The Transcriptional Repressor BS69 is a Conserved Target of the E1A Proteins from Several Human Adenovirus Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid Constructs
2.2. Yeast Two-Hybrid Assays
2.3. Western Blots
2.4. Cell Culture
2.5. Coimmunoprecipitation
2.6. Luciferase Assays
2.7. Statistical Analysis
3. Results
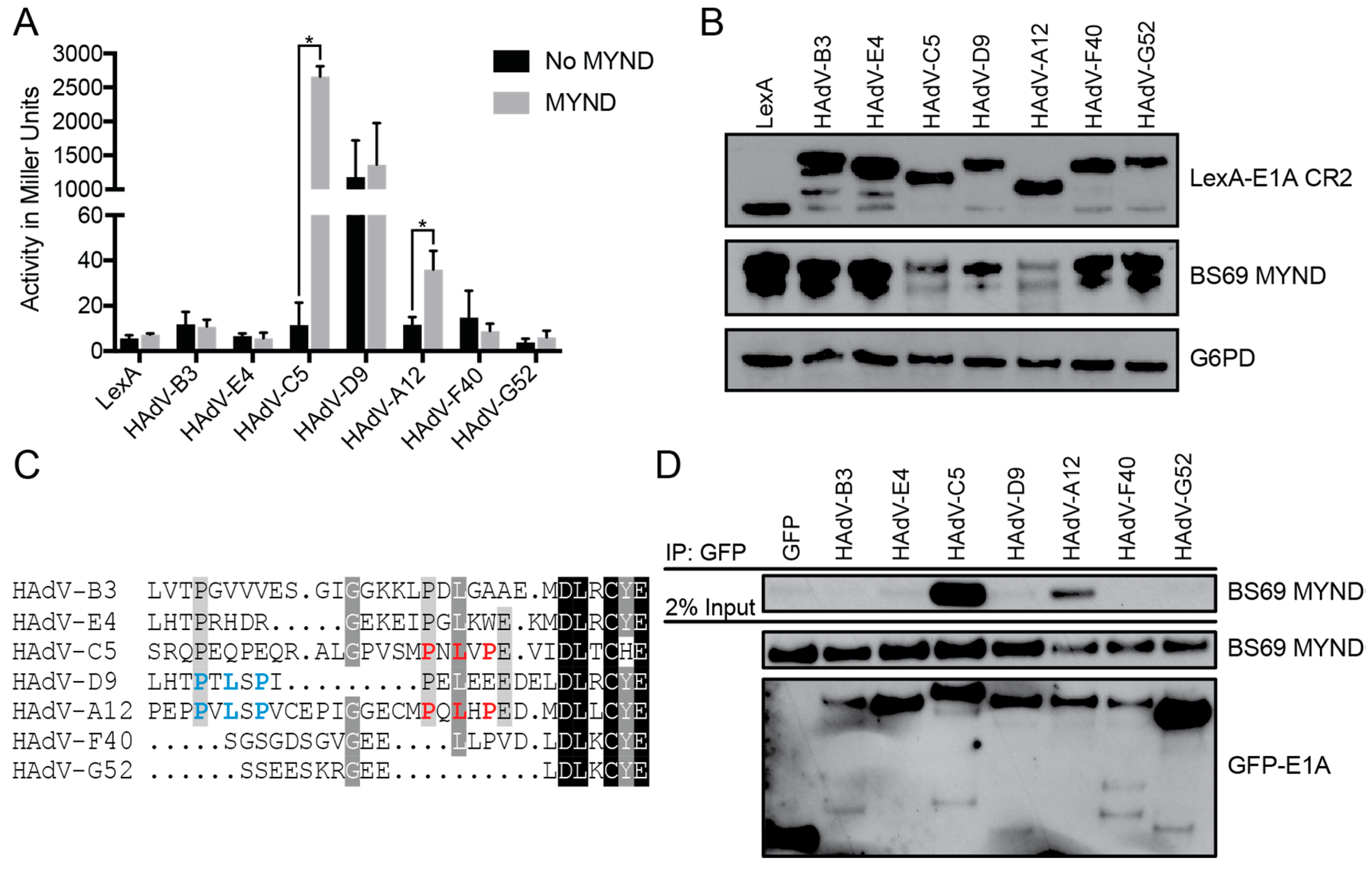
3.1. The Interaction between E1A and BS69 is Conserved Amongst HAdV Species C and A
3.2. Characterizing the Molecular Determinants Needed for HAdV-C5 E1A to Bind to BS69
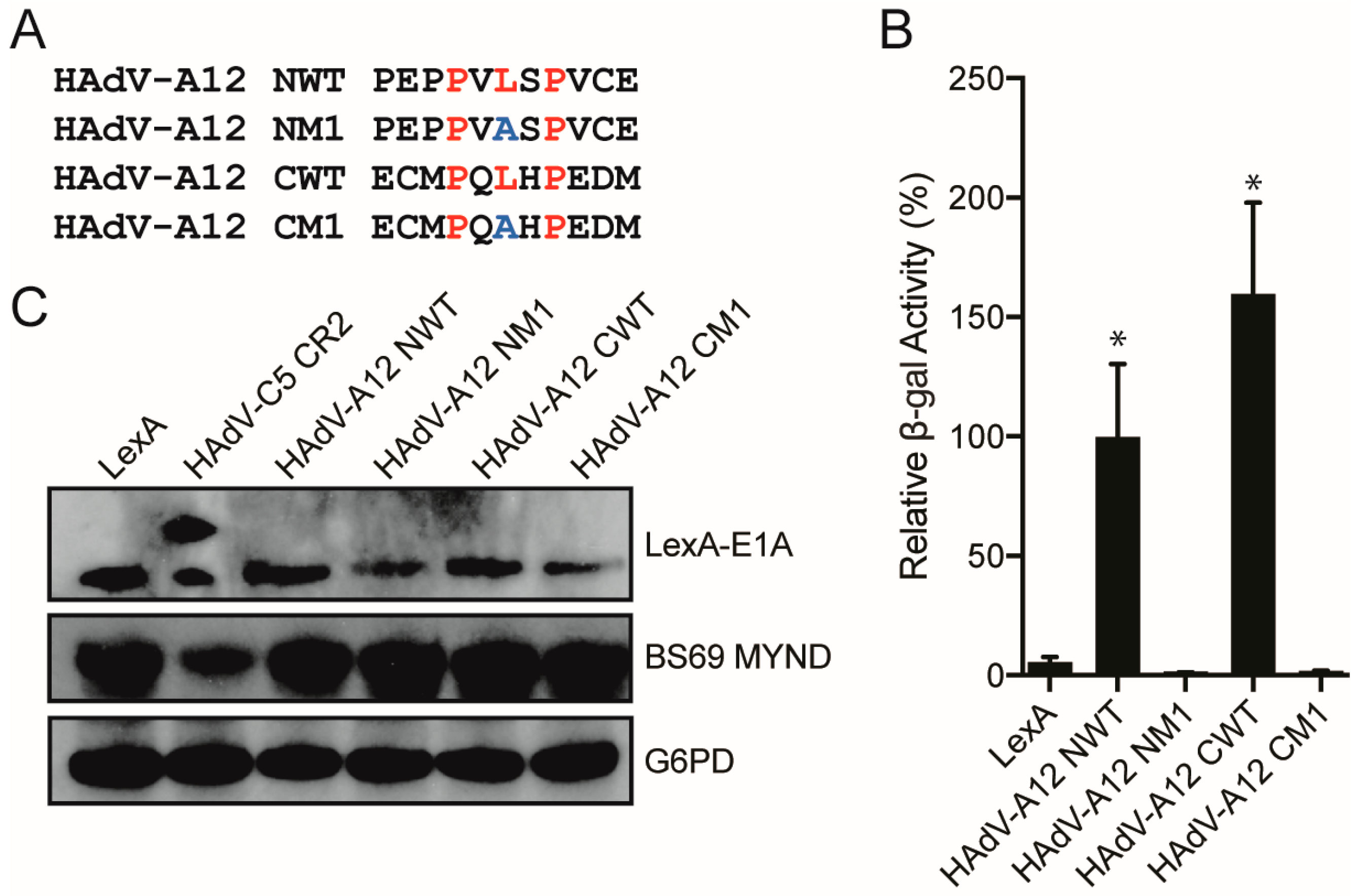
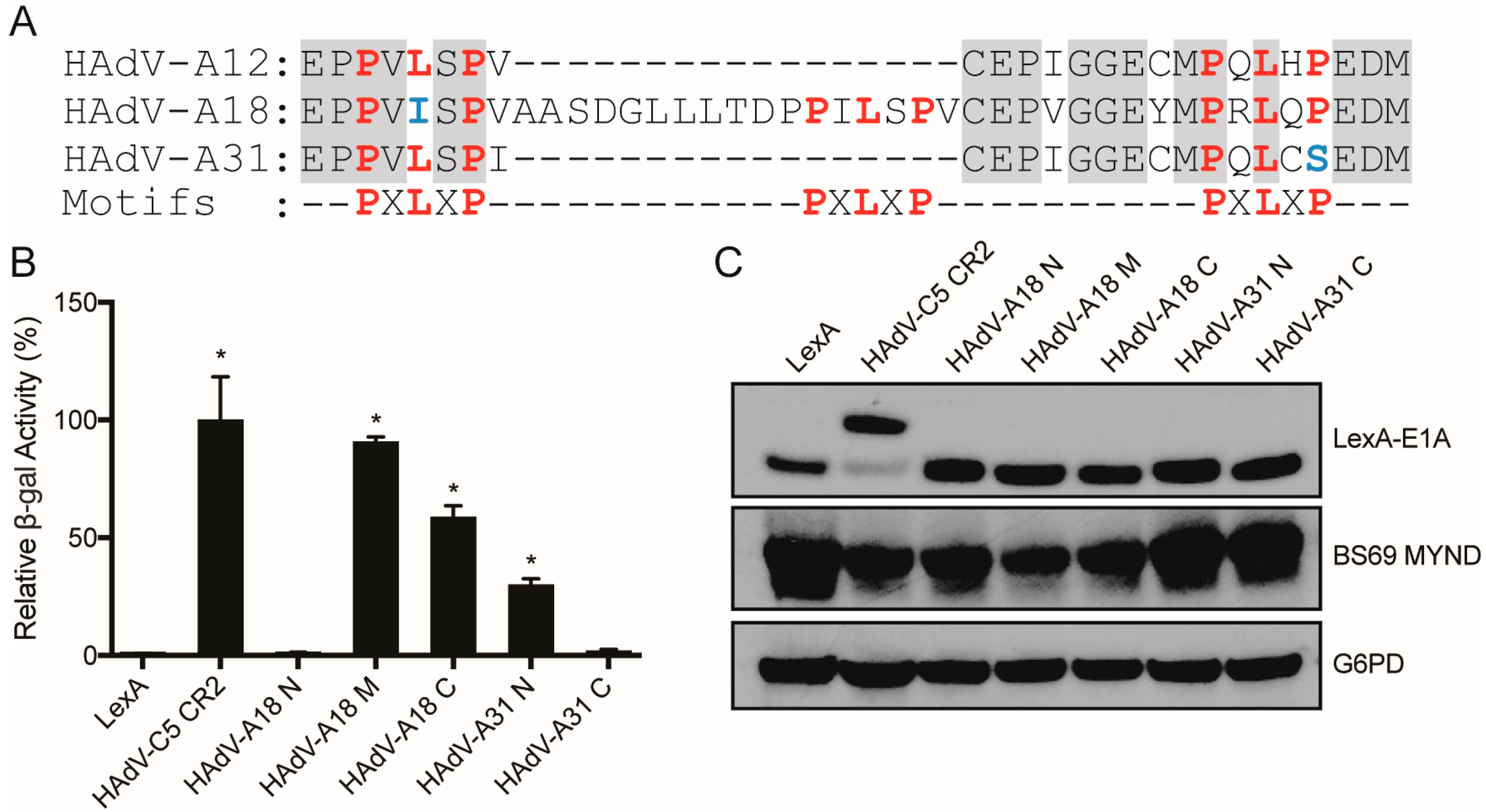
3.3. Characterizing the Molecular Determinants Needed for Species A HAdV E1A Interactions with BS69
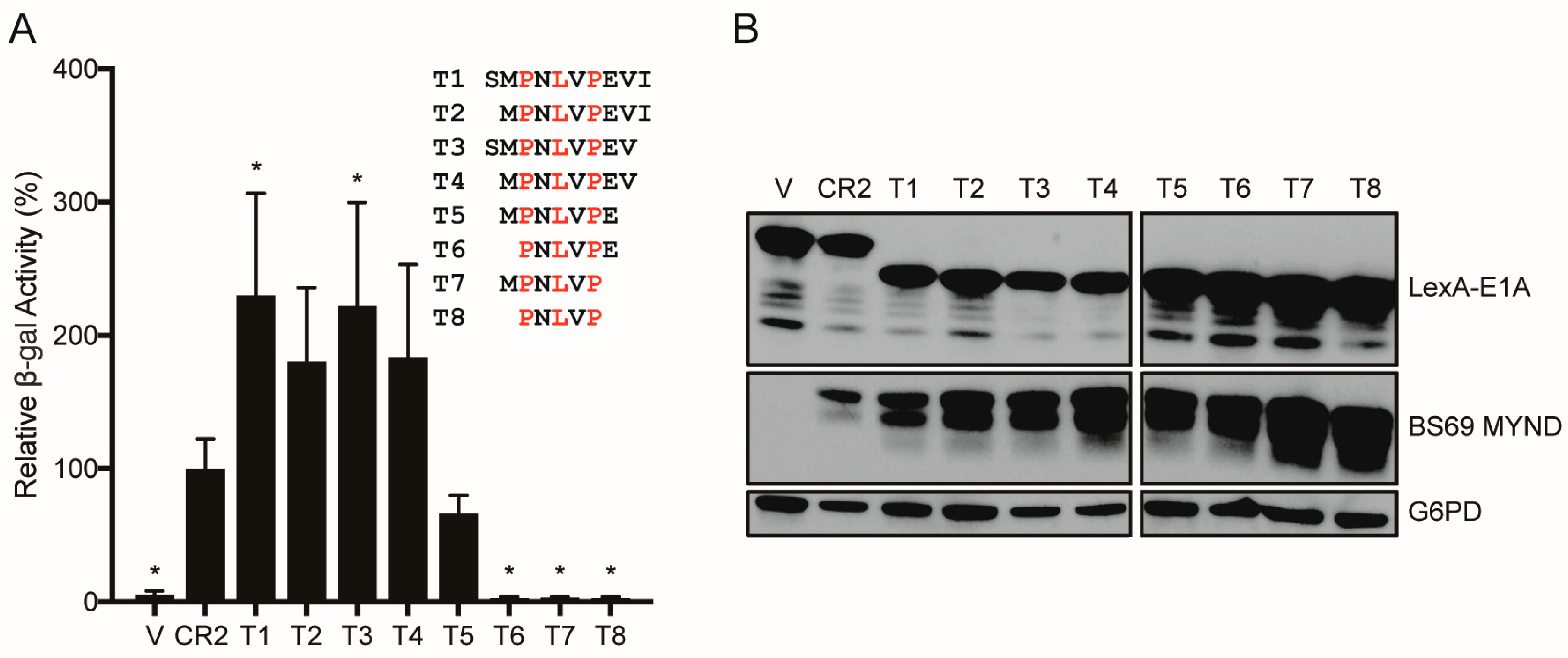
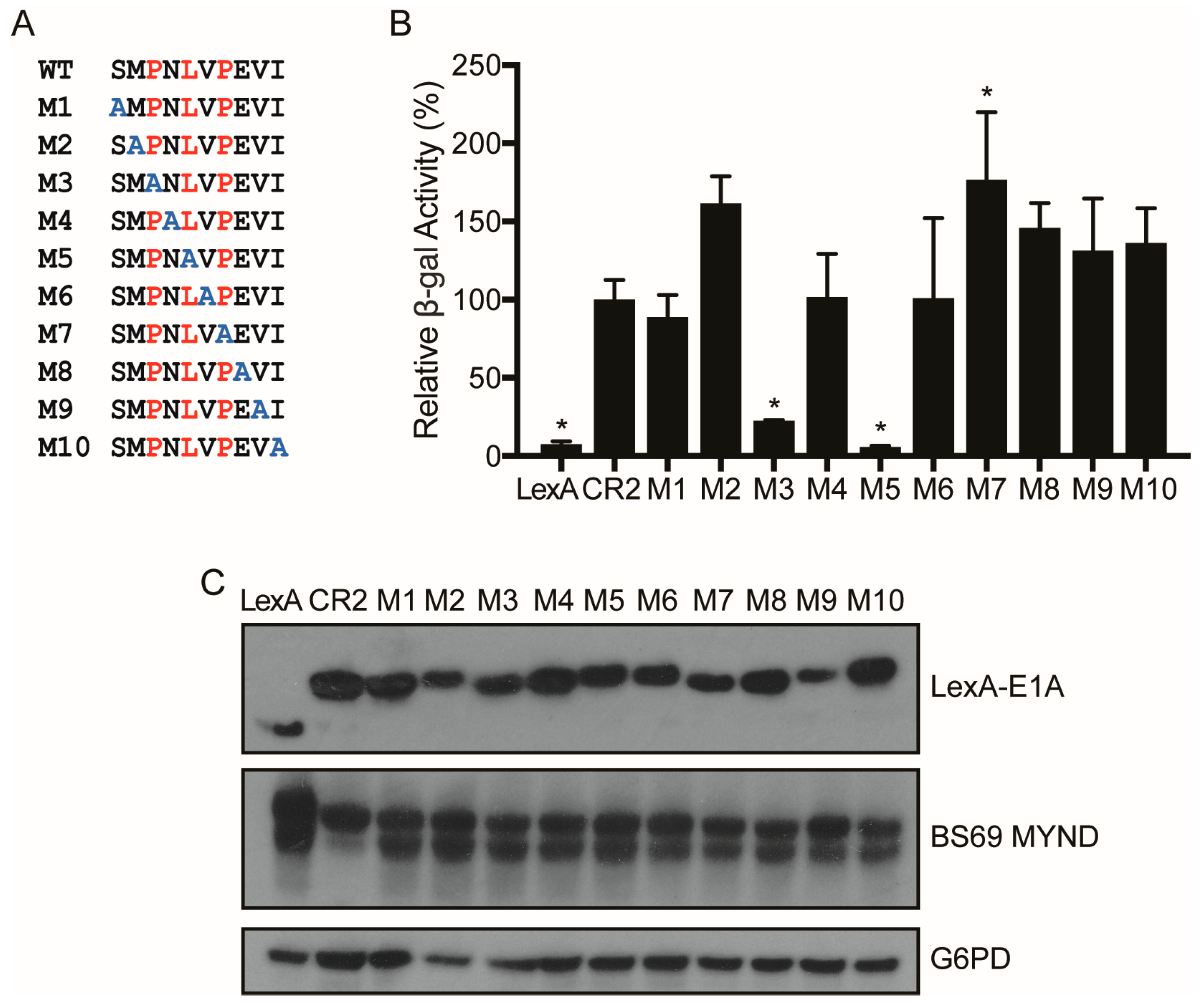
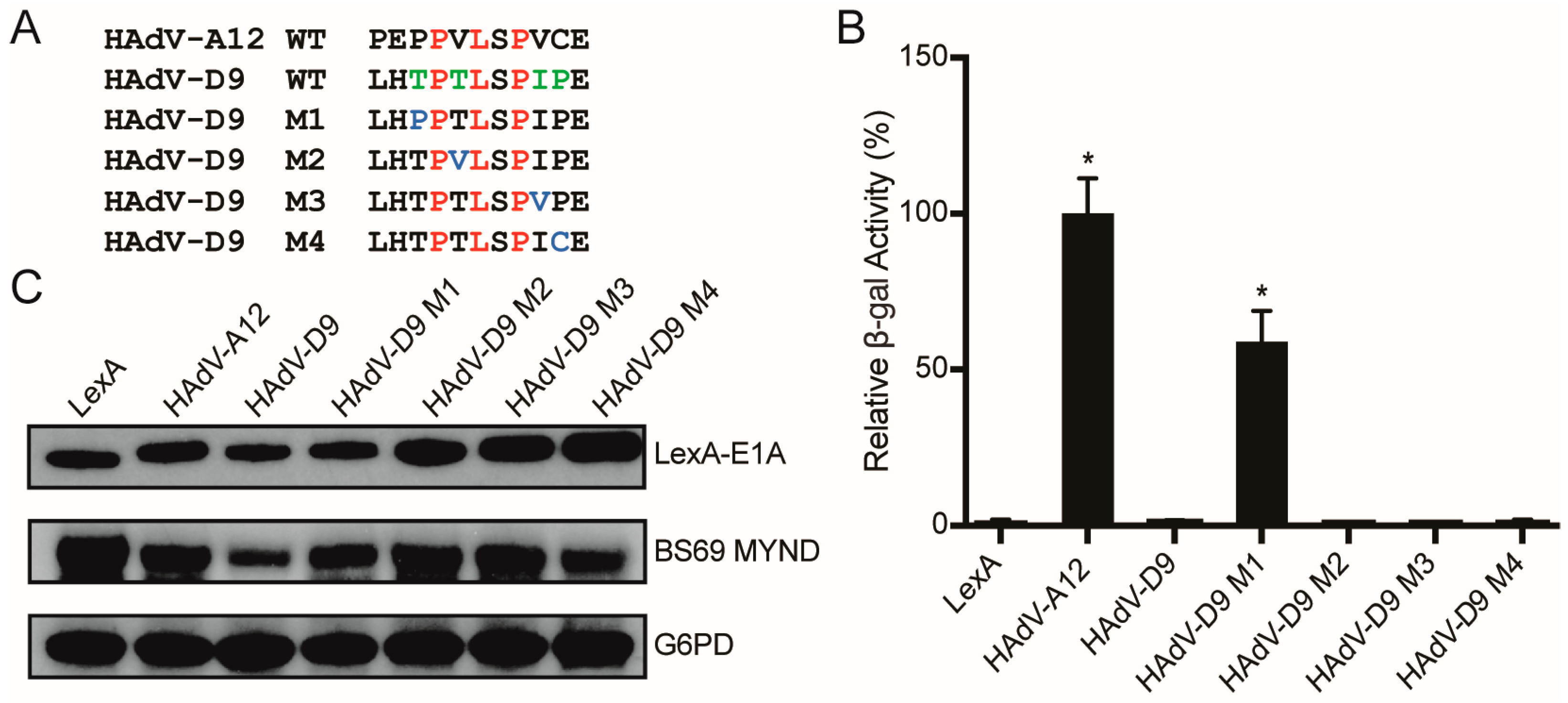
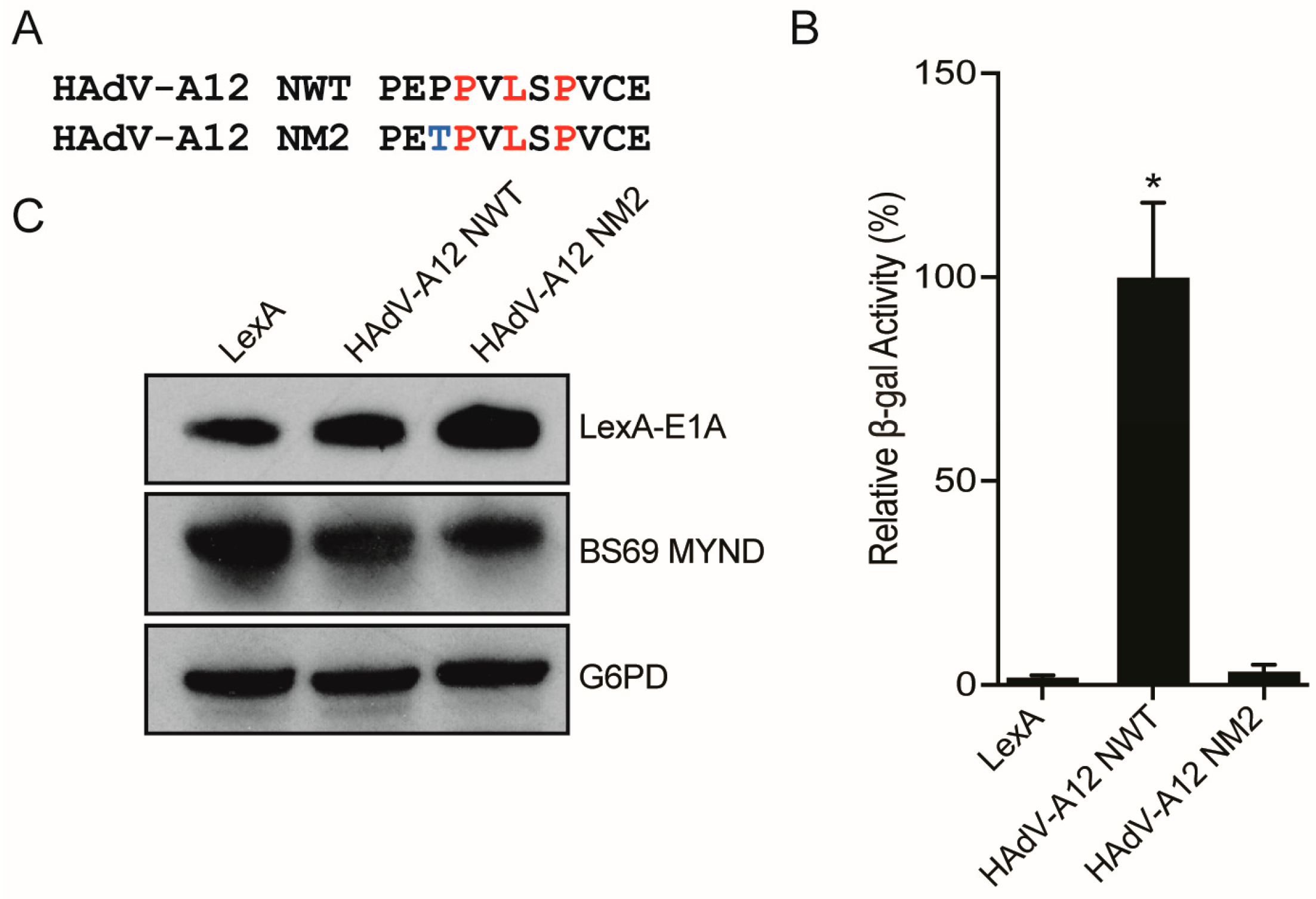
3.4. Conversion of a Non-Functional PXLXP SLiM into a Functional BS69-Binding Domain
3.5. BS69-Mediated Transcriptional Repression of E1A is Dependent on the PXLXP Motif in E1A and the MYND Domain of BS69
3.6. Interaction with BS69 Represses Transactivation by HAdV-C5 and HAdV-A12 E1A
4. Discussion
5. Conclusion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nevins, J.R. Regulation of early adenovirus gene expression. Microbiol. Rev. 1987, 51, 419–430. [Google Scholar] [PubMed]
- Pelka, P.; Ablack, J.N.G.; Fonseca, G.J.; Yousef, A.F.; Mymryk, J.S. Intrinsic structural disorder in adenovirus E1A: A viral molecular hub linking multiple diverse processes. J. Virol. 2008, 82, 7252–7263. [Google Scholar] [CrossRef] [PubMed]
- King, C.R.; Zhang, A.; Tessier, T.M.; Gameiro, S.F.; Mymryk, J.S. Hacking the Cell: Network Intrusion and Exploitation by Adenovirus E1A. MBio 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Green, M.R. Promoter targeting by adenovirus E1A through interaction with different cellular DNA-binding domains. Nature 1994, 368, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Shenk, T.; Flint, J. Transcriptional and transforming activities of the adenovirus E1A proteins. Adv. Cancer Res. 1991, 57, 47–85. [Google Scholar] [PubMed]
