Elements Involved in the Rsv3-Mediated Extreme Resistance against an Avirulent Strain of Soybean Mosaic Virus
Abstract
1. Introduction
2. Materials and Methods
2.1. Soybean RNA-Seq Data
2.2. Annotation of Gene Functions
2.3. Pathway Analyses
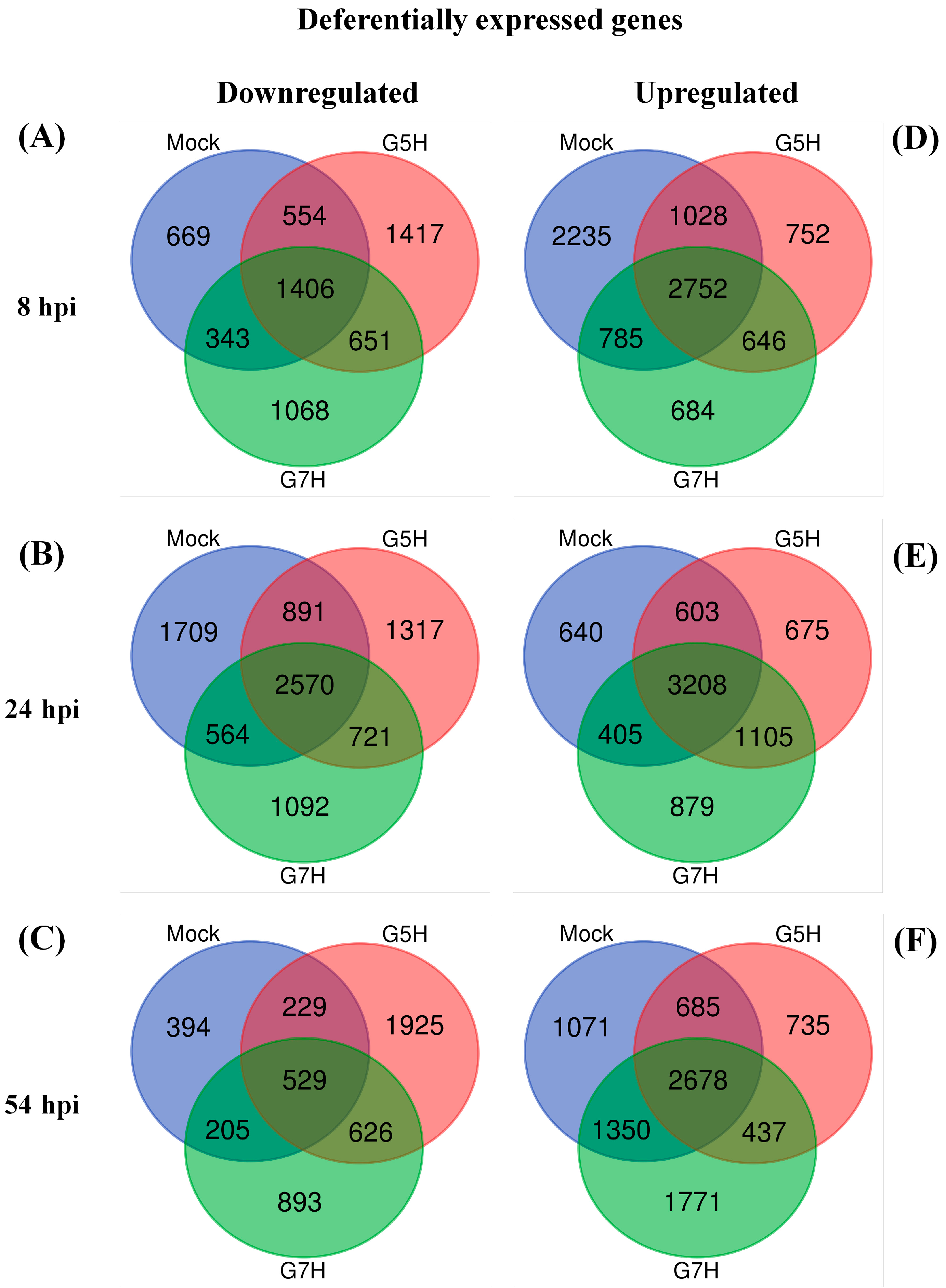
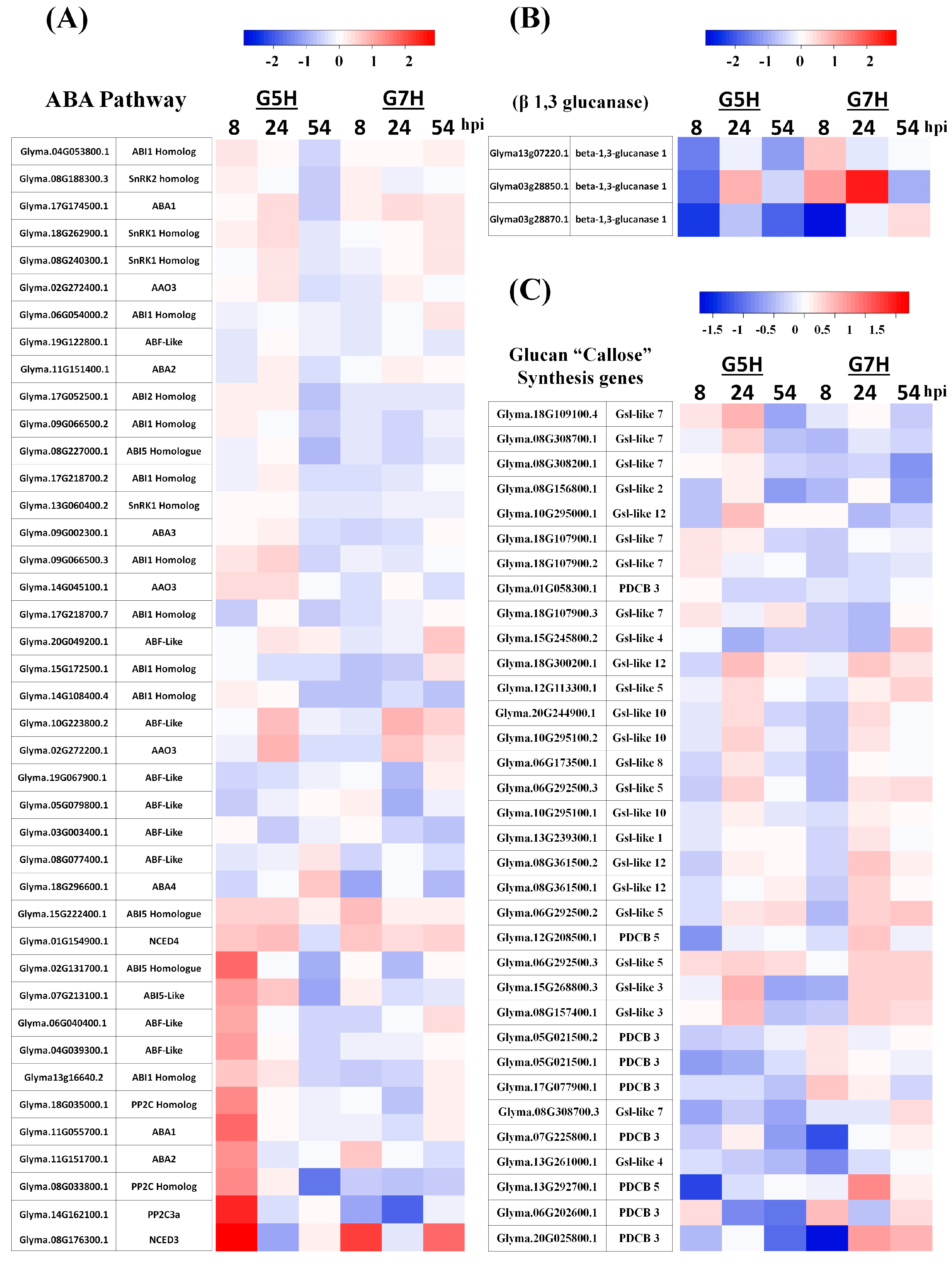
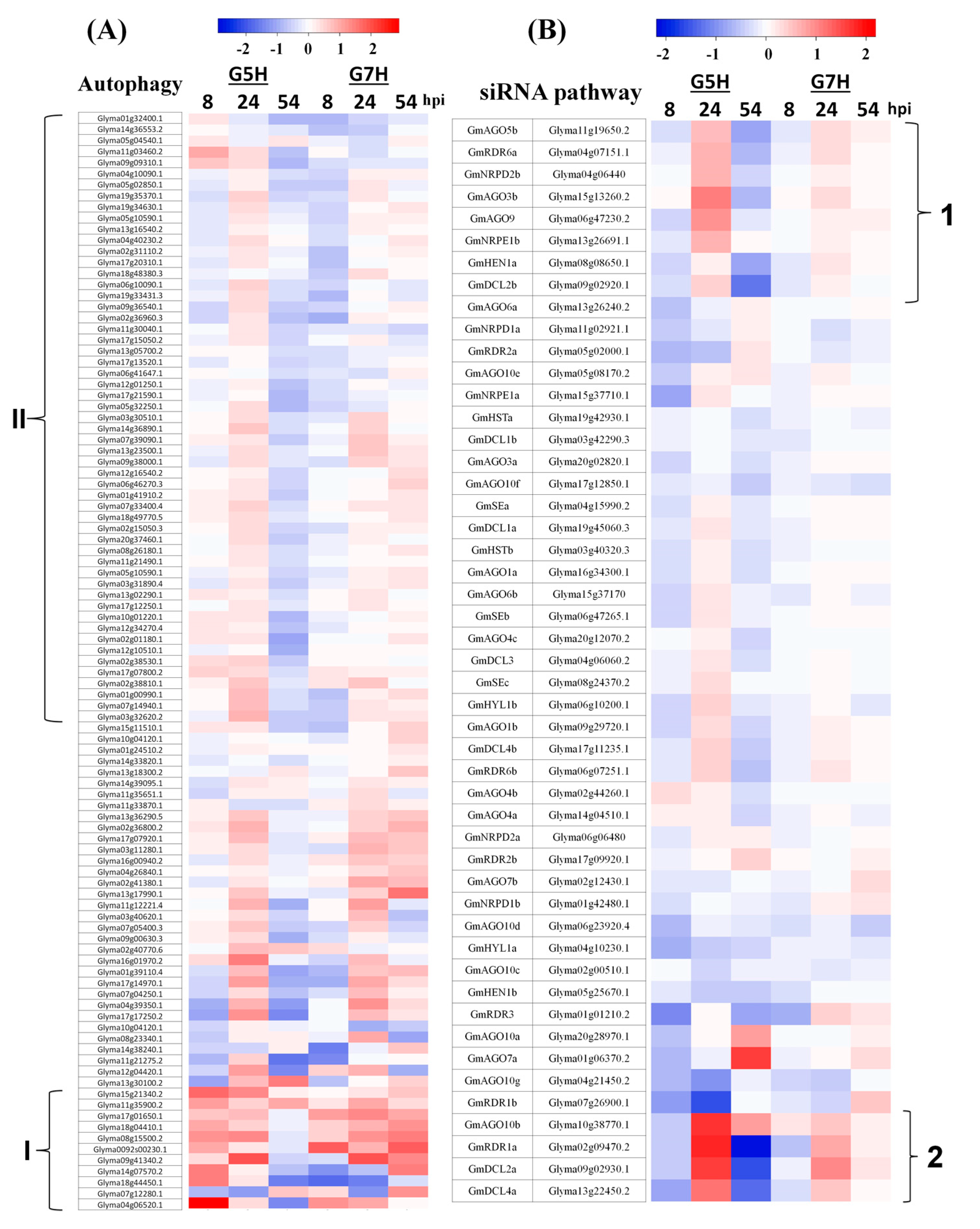
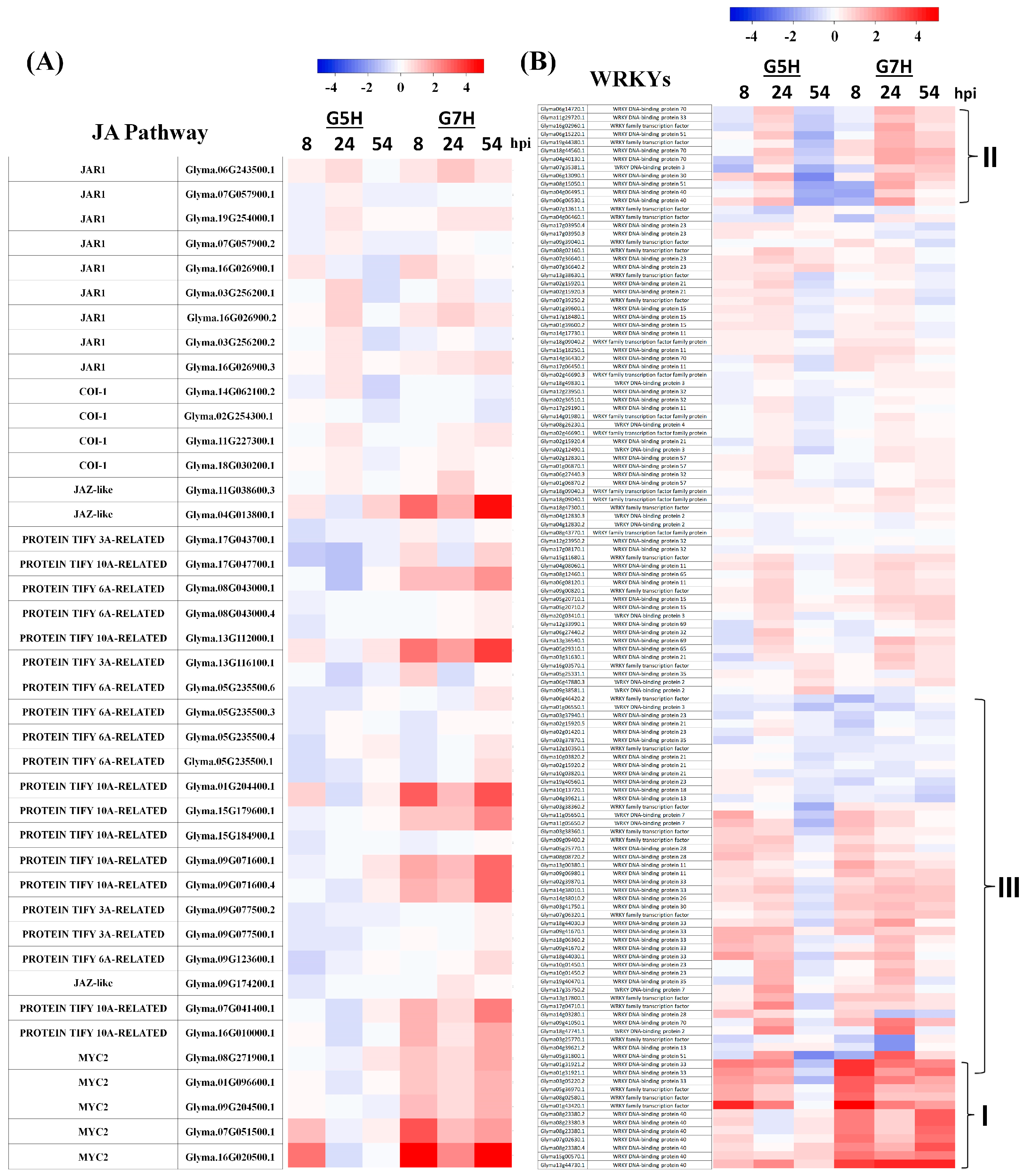
2.4. Venn Diagrams and Heatmaps
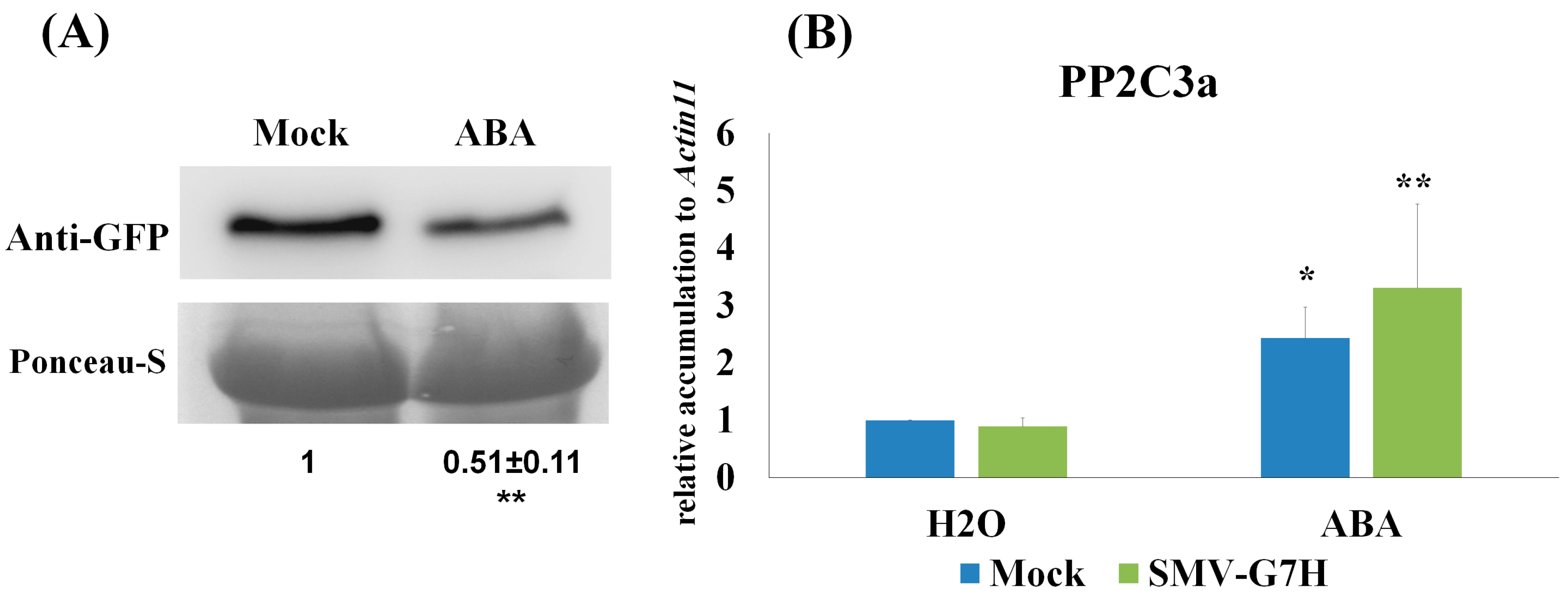
2.5. ABA Treatment
2.6. Virus Infection
2.7. RNA Analysis
2.8. Protein Blot
3. Results
3.1. Deferentially Expressed Genes Indicate Rapid Induction of the ABA Pathway
3.2. Autophagy and the Antiviral siRNA Pathway Are Regulated in G5H-Infected Plants
3.3. Other Defence Pathways May Have Opposite Effects on ER Against SMV
3.4. SA, CKs, and BRs May Not Be Involved in ER Against G5H
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kang, B.C.; Yeam, I.; Jahn, M.M. Genetics of plant virus resistance. Annu. Rev. Phytopathol. 2005, 43, 581–621. [Google Scholar] [CrossRef] [PubMed]
- Alazem, M.; Lin, N.S. Roles of plant hormones in the regulation of host-virus interactions. Mol. Plant Pathol. 2015, 16, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Soosaar, J.L.; Burch-Smith, T.M.; Dinesh-Kumar, S.P. Mechanisms of plant resistance to viruses. Nat. Rev. Microbiol. 2005, 3, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Carr, J.P.; Murphy, A.M.; Tungadi, T.; Yoon, J.-Y. Plant defense signals: Players and pawns in plant-virus-vector interactions. Plant Sci. 2018. [Google Scholar] [CrossRef]
