Constructing TC-1-GLUC-LMP2 Model Tumor Cells to Evaluate the Anti-Tumor Effects of LMP2-Related Vaccines
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Mice
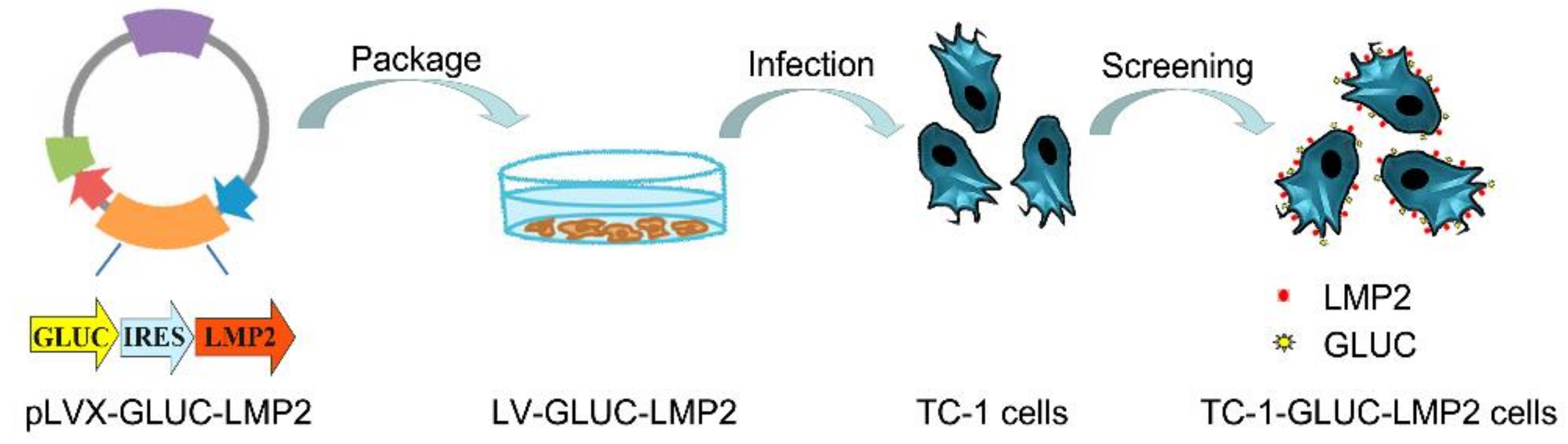
2.2. TC-1-GLUC-LMP2 Cell Line Construction
2.3. PCR and Reverse-Transcription PCR (RT-PCR)
2.4. Confirming the Stability of Protein
2.5. The Purity Detection of TC-1-GLUC-LMP2 Cell Line
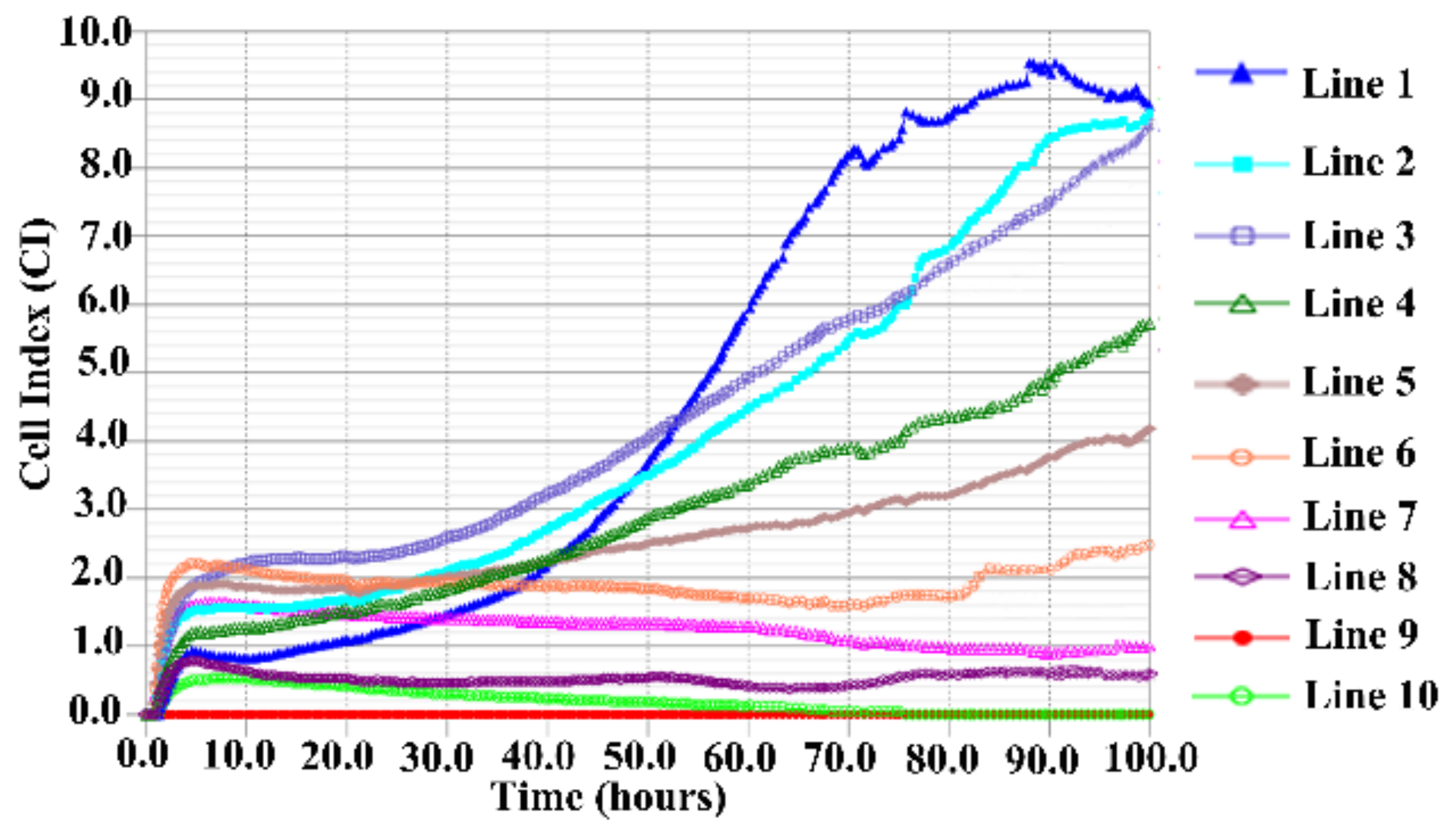
2.6. TC-1-GLUC-LMP2 Growth Curve
2.7. In Vitro Killing of LMP2-Targeted Model Cells
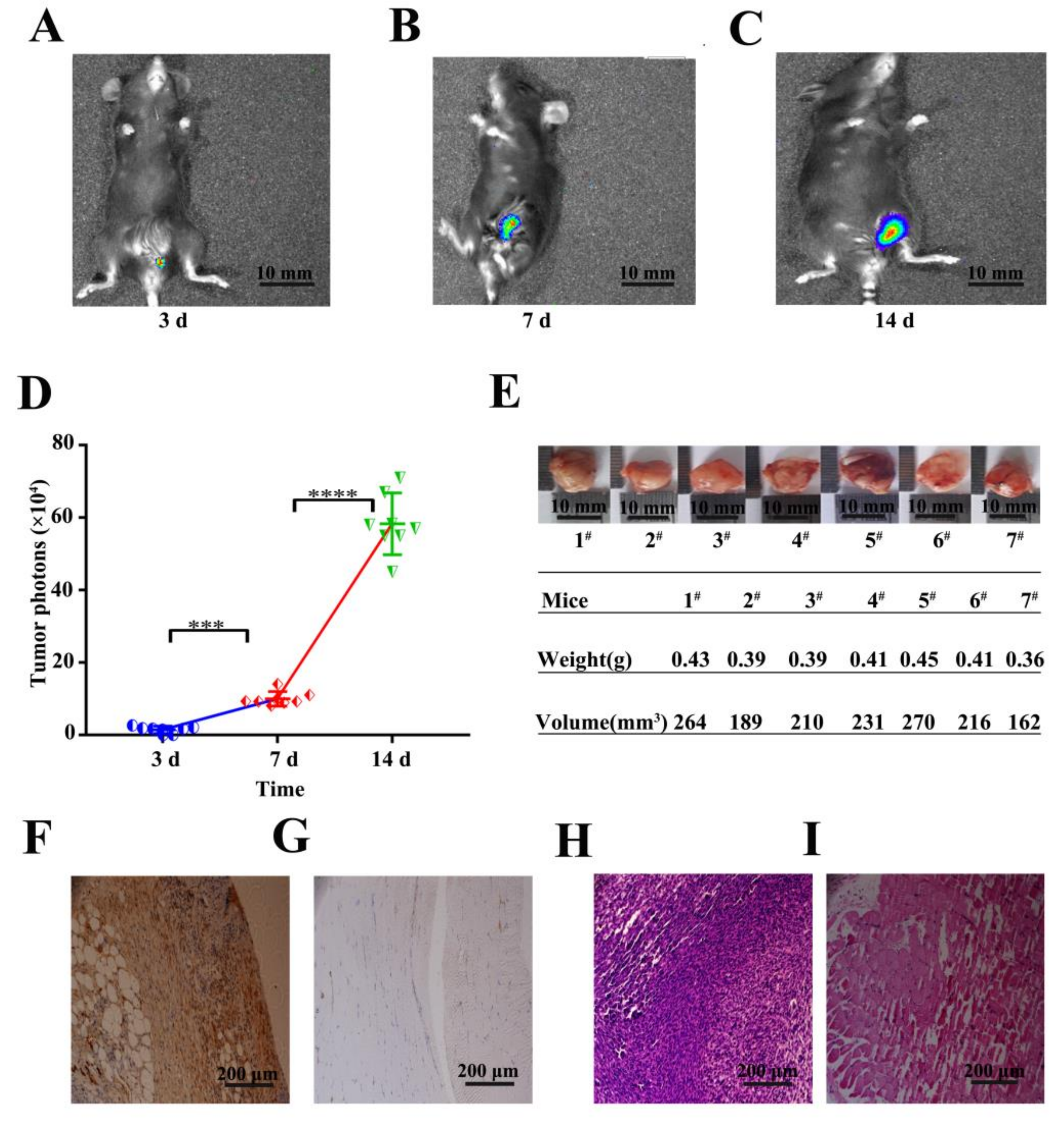
2.8. In Vivo Detection of Tumor Cells
2.9. Immunohistochemistry and Hematoxylin-Eosin (HE) Staining
2.10. TC-1-GLUC-LMP2 Tumor Challenge
2.11. Statistical Analysis
3. Results
3.1. TC-1-GLUC-LMP2 Construction
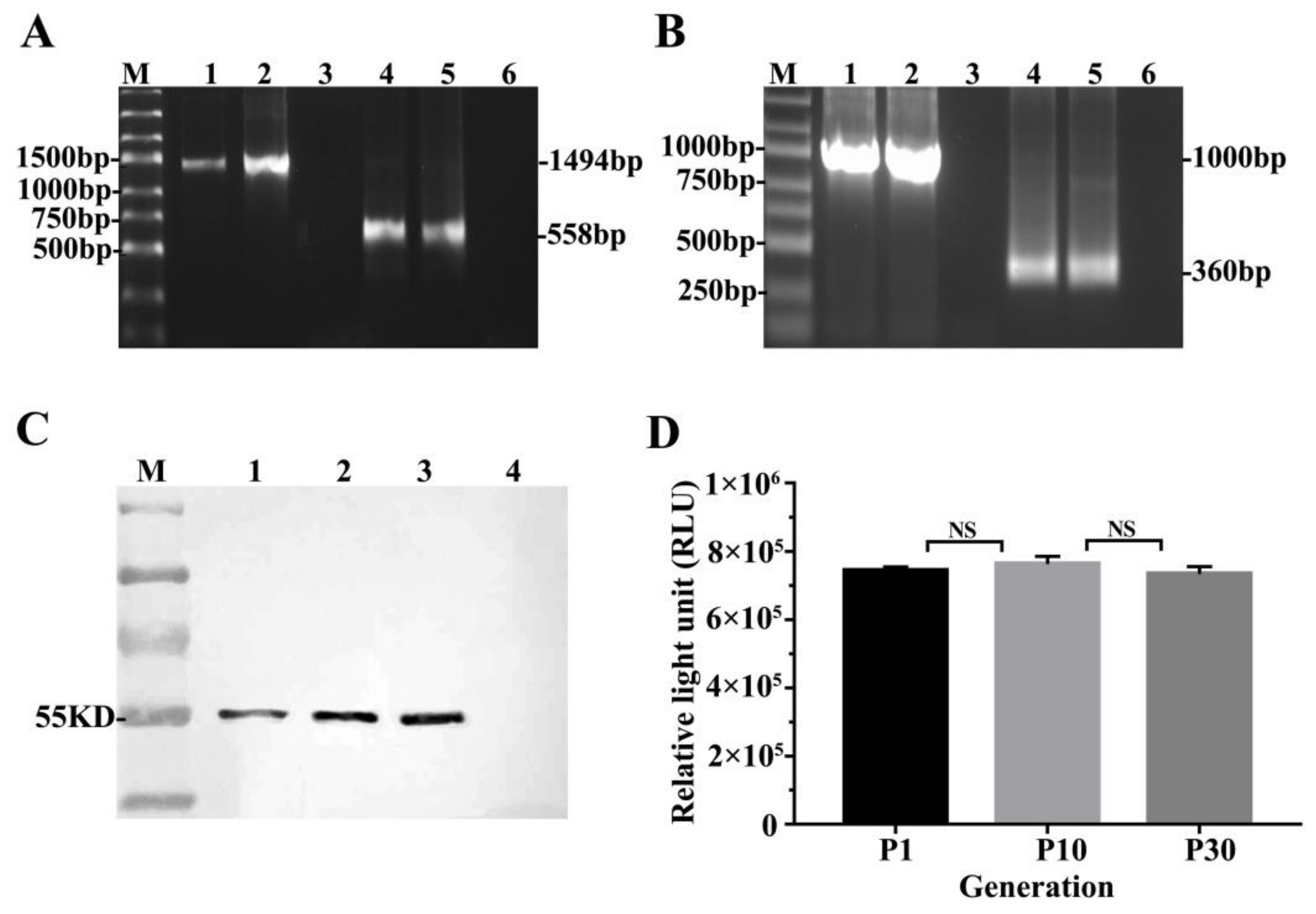
3.2. LMP2 and GLuc Genes Validation in TC-1-GLUC-LMP2 Cells
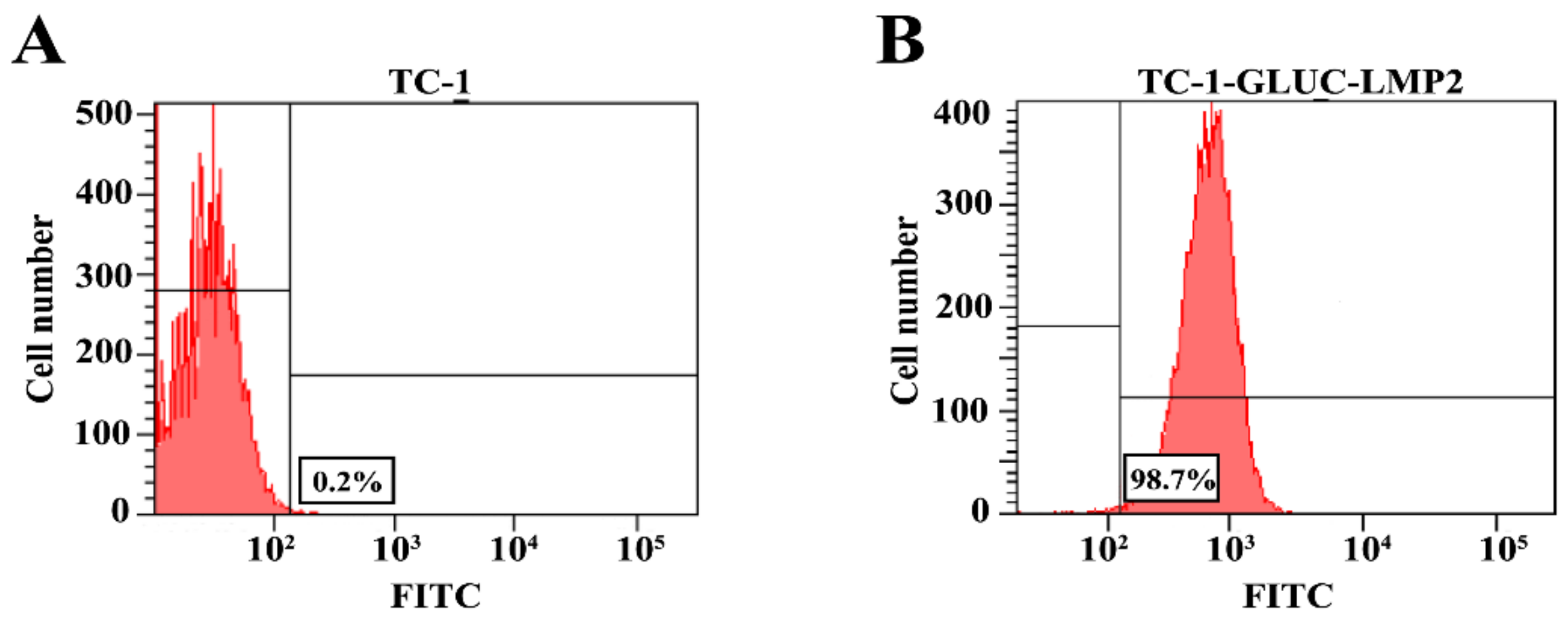
3.3. The Purity of TC-1-GLUC-LMP2 Cell Line
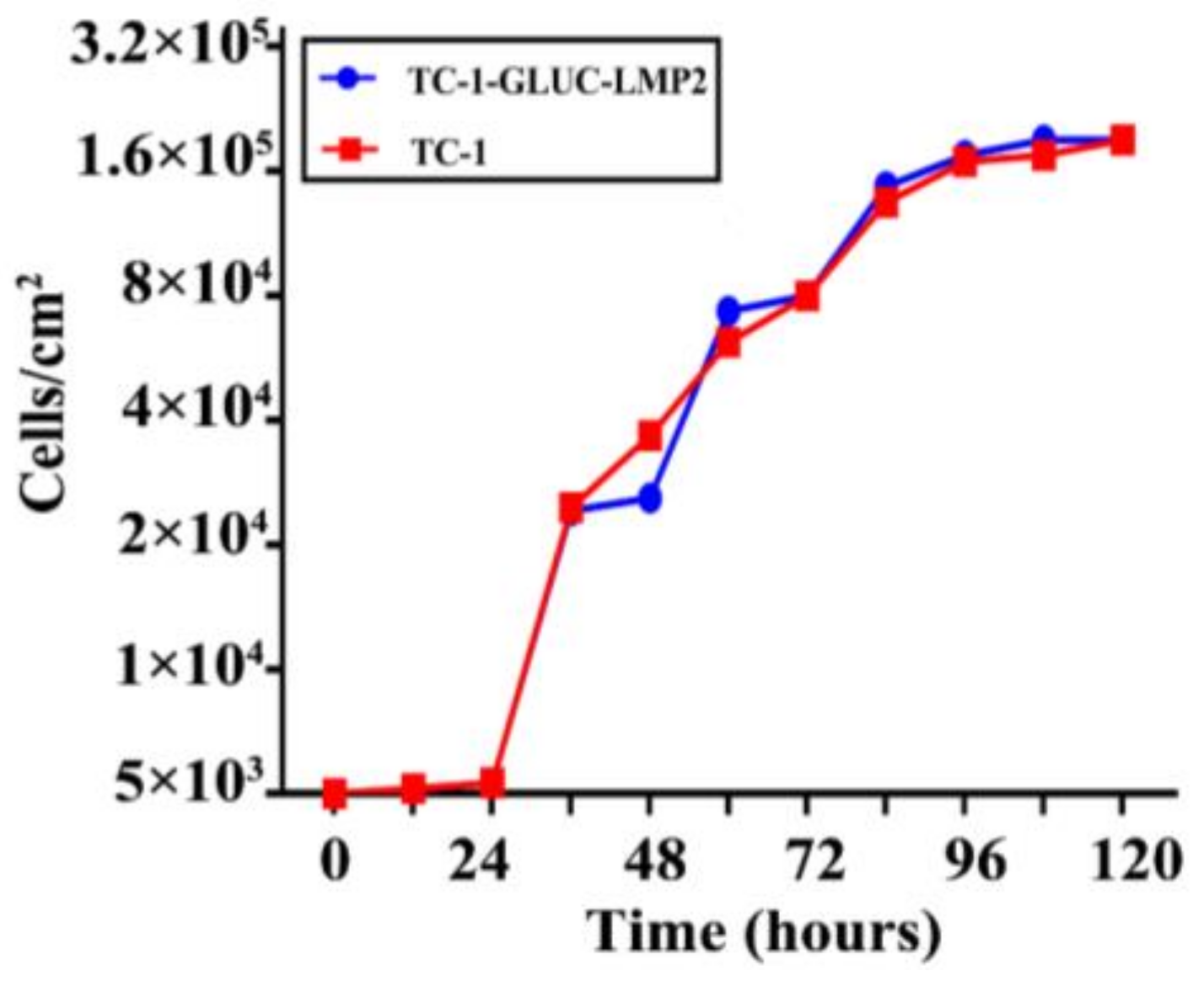
3.4. TC-1-GLUC-LMP2 Growth Curve
3.5. Immunogenicity of TC-1-GLUC-LMP2 Cells
3.6. In Vivo Detection of Tumor Formation
3.7. Tumor Challenge
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Young, L.S.; Rickinson, A.B. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 2004, 4, 757–768. [Google Scholar] [CrossRef] [PubMed]
- Grywalska, E.; Rolinski, J. Epstein-Barr virus-associated lymphomas. Semin. Oncol. 2015, 42, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Tashiro, H.; Brenner, M.K. Immunotherapy against cancer-related viruses. Cell Res. 2017, 27, 59–73. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.P.; Kurzrock, R. Epstein-Barr virus and cancer. Clin. Cancer Res. 2004, 10, 803–821. [Google Scholar] [CrossRef] [PubMed]