- Mymryk, J.S. Tumour suppressive properties of the adenovirus 5 E1A oncogene. Oncogene 1996, 13, 1581–1589. [Google Scholar] [PubMed]
- Gallimore, P.H.; Turnell, A.S. Adenovirus E1A: Remodelling the host cell, a life or death experience. Oncogene 2001, 20, 7824–7835. [Google Scholar] [CrossRef] [PubMed]
- Frisch, S.M.; Mymryk, J.S. Adenovirus-5 E1A: Paradox and paradigm. Nat. Rev. Mol. Cell Biol. 2002, 3, 441–452. [Google Scholar] [CrossRef] [PubMed]
- King, C.R.; Cohen, M.J.; Fonseca, G.J.; Dirk, B.S.; Dikeakos, J.D.; Mymryk, J.S. Functional and Structural Mimicry of Cellular Protein Kinase A Anchoring Proteins by a Viral Oncoprotein. PLoS Pathog. 2016, 12, e1005621. [Google Scholar] [CrossRef] [PubMed]
- Avvakumov, N.; Wheeler, R.; D’Halluin, J.C.; Mymryk, J.S. Comparative sequence analysis of the largest E1A proteins of human and simian adenoviruses. J. Virol. 2002, 76, 7968–7975. [Google Scholar] [CrossRef] [PubMed]
- Lillie, J.W.; Green, M.R. Transcription activation by the adenovirus E1A protein. Nature 1989, 338, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Martin, K.J.; Lillie, J.W.; Green, M.R. Evidence for interaction of different eukaryotic transcriptional activators with distinct cellular targets. Nature 1990, 346, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Davey, N.E.; Travé, G.; Gibson, T.J. How viruses hijack cell regulation. Trends Biochem. Sci. 2011, 36, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Via, A.; Uyar, B.; Brun, C.; Zanzoni, A. How pathogens use linear motifs to perturb host cell networks. Trends Biochem. Sci. 2015, 40, 36–48. [Google Scholar] [CrossRef] [PubMed]
- Van Roey, K.; Uyar, B.; Weatheritt, R.J.; Dinkel, H.; Seiler, M.; Budd, A.; Gibson, T.J.; Davey, N.E. Short linear motifs: ubiquitous and functionally diverse protein interaction modules directing cell regulation. Chem. Rev. 2014, 114, 6733–6778. [Google Scholar] [CrossRef] [PubMed]