- de Ronde, D.; Butterbach, P.; Kormelink, R. Dominant resistance against plant viruses. Front. Plant Sci. 2014, 5, 307. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, K.; Masuda, K.; Naito, S.; Meshi, T.; Ishikawa, M. An inhibitor of viral RNA replication is encoded by a plant resistance gene. Proc. Natl. Acad. Sci. USA 2007, 104, 13833–13838. [Google Scholar] [CrossRef] [PubMed]
- Conti, G.; Rodriguez, M.C.; Venturuzzi, A.L.; Asurmendi, S. Modulation of host plant immunity by Tobamovirus proteins. Ann. Bot. 2017, 119, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Tsuda, K.; Parker, J.E. Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 2015, 66, 487–511. [Google Scholar] [CrossRef] [PubMed]
- Baebler, S.; Witek, K.; Petek, M.; Stare, K.; Tusek-Znidaric, M.; Pompe-Novak, M.; Renaut, J.; Szajko, K.; Strzelczyk-Zyta, D.; Marczewski, W.; et al. Salicylic acid is an indispensable component of the Ny-1 resistance-gene-mediated response against Potato virus Y infection in potato. J. Exp. Bot. 2014, 65, 1095–1109. [Google Scholar] [CrossRef] [PubMed]
- Moffett, P. Transfer and modification of NLR proteins for virus resistance in plants. Curr. Opin. Virol. 2017, 26, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Alamillo, J.M.; Saenz, P.; Garcia, J.A. Salicylic acid-mediated and RNA-silencing defense mechanisms cooperate in the restriction of systemic spread of plum pox virus in tobacco. Plant. J. 2006, 48, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.K.; Kwon, S.J.; Cho, W.K.; Choi, H.S.; Kim, K.H. Type 2C Protein Phosphatase Is a Key Regulator of Antiviral Extreme Resistance Limiting Virus Spread. Sci. Rep. 2014, 4, 5905. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Z.; Fang, Y.; Pang, H.X. The Current Status of the Soybean-Soybean Mosaic Virus (SMV) Pathosystem. Front. Microbiol. 2016, 7, 1906. [Google Scholar] [CrossRef] [PubMed]
- Palukaitis, P. Resistance to Viruses of Potato and their Vectors. Plant Pathology J. 2012, 28, 248–258. [Google Scholar] [CrossRef]
