert Opin. Drug Saf. 2017, 16, 547–559. [Google Scholar] [CrossRef] [PubMed]
- De Wispelaere, M.; LaCroix, A.J.; Yang, P.L. The small molecules AZD0530 and dasatinib inhibit dengue virus RNA replication via Fyn kinase. J. Virol. 2013, 87, 7367–7381. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Agrawal, T.; Khan, N.A.; Nakayama, Y.; Medigeshi, G.R. Identification and characterization of the role of c-terminal Src kinase in dengue virus replication. Sci. Rep. 2016, 6, 30490. [Google Scholar] [CrossRef] [PubMed]
- Newsome, T.P.; Weisswange, I.; Frischknecht, F.; Way, M. Abl collaborates with Src family kinases to stimulate actin-based motility of vaccinia virus. Cell. Microbiol. 2006, 8, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Sisk, J.M.; Frieman, M.B.; Machamer, C.E. Coronavirus S protein-induced fusion is blocked prior to hemifusion by Abl kinase inhibitors. J. Gen. Virol. 2018, 99, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.S. Targeting Src in breast cancer. Ann. Oncol. 2008, 19, 1379–1386. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Liu, Y.; Zhang, X. Suppression of coronavirus replication by inhibition of the MEK signaling pathway. J. Virol. 2007, 81, 446–456. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Liu, Y.; Zhang, X. Induction of transcription factor Egr-1 gene expression in astrocytoma cells by Murine coronavirus infection. Virology 2006, 355, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Sierra, J.R.; Cepero, V.; Giordano, S. Molecular mechanisms of acquired resistance to tyrosine kinase targeted therapy. Mol. Cancer 2010, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Mini, E.; Nobili, S.; Caciagli, B.; Landini, I.; Mazzei, T. Cellular pharmacology of gemcitabine. Ann. Oncol. 2006, 17 (Suppl. 5), v7–v12. [Google Scholar] [CrossRef] [PubMed]
- Denisova, O.V.; Kakkola, L.; Feng, L.; Stenman, J.; Nagaraj, A.; Lampe, J.; Yadav, B.; Aittokallio, T.; Kaukinen, P.; Ahola, T.; et al. Obatoclax, saliphenylhalamide, and gemcitabine inhibit influenza a virus infection. J. Biol. Chem. 2012, 287, 35324–35332. [Google Scholar] [CrossRef] [PubMed]
- Clouser, C.L.; Patterson, S.E.; Mansky, L.M. Exploiting drug repositioning for discovery of a novel HIV combination therapy. J. Virol. 2010, 84, 9301–9309. [Google Scholar] [CrossRef] [PubMed]
- Song, J.H.; Kim, S.R.; Heo, E.Y.; Lee, J.Y.; Kim, D.E.; Cho, S.; Chang, S.Y.; Yoon, B.I.; Seong, J.; Ko, H.J. Antiviral activity of gemcitabine against human rhinovirus in vitro and in vivo. Antivir. Res. 2017, 145, 6–13. [Google Scholar] [CrossRef] [PubMed]
| No. | Compound | EC50 (µM) † | CC50 (μM) † | Pharmaceutical Class | Mode of Action |
|---|---|---|---|---|---|
| 1 | Nutlin-3 | 6.9 ± 1.4 | 26.8 ± 1.6 | Antiviral, anticancer | MDM2 inhibitor |
| 2 | Hydroxyzine pamoate | 14.4 ± 3.4 | >50 | Antihistamine | Cholinergic system modulator |
| 3 | Amodiaquine dihydrochloride § | 2.1 ± 0.7 | 12.3 ± 5.9 | Antiprotozoal | Histamine N-methyltransferase inhibitor |
| 4 | Amodiaquine dihydrochloride dehydrate § | 2.4 ± 0.7 | 13.2 ± 5.0 | Antiprotozoal | Histamine N-methyltransferase inhibitor |
| 5 | Chloroquine diphosphate § | 12.0 ± 3.0 | >50 | Antiprotozoal | Hemozoin formation inhibitor |
| 6 | Hydroxychloroquine sulfate § | 13.3 ± 2.1 | >50 | Antiprotozoal | Hemozoin formation inhibitor |
| 7 | Saracatinib | 2.9 ± 0.6 | 57 ± 5.5 | Anticancer | Tyrosine kinase inhibitor |
| 8 | Sotrastaurin | 9.7 ± 3.3 | >50 | Anticancer | Protein kinase C inhibitor |
| 9 | Acetophenazine maleate | 11.2 ± 5.0 | 23.6 ± 3.8 | Antipsychotic | Dopamine receptor antagonist |
| 10 | Dosulepin hydrochloride | 3.4 ± 0.0 | 28.9 ± 0.0 | Antidepressant | Serotonin/noradrenaline reuptake inhibitor |
| 11 | Methotrimeprazine maleate salt | 2.5 ± 0.0 | 24.5 ± 0.0 | Antipsychotic | Dopamine receptor antagonist |
| 12 | N1-(4-pyridyl)-2-chloro-5-nitrobenzamide | 10.5 ± 0.3 | >50 |
| Drug Combination | Combination Ratio | a CI Values at Inhibition of | b Weighted Average CI Values | c Graded Symbols | Description | |||
|---|---|---|---|---|---|---|---|---|
| 50% | 75% | 90% | 95% | |||||
| Saracatinib + Gemcitabine | 1:1 | 1.260 | 0.696 | 0.434 | 0.334 | 0.529 | +++ | Synergism |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, J.S.; Jung, E.; Kim, M.; Baric, R.S.; Go, Y.Y. Saracatinib Inhibits Middle East Respiratory Syndrome-Coronavirus Replication In Vitro. Viruses 2018, 10, 283. https://doi.org/10.3390/v10060283
Shin JS, Jung E, Kim M, Baric RS, Go YY. Saracatinib Inhibits Middle East Respiratory Syndrome-Coronavirus Replication In Vitro. Viruses. 2018; 10(6):283. https://doi.org/10.3390/v10060283
Chicago/Turabian StyleShin, Jin Soo, Eunhye Jung, Meehyein Kim, Ralph S. Baric, and Yun Young Go. 2018. "Saracatinib Inhibits Middle East Respiratory Syndrome-Coronavirus Replication In Vitro" Viruses 10, no. 6: 283. https://doi.org/10.3390/v10060283
APA StyleShin, J. S., Jung, E., Kim, M., Baric, R. S., & Go, Y. Y. (2018). Saracatinib Inhibits Middle East Respiratory Syndrome-Coronavirus Replication In Vitro. Viruses, 10(6), 283. https://doi.org/10.3390/v10060283