- Babcock, G.J.; Hochberg, D.; Thorley-Lawson, A.D. The expression pattern of Epstein-Barr virus latent genes in vivo is dependent upon the differentiation stage of the infected B cell. Immunity 2000, 13, 497–506. [Google Scholar] [CrossRef]
- Rowe, M.; Lear, A.L.; Croom-Carter, D.; Davies, A.H.; Rickinson, A.B. Three pathways of Epstein-Barr virus gene activation from EBNA1-positive latency in B lymphocytes. J. Virol. 1992, 66, 122–131. [Google Scholar] [PubMed]
- Wei, W.I.; Sham, J.S. Nasopharyngeal carcinoma. Lancet 2005, 365, 2041–2054. [Google Scholar] [CrossRef]
- Raab-Traub, N. Epstein-Barr virus in the pathogenesis of NPC. Semin. Cancer Biol. 2002, 12, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Brooks, L.; Yao, Q.Y.; Rickinson, A.B.; Young, L.S. Epstein-barr virus latent gene transcription in nasopharyngeal carcinoma cells: Coexpression of EBNA1, LMP1, and LMP2 transcripts. J. Virol. 1992, 66, 2689–2697. [Google Scholar] [PubMed]
- Sam, C.K.; Brooks, L.A.; Niedobitek, G.; Young, L.S.; Prasad, U.; Rickinson, A.B. Analysis of Epstein-Barr virus infection in nasopharyngeal biopsies from a group at high risk of nasopharyngeal carcinoma. Int. J. Cancer 1993, 53, 957–962. [Google Scholar] [CrossRef] [PubMed]
- Young, L.S.; Dawson, C.W.; Clark, D.; Rupani, H.; Busson, P.; Tursz, T.; Johnson, A.; Rickinson, A.B. Epstein-Barr virus gene expression in nasopharyngeal carcinoma. J. Gen. Virol. 1988, 69 Pt 5, 1051–1065. [Google Scholar] [CrossRef] [PubMed]
- Decaussin, G.; Sbih-Lammali, F.; de Turenne-Tessier, M.; Bouguermouh, A.; Ooka, T. Expression of BARF1 gene encoded by Epstein-Barr virus in nasopharyngeal carcinoma biopsies. Cancer Res. 2000, 60, 5584–5588. [Google Scholar] [PubMed]