- Bendahmane, A.; Kanyuka, K.; Baulcombe, D.C. The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 1999, 11, 781–792. [Google Scholar] [CrossRef] [PubMed]
- Townsend, P.D.; Dixon, C.H.; Slootweg, E.J.; Sukarta, O.C.A.; Yang, A.W.H.; Hughes, T.R.; Sharples, G.J.; Palsson, L.O.; Takken, F.L.W.; Goverse, A.; et al. The intracellular immune receptor Rx1 regulates the DNA-binding activity of a Golden2-like transcription factor. J. Biol. Chem. 2018, 293, 3218–3233. [Google Scholar] [CrossRef] [PubMed]
- Sekine, K.T.; Kawakami, S.; Hase, S.; Kubota, M.; Ichinose, Y.; Shah, J.; Kang, H.G.; Klessig, D.F.; Takahashi, H. High Level Expression of a Virus Resistance Gene, RCY1, Confers Extreme Resistance to Cucumber mosaic virus in Arabidopsis thaliana. Mol. Plant-Microbe Interact. 2008, 21, 1398–1407. [Google Scholar] [CrossRef] [PubMed]
- Tran, P.T.; Widyasari, K.; Seo, J.K.; Kim, K.H. Isolation and validation of a candidate Rsv3 gene from a soybean genotype that confers strain-specific resistance to soybean mosaic virus. Virology 2018, 513, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.K.; Lee, S.H.; Kim, K.H. Strain-Specific Cylindrical Inclusion Protein of Soybean mosaic virus Elicits Extreme Resistance and a Lethal Systemic Hypersensitive Response in Two Resistant Soybean Cultivars. Mol. Plant-Microbe Interact. 2009, 22, 1151–1159. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Kanayama, Y.; Zheng, M.S.; Kusano, T.; Hase, S.; Ikegami, M.; Shah, J. Antagonistic interactions between the SA and JA signaling pathways in Arabidopsis modulate expression of defense genes and gene-for-gene resistance to cucumber mosaic virus. Plant Cell Physiol. 