- Neduva, V.; Russell, R.B. Linear motifs: evolutionary interaction switches. FEBS Lett. 2005, 579, 3342–3345. [Google Scholar] [CrossRef] [PubMed]
- DeCaprio, J.A. How the RB tumor suppressor structure and function was revealed by the study of Adenovirus and SV40. Virology 2009, 384, 274–284. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, A.; Gavin, M.R.; Luo, R.X.; Dean, D.C. Role of the LXCXE binding site in RB function. Mol. Cell Biol. 2000, 20, 6799–6805. [Google Scholar] [CrossRef] [PubMed]
- Dyson, N.; Guida, P.; Münger, K.; Harlow, E. Homologous sequences in adenovirus E1A and human papillomavirus E7 proteins mediate interaction with the same set of cellular proteins. J. Virol. 1992, 66, 6893–6902. [Google Scholar] [PubMed]
- Yousef, A.F.; Fonseca, G.J.; Pelka, P.; Ablack, J.N.G.; Walsh, C.; Dick, F.A.; Bazett-Jones, D.P.; Shaw, G.S.; Mymryk, J.S. Identification of a molecular recognition feature in the E1A oncoprotein that binds the SUMO conjugase UBC9 and likely interferes with polySUMOylation. Oncogene 2010, 29, 4693–4704. [Google Scholar] [CrossRef] [PubMed]
- Lau, L.; Gray, E.E.; Brunette, R.L.; Stetson, D.B. DNA tumor virus oncogenes antagonize the cGAS-STING DNA-sensing pathway. Science 2015, 350, 568–571. [Google Scholar] [CrossRef] [PubMed]
- Hateboer, G.; Gennissen, A.; Ramos, Y.F.; Kerkhoven, R.M.; Sonntag-Buck, V.; Stunnenberg, H.G.; Bernards, R. BS69, a novel adenovirus E1A-associated protein that inhibits E1A transactivation. EMBO J. 1995, 14, 3159–3169. [Google Scholar] [CrossRef] [PubMed]
- Ansieau, S.; Leutz, A. The conserved MYND domain of BS69 binds cellular and oncoviral proteins through a common PXLXP motif. J. Biol. Chem. 2002, 277, 4906–4910. [Google Scholar] [CrossRef] [PubMed]
- Avvakumov, N.; Kajon, A.E.; Hoeben, R.C.; Mymryk, J.S. Comprehensive sequence analysis of the E1A proteins of human and simian adenoviruses. Virology 2004, 329, 477–492. [Google Scholar] [CrossRef] [PubMed]