- Nakada, R.; Hirano, H.; Matsuura, Y. Structural basis for the regulation of nuclear import of Epstein-Barr virus nuclear antigen 1 (EBNA1) by phosphorylation of the nuclear localization signal. Biochem. Biophys. Res. Commun. 2017, 484, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Manoury, B.; Fahraeus, R. Self-inhibition of synthesis and antigen presentation by Epstein-Barr virus-encoded EBNA1. Science 2003, 301, 1371–1374. [Google Scholar] [PubMed]
- Li, H.P.; Chang, Y.S. Epstein-Barr virus latent membrane protein 1: Structure and functions. J. Biomed. Sci. 2003, 10, 490–504. [Google Scholar] [CrossRef] [PubMed]
- Rider, M.A.; Cheerathodi, M.R.; Hurwitz, S.N.; Nkosi, D.; Howell, L.A.; Tremblay, D.C.; Liu, X.; Zhu, F.; Meckes, D.G., Jr. The interactome of EBV LMP1 evaluated by proximity-based bioid approach. Virology 2018, 516, 55–70. [Google Scholar] [PubMed]
- Lee, S.P. Nasopharyngeal carcinoma and the EBV-specific T cell response: Prospects for immunotherapy. Semin. Cancer Biol. 2002, 12, 463–471. [Google Scholar] [CrossRef]
- Hislop, A.D.; Taylor, G.S.; Sauce, D.; Rickinson, A.B. Cellular responses to viral infection in humans: Lessons from Epstein-Barr virus. Annu. Rev. Immunol. 2007, 25, 587–617. [Google Scholar] [CrossRef] [PubMed]
- Khanna, R.; Tellam, J.; Duraiswamy, J.; Cooper, L. Immunotherapeutic strategies for EBV-associated malignancies. Trends Mol. Med. 2001, 7, 270–276. [Google Scholar] [PubMed]
- Redchenko, I.V.; Rickinson, A.B. Accessing Epstein-Barr virus-specific T-cell memory with peptide-loaded dendritic cells. J. Virol. 1999, 73, 334–342. [Google Scholar] [PubMed]