2004, 45, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.Q.; Wu, N.Y.; Chow, C.N.; Tseng, K.C.; Chien, C.H.; Hung, Y.C.; Li, G.Z.; Chang, W.C. EXPath tool-a system for comprehensively analyzing regulatory pathways and coexpression networks from high-throughput transcriptome data. DNA Res. 2017, 24, 371–375. [Google Scholar] [CrossRef] [PubMed]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.J.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic. Acids. Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.K.; Choi, H.S.; Kim, K.H. Engineering of soybean mosaic virus as a versatile tool for studying protein-protein interactions in soybean. Sci. Rep. 2016, 6, 22436. [Google Scholar] [CrossRef] [PubMed]
- Rezzonico, E.; Flury, N.; Meins, F., Jr.; Beffa, R. Transcriptional down-regulation by abscisic acid of pathogenesis-related beta-1,3-glucanase genes in tobacco cell cultures. Plant Physiol. 1998, 117, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Oide, S.; Bejai, S.; Staal, J.; Guan, N.; Kaliff, M.; Dixelius, C. A novel role of PR2 in abscisic acid (ABA) mediated, pathogen-induced callose deposition in Arabidopsis thaliana. New Phytol. 2013, 200, 1187–1199. [Google Scholar] [CrossRef] [PubMed]
- Ellinger, D.; Voigt, C.A. Callose biosynthesis in Arabidopsis with a focus on pathogen response: What we have learned within the last decade. Ann. Bot. 2014, 114, 1349–1358. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Hong, Z.; Chatterjee, J.; Kim, S.; Verma, D.P. Expression of callose synthase genes and its connection with Npr1 signaling pathway during pathogen infection. Planta 2008, 229, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Simpson, C.; Thomas, C.; Findlay, K.; Bayer, E.; Maule, A.J. An Arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell 2009, 21, 581–594. [Google Scholar] [CrossRef] [PubMed]