- Harter, M.R.; Liu, C.-D.; Shen, C.-L.; Gonzalez-Hurtado, E.; Zhang, Z.-M.; Xu, M.; Martinez, E.; Peng, C.-W.; Song, J. BS69/ZMYND11 C-Terminal Domains Bind and Inhibit EBNA2. PLoS Pathog. 2016, 12, e1005414. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Zheng, L.; Park, J.W.; Lv, R.; Chen, H.; Jiao, F.; Xu, W.; Mu, S.; Wen, H.; Qiu, J.; et al. BS69/ZMYND11 reads and connects histone H3.3 lysine 36 trimethylation-decorated chromatin to regulated pre-mRNA processing. Mol. Cell 2014, 56, 298–310. [Google Scholar] [CrossRef] [PubMed]
- Wen, H.; Li, Y.; Xi, Y.; Jiang, S.; Stratton, S.; Peng, D.; Tanaka, K.; Ren, Y.; Xia, Z.; Wu, J.; et al. ZMYND11 links histone H3.3K36ME3 to transcription elongation and tumour suppression. Nature 2014, 508, 263–268. [Google Scholar] [CrossRef] [PubMed]
- Velasco, G.; Grkovic, S.; Ansieau, S. New insights into BS69 functions. J. Biol. Chem. 2006, 281, 16546–16550. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Qin, S.; Li, F.; Li, S.; Zhang, W.; Peng, J.; Zhang, Z.; Gong, Q.; Wu, J.; Shi, Y. Crystal structure of human BS69 Bromo-ZnF-PWWP reveals its role in H3K36ME3 nucleosome binding. Cell Res. 2014, 24, 890–893. [Google Scholar] [CrossRef] [PubMed]
- Kateb, F.; Perrin, H.; Tripsianes, K.; Zou, P.; Spadaccini, R.; Bottomley, M.; Franzmann, T.M.; Buchner, J.; Ansieau, S.; Sattler, M. Structural and functional analysis of the DEAF-1 and BS69 MYND domains. PLoS ONE 2013, 8, e54715. [Google Scholar] [CrossRef] [PubMed]
- Ogata-Kawata, H.; Yamada, K.; Uesaka-Yoshino, M.; Kagawa, N.; Miyamoto, K. BS69, a corepressor interacting with ZHX1, is a bifunctional transcription factor. Front. Biosci. 2007, 12, 1911–1926. [Google Scholar] [CrossRef] [PubMed]