- Lin, C.L.; Lo, W.F.; Lee, T.H.; Ren, Y.; Hwang, S.L.; Cheng, Y.F.; Chen, C.L.; Chang, Y.S.; Lee, S.P.; Rickinson, A.B.; et al. Immunization with Epstein-Barr virus (EBV) peptide-pulsed dendritic cells induces functional CD8+ T-cell immunity and may lead to tumor regression in patients with EBV-positive nasopharyngeal carcinoma. Cancer Res. 2002, 62, 6952–6958. [Google Scholar] [PubMed]
- Sample, J.; Liebowitz, D.; Kieff, E. Two related Epstein-Barr virus membrane proteins are encoded by separate genes. J. Virol. 1989, 63, 933–937. [Google Scholar] [PubMed]
- Longnecker, R.; Miller, C.L. Regulation of Epstein-Barr virus latency by latent membrane protein 2. Trends Microbiol. 1996, 4, 38–42. [Google Scholar] [CrossRef]
- Songyang, Z.; Shoelson, S.E.; McGlade, J.; Olivier, P.; Pawson, T.; Bustelo, X.R.; Barbacid, M.; Sabe, H.; Hanafusa, H.; Yi, T.; et al. Specific motifs recognized by the SH2 domains of CSK, 3BP2, FPS/FES, GRB-2, HCP, SHC, SYK, and VAV. Mol. Cell Biol. 1994, 14, 2777–2785. [Google Scholar] [CrossRef] [PubMed]
- Steven, N.M. Epstein-Barr virus latent infection in vivo. Rev. Med. Virol. 1997, 7, 97–106. [Google Scholar] [CrossRef]
- Rickinson, A.B.; Moss, D.J. Human cytotoxic T lymphocyte responses to Epstein-Barr virus infection. Annu. Rev. Immunol. 1997, 15, 405–431. [Google Scholar] [CrossRef] [PubMed]
- Lautscham, G.; Mayrhofer, S.; Taylor, G.; Haigh, T.; Leese, A.; Rickinson, A.; Blake, N. Processing of a multiple membrane spanning epstein-barr virus protein for CD8(+) T cell recognition reveals a proteasome-dependent, transporter associated with antigen processing-independent pathway. J. Exp. Med. 2001, 194, 1053–1068. [Google Scholar] [CrossRef] [PubMed]