- Kline, K.G.; Sussman, M.R.; Jones, A.M. Abscisic acid receptors. Plant Physiol. 2010, 154, 479–482. [Google Scholar] [CrossRef] [PubMed]
- Hafren, A.; Macia, J.L.; Love, A.J.; Milner, J.J.; Drucker, M.; Hofius, D. Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc. Natl. Acad. Sci. USA 2017, 114, E2026–E2035. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Li, Y.; Ding, S.W. Small RNA-based antimicrobial immunity. Nat. Rev. Immunol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.H.; Ro, S.H.; Cao, J.; Otto, N.M.; Kim, D.H. mTOR regulation of autophagy. FEBS Lett. 2010, 584, 1287–1295. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Sheen, J. The role of target of rapamycin signaling networks in plant growth and metabolism. Plant Physiol. 2014, 164, 499–512. [Google Scholar] [CrossRef] [PubMed]
- Alazem, M.; He, M.H.; Moffett, P.; Lin, N.S. Abscisic Acid Induces Resistance against Bamboo Mosaic Virus through Argonaute2 and 3. Plant Physiol. 2017, 174, 339–355. [Google Scholar] [CrossRef] [PubMed]
- Alazem, M.; Lin, N.S. Antiviral Roles of Abscisic Acid in Plants. Front. Plant Sci. 2017, 8, 1760. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Lu, T.; Dou, Y.; Yu, B.; Zhang, C. Identification of RNA silencing components in soybean and sorghum. BMC Bioinform. 2014, 15, 4. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, A.; Carrington, J.C. Antiviral roles of plant ARGONAUTES. Curr. Opin. Plant Biol. 2015, 27, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Musidlak, O.; Nawrot, R.; Gozdzicka-Jozefiak, A. Which Plant Proteins Are Involved in Antiviral Defense? Review on In Vivo and In Vitro Activities of Selected Plant Proteins against Viruses. Int. J. Mol. Sci. 2017, 18, 2300. [Google Scholar] [CrossRef] [PubMed]
