Creating Disease Resistant Chickens: A Viable Solution to Avian Influenza?
Abstract
1. Introduction
2. Avian Influenza Viruses
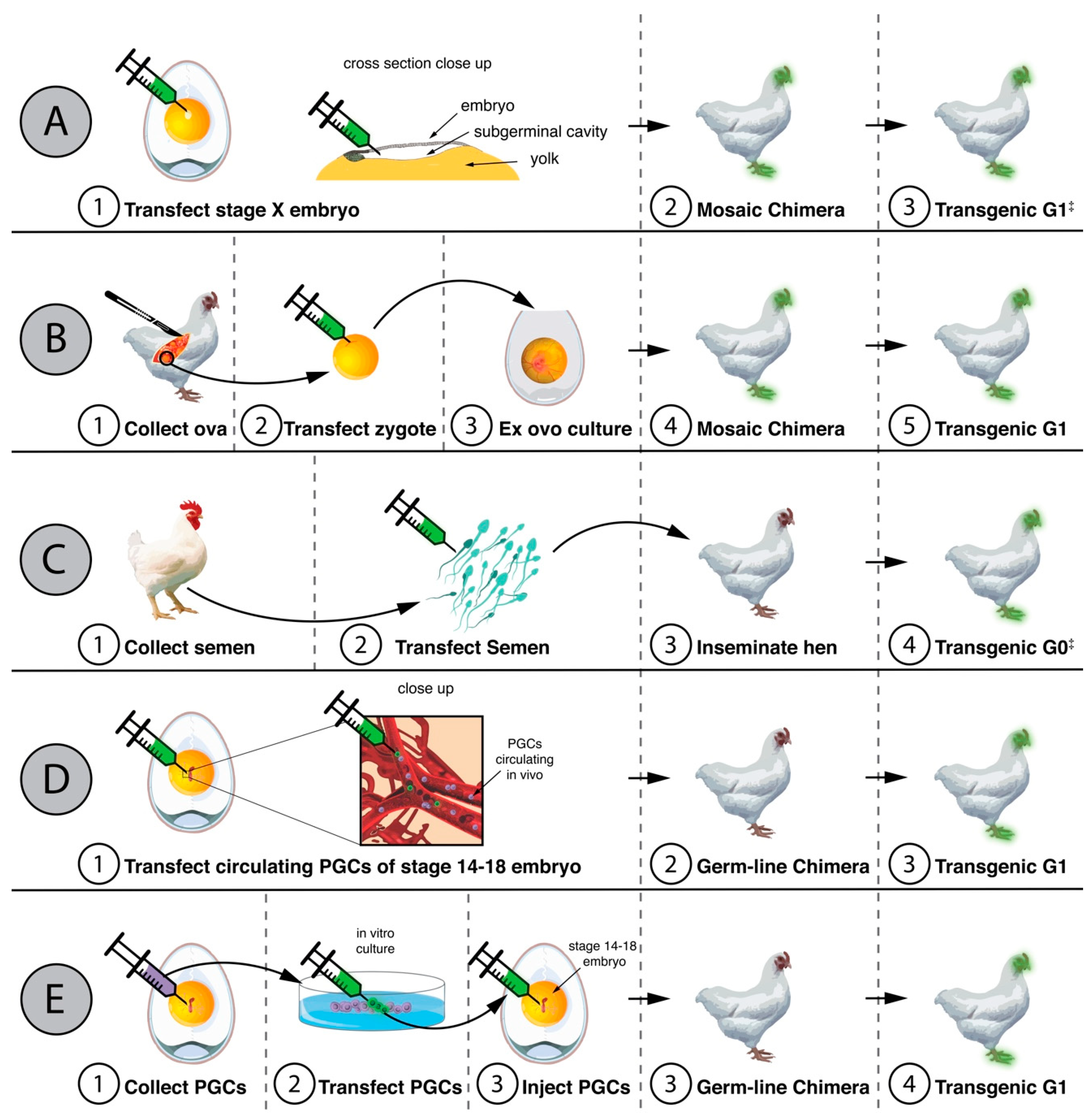
3. Genetically Modified Chickens Resistant to Avian Influenza Viruses
4. RIG-I: A Transgene of Interest?
5. Other Possible Immune Transgenes
6. Limitations of Transgenics
7. Alternative Approaches to Genetic Modification: Selective Breeding
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Waltz, E. First genetically engineered salmon sold in Canada. Nature 2017, 548, 148. [Google Scholar] [CrossRef] [PubMed]
- Wall, R.J.; Powell, A.M.; Paape, M.J.; Kerr, D.E.; Bannermann, D.D.; Pursel, V.G.; Wells, K.D.; Talbot, N.; Hawk, H.W. Genetically enhanced cows resist intramammary Staphylococcus aureus infection. Nat. Biotechnol. 2005, 23, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Lillico, S.G.; Proudfoot, C.; King, T.J.; Tan, W.; Zhang, L.; Mardjuki, R.; Paschon, D.E.; Rebar, E.J.; Urnov, F.D.; Mileham, A.J.; et al. Mammalian interspecies substitution of immune modulatory alleles by genome editing. Sci. Rep. 2016, 6, 21645. [Google Scholar] [CrossRef] [PubMed]
- Magdelaine, P.; Spiess, M.; Valceschini, E. Poultry meat consumption trends in Europe. Worlds Poult. Sci. J. 2008, 64, 53–64. [Google Scholar] [CrossRef]
- Si, Y.J.; Lee, I.W.; Kim, E.H.; Kim, Y.I.; Kwon, H.I.; Park, S.J.; Nguyen, H.D.; Kim, S.M.; Kwon, J.J.; Choi, W.S.; et al. Genetic characterisation of novel, highly pathogenic avian influenza (HPAI) H5N6 viruses isolated in birds, South Korea, November 2016. Euro Surveill. 2017, 22, 30434. [Google Scholar] [CrossRef] [PubMed]
- Drobik-Czwarno, W.; Wolc, A.; Fulton, J.E.; Jankowski, T.; Arango, J.; O’Sullivan, N.P.; Dekkers, J.C.M. Genetic basis of resistance to avian influenza in different commercial varieties of layer chickens. Poult. Sci. 2018, 97, 3421–3428. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.J.; Kwon, T.; Lyoo, Y.S. Challenges of influenza A viruses in humans and animals and current animal vaccines as an effective control measure. Clin. Exp. Vaccine Res. 2018, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Short, K.R.; Veldhuis Kroeze, E.J.; Reperant, L.A.; Richard, M.; Kuiken, T. Influenza virus and endothelial cells: A species specific relationship. Front. Microbiol. 2014, 5, 653. [Google Scholar] [CrossRef] [PubMed]