- Rovedo, M.; Longnecker, R. Epstein-Barr virus latent membrane protein 2B (LMP2B) modulates LMP2A activity. J. Virol. 2007, 81, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Rechsteiner, M.P.; Berger, C.; Zauner, L.; Sigrist, J.A.; Weber, M.; Longnecker, R.; Bernasconi, M.; Nadal, D. Latent membrane protein 2B regulates susceptibility to induction of lytic Epstein-Barr virus infection. J. Virol. 2008, 82, 1739–1747. [Google Scholar] [CrossRef] [PubMed]
- Tannous, B.A.; Kim, D.E.; Fernandez, J.L.; Weissleder, R.; Breakefield, X.O. Codon-optimized gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol. Ther. 2005, 11, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Eckert, N.; Wrensch, F.; Gartner, S.; Palanisamy, N.; Goedecke, U.; Jager, N.; Pohlmann, S.; Winkler, M. Influenza a virus encoding secreted gaussia luciferase as useful tool to analyze viral replication and its inhibition by antiviral compounds and cellular proteins. PLoS ONE 2014, 9, e97695. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, S.; Zhou, L.; Du, H.; Mo, W.; Zeng, Y. Specific cellular immune responses in mice immunized with DNA, adeno-associated virus and adenoviral vaccines of Epstein-Barr virus-LMP2 alone or in combination. Sci. China Life Sci. 2011, 54, 263–266. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kochneva, G.; Zonov, E.; Grazhdantseva, A.; Yunusova, A.; Sibolobova, G.; Popov, E.; Taranov, O.; Netesov, S.; Chumakov, P.; Ryabchikova, E. Apoptin enhances the oncolytic properties of vaccinia virus and modifies mechanisms of tumor regression. Oncotarget 2014, 5, 11269–11282. [Google Scholar] [CrossRef] [PubMed]