- McLoughlin, F.; Basha, E.; Fowler, M.E.; Kim, M.; Bordowitz, J.; Katiyar-Agarwal, S.; Vierling, E. Class I and II Small Heat Shock Proteins Together with HSP101 Protect Protein Translation Factors during Heat Stress. Plant Physiol. 2016, 172, 1221–1236. [Google Scholar] [PubMed]
- Hernandez, J.A.; Gullner, G.; Clemente-Moreno, M.J.; Kunstler, A.; Juhasz, C.; Diaz-Vivancos, P.; Kiraly, L. Oxidative stress and antioxidative responses in plant-virus interactions. Physiol. Mol. Plant Pathol. 2016, 94, 134–148. [Google Scholar] [CrossRef]
- Gaufichon, L.; Reisdorf-Cren, M.; Rothstein, S.J.; Chardon, F.; Suzuki, A. Biological functions of asparagine synthetase in plants. Plant Sci. 2010, 179, 141–153. [Google Scholar] [CrossRef]
- Hakmaoui, A.; Perez-Bueno, M.L.; Garcia-Fontana, B.; Camejo, D.; Jimenez, A.; Sevilla, F.; Baron, M. Analysis of the antioxidant response of Nicotiana benthamiana to infection with two strains of Pepper mild mottle virus. J. Exp. Bot. 2012, 63, 5487–5496. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.P.; Somssich, I.E. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009, 150, 1648–1655. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Grosic, S.; Whitham, S.A.; Hill, J.H. The requirement of multiple defense genes in soybean Rsv1-mediated extreme resistance to soybean mosaic virus. Mol. Plant-Microbe Interact. 2012, 25, 1307–1313. [Google Scholar] [CrossRef] [PubMed]
- Walling, L.L. Extreme resistance: The GLK-Rx1 alliance. J. Biol. Chem. 2018, 293, 3234–3235. [Google Scholar] [CrossRef] [PubMed]