- Kawata, H.; Yamada, K.; Shou, Z.; Mizutani, T.; Yazawa, T.; Yoshino, M.; Sekiguchi, T.; Kajitani, T.; Miyamoto, K. Zinc-fingers and homeoboxes (ZHX) 2, a novel member of the ZHX family, functions as a transcriptional repressor. Biochem. J. 2003, 373, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Sims, R.J.; Weihe, E.K.; Zhu, L.; O’Malley, S.; Harriss, J.V.; Gottlieb, P.D. M-Bop, a repressor protein essential for cardiogenesis, interacts with skNAC, a heart- and muscle-specific transcription factor. J. Biol. Chem. 2002, 277, 26524–26529. [Google Scholar] [CrossRef] [PubMed]
- Plotnik, J.P.; Budka, J.A.; Ferris, M.W.; Hollenhorst, P.C. ETS1 is a genome-wide effector of RAS/ERK signaling in epithelial cells. Nucleic Acids Res. 2014, 42, 11928–11940. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.; Schaffner, A.E.; Baker, K.M.; Mansky, K.C.; Ostrowski, M.C. ETS-2 interacts with co-repressor BS69 to repress target gene expression. Anticancer Res. 2003, 23, 2173–2178. [Google Scholar] [PubMed]
- Plotnik, J.P.; Hollenhorst, P.C. Interaction with ZMYND11 mediates opposing roles of Ras-responsive transcription factors ETS1 and ETS2. Nucleic Acids Res. 2017, gkx039. [Google Scholar] [CrossRef] [PubMed]
- Isobe, T.; Uchida, C.; Hattori, T.; Kitagawa, K.; Oda, T.; Kitagawa, M. Ubiquitin-dependent degradation of adenovirus E1A protein is inhibited by BS69. Biochem. Biophys. Res. Commun. 2006, 339, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Ladendorff, N.E.; Wu, S.; Lipsick, J.S. BS69, an adenovirus E1A-associated protein, inhibits the transcriptional activity of c-MYB. Oncogene 2001, 20, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Masselink, H.; Bernards, R. The adenovirus E1A binding protein BS69 is a corepressor of transcription through recruitment of N-CoR. Oncogene 2000, 19, 1538–1546. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Smith, M.M.; Mymryk, J.S. Interaction of the E1A oncoprotein with Yak1p, a novel regulator of yeast pseudohyphal differentiation, and related mammalian kinases. Mol. Biol. Cell 2001, 12, 699–710. [Google Scholar] [CrossRef] [PubMed]
- Ho, S.N.; Hunt, H.D.; Horton, R.M.; Pullen, J.K.; Pease, L.R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 1989, 77, 51–59. [Google Scholar] [CrossRef]