- Niedobitek, G.; Meru, N.; Delecluse, H.J. Epstein-Barr virus infection and human malignancies. Int. J. Exp. Pathol. 2001, 82, 149–170. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.G.; Young, L.S. Epstein-Barr virus infection: Basis of malignancy and potential for therapy. Expert Rev. Mol. Med. 2001, 3, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Vickers, M.A.; Wilkie, G.M.; Robinson, N.; Rivera, N.; Haque, T.; Crawford, D.H.; Barry, J.; Fraser, N.; Turner, D.M.; Robertson, V.; et al. Establishment and operation of a good manufacturing practice-compliant allogeneic Epstein-Barr virus (EBV)-specific cytotoxic cell bank for the treatment of EBV-associated lymphoproliferative disease. Br. J. Haematol. 2014, 167, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.P.; Chan, A.T.; Cheung, S.T.; Thomas, W.A.; CroomCarter, D.; Dawson, C.W.; Tsai, C.H.; Leung, S.F.; Johnson, P.J.; Huang, D.P. CTL control of ebv in nasopharyngeal carcinoma (NPC): EBV-specific CTL responses in the blood and tumors of npc patients and the antigen-processing function of the tumor cells. J. Immunol. 2000, 165, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Straathof, K.C.; Bollard, C.M.; Popat, U.; Huls, M.H.; Lopez, T.; Morriss, M.C.; Gresik, M.V.; Gee, A.P.; Russell, H.V.; Brenner, M.K.; et al. Treatment of nasopharyngeal carcinoma with Epstein-Barr virus--specific T lymphocytes. Blood 2005, 105, 1898–1904. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.P.; Tierney, R.J.; Thomas, W.A.; Brooks, J.M.; Rickinson, A.B. Conserved CTL epitopes within EBV latent membrane protein 2: A potential target for CTL-based tumor therapy. J. Immunol. 1997, 158, 3325–3334. [Google Scholar] [PubMed]