- Alazem, M.; Lin, K.Y.; Lin, N.S. The Abscisic Acid Pathway Has Multifaceted Effects on the Accumulation of Bamboo mosaic virus. Mol. Plant-Microbe Interact. 2014, 27, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Li, L.; Zhang, H.; Wang, R.; Tan, X.; He, Y.; Hong, G.; Li, J.; Ming, F.; Yao, X.; et al. Abscisic Acid Negatively Modulates Plant Defense against Rice Black-Streaked Dwarf Virus Infection by Suppressing the Jasmonate Pathway and Regulating ROS Levels in Rice. Plant Cell Environ. 2018, 41, 2504–2514. [Google Scholar] [CrossRef] [PubMed]
- Clavel, M.; Michaeli, S.; Genschik, P. Autophagy: A Double-Edged Sword to Fight Plant Viruses. Trends Plant Sci. 2017, 22, 646–648. [Google Scholar] [CrossRef] [PubMed]
- Parent, J.S.; Bouteiller, N.; Elmayan, T.; Vaucheret, H. Respective contributions of Arabidopsis DCL2 and DCL4 to RNA silencing. Plant J. 2015, 81, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Li, B.; Fan, Y.; Zhang, X.; Yu, Z.; Ryabov, E.; Zhao, M.; Wang, H.; Shi, N.; Zhang, P.; et al. Roles of Dicer-Like Proteins 2 and 4 in Intra- and Intercellular Antiviral Silencing. Plant Physiol. 2017, 174, 1067–1081. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.B.; Wu, Q.; Ito, T.; Cillo, F.; Li, W.X.; Chen, X.; Yu, J.L.; Ding, S.W. RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 484–489. [Google Scholar] [CrossRef] [PubMed]
- Aliyari, R.; Ding, S.W. RNA-based viral immunity initiated by the Dicer family of host immune receptors. Immunol. Rev. 2009, 227, 176–188. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, R.; Garcia-Marcos, A.; Manzano, A.; de Lacoba, M.G.; Camanes, G.; Garcia-Agustin, P.; Diaz-Ruiz, J.R.; Tenllado, F. Comparative analysis of transcriptomic and hormonal responses to compatible and incompatible plant-virus interactions that lead to cell death. Mol. Plant-Microbe Interact. 2012, 25, 709–723. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Marcos, A.; Pacheco, R.; Manzano, A.; Aguilar, E.; Tenllado, F. Oxylipin biosynthesis genes positively regulate programmed cell death during compatible infections with the synergistic pair potato virus X-potato virus Y and Tomato spotted wilt virus. J. Virol. 2013, 87, 5769–5783. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Yeakley, J.M.; Garcia, E.W.; Holdridge, J.D.; Fan, J.B.; Whitham, S.A. Salicylic acid-dependent expression of host genes in compatible Arabidopsis-virus interactions. Plant Physiol. 2005, 137, 1147–1159. [Google Scholar] [CrossRef] [PubMed]
- Oka, K.; Kobayashi, M.; Mitsuhara, I.; Seo, S. Jasmonic acid negatively regulates resistance to Tobacco mosaic virus in tobacco. Plant Cell Physiol. 2013, 54, 1999–2010. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alazem, M.; Tseng, K.-C.; Chang, W.-C.; Seo, J.-K.; Kim, K.-H. Elements Involved in the Rsv3-Mediated Extreme Resistance against an Avirulent Strain of Soybean Mosaic Virus. Viruses 2018, 10, 581. https://doi.org/10.3390/v10110581
Alazem M, Tseng K-C, Chang W-C, Seo J-K, Kim K-H. Elements Involved in the Rsv3-Mediated Extreme Resistance against an Avirulent Strain of Soybean Mosaic Virus. Viruses. 2018; 10(11):581. https://doi.org/10.3390/v10110581
Chicago/Turabian StyleAlazem, Mazen, Kuan-Chieh Tseng, Wen-Chi Chang, Jang-Kyun Seo, and Kook-Hyung Kim. 2018. "Elements Involved in the Rsv3-Mediated Extreme Resistance against an Avirulent Strain of Soybean Mosaic Virus" Viruses 10, no. 11: 581. https://doi.org/10.3390/v10110581
APA StyleAlazem, M., Tseng, K.-C., Chang, W.-C., Seo, J.-K., & Kim, K.-H. (2018). Elements Involved in the Rsv3-Mediated Extreme Resistance against an Avirulent Strain of Soybean Mosaic Virus. Viruses, 10(11), 581. https://doi.org/10.3390/v10110581