- Adams, A.; Gottschling, D.E.; Kaiser, C.A. Methods in Yeast Genetics; Cold Spring Harbor Press: New York, NY, USA, 1997. [Google Scholar]
- Gietz, R.D.; Schiestl, R.H.; Willems, A.R.; Woods, R.A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 1995, 11, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Von der Haar, T. Optimized protein extraction for quantitative proteomics of yeasts. PLoS ONE 2007, 2, e1078. [Google Scholar] [CrossRef] [PubMed]
- Shuen, M.; Avvakumov, N.; Torchia, J.; Mymryk, J.S. The E1A proteins of all six human adenovirus subgroups target the p300/CBP acetyltransferases and the SAGA transcriptional regulatory complex. Virology 2003, 316, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Krajewski, M.; Mikolajka, A.; Holak, T.A. Molecular Determinants for the Complex Formation between the Retinoblastoma Protein and LXCXE Sequences. J. Biol. Chem. 2005, 280, 37868–37876. [Google Scholar] [CrossRef] [PubMed]
- Nevins, J.R. E2F: A link between the RB tumor suppressor protein and viral oncoproteins. Science 1992, 258, 424–429. [Google Scholar] [CrossRef] [PubMed]
- Zamanian, M.; La Thangue, N.B. Adenovirus E1A prevents the retinoblastoma gene product from repressing the activity of a cellular transcription factor. EMBO J. 1992, 11, 2603–2610. [Google Scholar] [CrossRef] [PubMed]
- Cobrinik, D. Pocket proteins and cell cycle control. Oncogene 2005, 24, 2796–2809. [Google Scholar] [CrossRef] [PubMed]
- Frolov, M.V.; Dyson, N.J. Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J. Cell Sci. 2004, 117, 2173–2181. [Google Scholar] [CrossRef] [PubMed]
- Bagchi, S.; Raychaudhuri, P.; Nevins, J.R. Adenovirus E1A proteins can dissociate heteromeric complexes involving the E2F transcription factor: A novel mechanism for E1A trans-activation. Cell 1990, 62, 659–669. [Google Scholar] [CrossRef]
- Ferguson, B.; Krippl, B.; Andrisani, O.; Jones, N.; Westphal, H.; Rosenberg, M. E1A 13S and 12S mRNA products made in Escherichia coli both function as nucleus-localized transcription activators but do not directly bind DNA. Mol. Cell Biol. 1985, 5, 2653–2661. [Google Scholar] [CrossRef] [PubMed]