- Shen, J.; Wang, L.F.; Zou, Z.Y.; Kong, W.W.; Yan, J.; Meng, F.Y.; Chen, F.J.; Du, J.; Shao, J.; Xu, Q.P.; et al. Phase I clinical study of personalized peptide vaccination combined with radiotherapy for advanced hepatocellular carcinoma. World J. Gastroenterol. 2017, 23, 5395–5404. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Kunda, P.E.; Lin, J.; Zhou, M.; Huang, J.; Zhang, H.; Liu, T. SYK-targeted dendritic cell-mediated cytotoxic T lymphocytes enhance the effect of immunotherapy on retinoblastoma. J. Cancer Res. Clin. Oncol. 2018, 144, 675–684. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.; Khanna, R. The development of prophylactic and therapeutic EBV vaccines. Curr. Top. Microbiol. Immunol. 2015, 391, 455–473. [Google Scholar] [PubMed]
- Cohen, J.I. Epstein-Barr virus vaccines. Clin. Transl. Immunol. 2015, 4, e32. [Google Scholar] [CrossRef] [PubMed]
- Taylor, G.S.; Jia, H.; Harrington, K.; Lee, L.W.; Turner, J.; Ladell, K.; Price, D.A.; Tanday, M.; Matthews, J.; Roberts, C.; et al. A recombinant Modified Vaccinia Ankara vaccine encoding Epstein-Barr virus (EBV) target antigens: A phase I trial in UK patients with EBV-positive cancer. Clin. Cancer Res. 2014, 20, 5009–5022. [Google Scholar] [CrossRef] [PubMed]
- Si, Y.; Deng, Z.; Lan, G.; Du, H.; Wang, Y.; Si, J.; Wei, J.; Weng, J.; Qin, Y.; Huang, B.; et al. The safety and immunological effects of RAD5-EBV-LMP2 vaccine in nasopharyngeal carcinoma patients: A phase I clinical trial and two-year follow-up. Chem. Pharm. Bull. (Tokyo) 2016, 64, 1118–1123. [Google Scholar] [CrossRef] [PubMed]
- Novalic, Z.; Verkuijlen, S.; Verlaan, M.; Eersels, J.L.H.; de Greeuw, I.; Molthoff, C.F.M.; Middeldorp, J.M.; Greijer, A.E. Cytolytic virus activation therapy and treatment monitoring for Epstein-Barr virus associated nasopharyngeal carcinoma in a mouse tumor model. J. Med. Virol. 2017, 89, 2207–2216. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Yang, A.; Peng, S.; Qiu, J.; Farmer, E.; Hung, C.F.; Wu, T.C. Characterization of HPV18 E6-specific T cell responses and establishment of HPV18 E6-expressing tumor model. Vaccine 2017, 35, 3850–3858. [Google Scholar] [CrossRef] [PubMed]
- Li, L.L.; Wang, H.R.; Zhou, Z.Y.; Luo, J.; Wang, X.L.; Xiao, X.Q.; Zhou, Y.B.; Zeng, Y. C3-LUC cells are an excellent model for evaluation of cellular immunity following HPV16L1 vaccination. PLoS ONE 2016, 11, e0149748. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, N.; Meng, D.; Hao, M.; Wei, L.; Chai, T. The mRNA and proteins expression levels analysis of TC-1 cells immune response to H9N2 avian influenza virus. Front. Microbiol. 2016, 7, 1039. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, L.; Hao, Y.; Wang, Z.; Zeng, Y. Constructing TC-1-GLUC-LMP2 Model Tumor Cells to Evaluate the Anti-Tumor Effects of LMP2-Related Vaccines. Viruses 2018, 10, 145. https://doi.org/10.3390/v10040145
Sun L, Hao Y, Wang Z, Zeng Y. Constructing TC-1-GLUC-LMP2 Model Tumor Cells to Evaluate the Anti-Tumor Effects of LMP2-Related Vaccines. Viruses. 2018; 10(4):145. https://doi.org/10.3390/v10040145
Chicago/Turabian StyleSun, Liying, Yanzhe Hao, Zhan Wang, and Yi Zeng. 2018. "Constructing TC-1-GLUC-LMP2 Model Tumor Cells to Evaluate the Anti-Tumor Effects of LMP2-Related Vaccines" Viruses 10, no. 4: 145. https://doi.org/10.3390/v10040145
APA StyleSun, L., Hao, Y., Wang, Z., & Zeng, Y. (2018). Constructing TC-1-GLUC-LMP2 Model Tumor Cells to Evaluate the Anti-Tumor Effects of LMP2-Related Vaccines. Viruses, 10(4), 145. https://doi.org/10.3390/v10040145