- Webster, L.C.; Ricciardi, R.P. Trans-dominant mutants of E1A provide genetic evidence that the zinc finger of the trans-activating domain binds a transcription factor. Mol. Cell Biol. 1991, 11, 4287–4296. [Google Scholar] [CrossRef] [PubMed]
- Boyer, T.G.; Martin, M.E.; Lees, E.; Ricciardi, R.P.; Berk, A.J. Mammalian SRB/Mediator complex is targeted by adenovirus E1A protein. Nature 1999, 399, 276–279. [Google Scholar] [CrossRef] [PubMed]
- Cantin, G.T.; Stevens, J.L.; Berk, A.J. Activation domain-mediator interactions promote transcription preinitiation complex assembly on promoter DNA. Proc. Natl. Acad. Sci. USA 2003, 100, 12003–12008. [Google Scholar] [CrossRef] [PubMed]
- Geisberg, J.V.; Lee, W.S.; Berk, A.J.; Ricciardi, R.P. The zinc finger region of the adenovirus E1A transactivating domain complexes with the TATA box binding protein. Proc. Natl. Acad. Sci. USA 1994, 91, 2488–2492. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Berk, A.J. In vivo association of adenovirus large E1A protein with the human mediator complex in adenovirus-infected and -transformed cells. J. Virol. 2002, 76, 9186–9193. [Google Scholar] [CrossRef] [PubMed]
- Ablack, J.N.G.; Pelka, P.; Yousef, A.F.; Turnell, A.S.; Grand, R.J.A.; Mymryk, J.S. Comparison of E1A CR3-dependent transcriptional activation across six different human adenovirus subgroups. J. Virol. 2010, 84, 12771–12781. [Google Scholar] [CrossRef] [PubMed]
- Berk, A.J.; Lee, F.; Harrison, T.; Williams, J.; Sharp, P.A. Pre-early adenovirus 5 gene product regulates synthesis of early viral messenger RNAs. Cell 1979, 17, 935–944. [Google Scholar] [CrossRef]
- Jones, N.; Shenk, T. An adenovirus type 5 early gene function regulates expression of other early viral genes. Proc. Natl. Acad. Sci. USA 1979, 76, 3665–3669. [Google Scholar] [CrossRef] [PubMed]
- King, C.R.; Gameiro, S.F.; Tessier, T.M.; Zhang, A.; Mymryk, J.S. Mimicry of Cellular A Kinase-Anchoring Proteins Is a Conserved and Critical Function of E1A across Various Human Adenovirus Species. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.J.; Yousef, A.F.; Massimi, P.; Fonseca, G.J.; Todorovic, B.; Pelka, P.; Turnell, A.S.; Banks, L.; Mymryk, J.S. Dissection of the C-terminal region of E1A redefines the roles of CtBP and other cellular targets in oncogenic transformation. J. Virol. 2013, 87, 10348–10355. [Google Scholar] [CrossRef] [PubMed]
- Davey, N.E.; Cyert, M.S.; Moses, A.M. Short linear motifs - ex nihilo evolution of protein regulation. Cell Commun. Signal. 2015, 13, 43. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, A.; Tessier, T.M.; Galpin, K.J.C.; King, C.R.; Gameiro, S.F.; Anderson, W.W.; Yousef, A.F.; Qin, W.T.; Li, S.S.C.; Mymryk, J.S. The Transcriptional Repressor BS69 is a Conserved Target of the E1A Proteins from Several Human Adenovirus Species. Viruses 2018, 10, 662. https://doi.org/10.3390/v10120662
Zhang A, Tessier TM, Galpin KJC, King CR, Gameiro SF, Anderson WW, Yousef AF, Qin WT, Li SSC, Mymryk JS. The Transcriptional Repressor BS69 is a Conserved Target of the E1A Proteins from Several Human Adenovirus Species. Viruses. 2018; 10(12):662. https://doi.org/10.3390/v10120662
Chicago/Turabian StyleZhang, Ali, Tanner M. Tessier, Kristianne J. C. Galpin, Cason R. King, Steven F. Gameiro, Wyatt W. Anderson, Ahmed F. Yousef, Wen T. Qin, Shawn S. C. Li, and Joe S. Mymryk. 2018. "The Transcriptional Repressor BS69 is a Conserved Target of the E1A Proteins from Several Human Adenovirus Species" Viruses 10, no. 12: 662. https://doi.org/10.3390/v10120662
APA StyleZhang, A., Tessier, T. M., Galpin, K. J. C., King, C. R., Gameiro, S. F., Anderson, W. W., Yousef, A. F., Qin, W. T., Li, S. S. C., & Mymryk, J. S. (2018). The Transcriptional Repressor BS69 is a Conserved Target of the E1A Proteins from Several Human Adenovirus Species. Viruses, 10(12), 662. https://doi.org/10.3390/v10120662