- Short, K.R.; Richard, M.; Verhagen, J.H.; van Riel, D.; Schrauwen, E.J.; van den Brand, J.M.; Mänz, B.; Bodewes, R.; Herfst, S. One health, multiple challenges: The inter-species transmission of influenza A virus. One Health 2015, 1, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Bertran, K.; Kwon, J.H.; Swayne, D.E. Evolution, global spread, and pathogenicity of highly pathogenic avian influenza H5Nx clade 2.3.4.4. J. Vet. Sci. 2017, 18, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Abdelwhab, E.; Hafez, H.M. Insight into alternative approaches for control of avian influenza in poultry, with emphasis on highly pathogenic H5N1. Viruses 2012, 4, 3179–3208. [Google Scholar] [CrossRef] [PubMed]
- Abdelwhab, E.-S.M.; Hafez, H.M. Control of Avian Influenza in Poultry with Antivirals and Molecular Manipulation. In Epidemiology II: Theory, Research and Practice; iConcept Press: Hong Kong, China, 2015; ISBN 978-1-922227-76-8. [Google Scholar]
- Domenech, J.; Dauphin, G.; Rushton, J.; McGrane, J.; Lubroth, J.; Tripodi, A.; Gilbert, J.; Sims, L.D. Experiences with vaccination in countries endemically infected with highly pathogenic avian influenza: The Food and Agriculture Organization perspective. Rev. Sci. Tech. 2009, 28, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Peyre, M.; Samaha, H.; Makonnen, Y.J.; Saad, A.; Abd-Elnabi, A.; Galal, S.; Ettel, T.; Dauphin, G.; Lubroth, J.; Roger, F.; et al. Avian influenza vaccination in Egypt: Limitations of the current strategy. J. Mol. Genet. Med. 2009, 3, 198–204. [Google Scholar] [PubMed]
- Spackman, E.; Pantin-Jackwood, M.J. Practical aspects of vaccination of poultry against avian influenza virus. Vet. J. 2014, 202, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Lee, D.H.; Fusaro, A.; Song, C.S.; Suarez, D.L.; Swayne, D.E. Poultry vaccination directed evolution of H9N2 low pathogenicity avian influenza viruses in Korea. Virology 2016, 488, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Lyall, J.; Irvine, R.M.; Sherman, A.; McKinley, T.J.; Núñez, A.; Purdie, A.; Outtrim, L.; Brown, I.H.; Rolleston-Smith, G.; Sang, H.; et al. Suppression of avian influenza transmission in genetically modified chickens. Science 2011, 331, 223–226. [Google Scholar] [CrossRef] [PubMed]
- Kishida, N.; Sakoda, Y.; Eto, M.; Sunaga, Y.; Kida, H. Co-infection of Staphylococcus aureus or Haemophilus paragallinarum exacerbates H9N2 influenza A virus infection in chickens. Arch. Virol. 2004, 149, 2095–2104. [Google Scholar] [CrossRef] [PubMed]
- SSlomka, M.J.; Seekings, A.H.; Mahmood, S.; Thomas, S.; Puranik, A.; Watson, S.; Byrne, A.M.P.; Hicks, D.; Nunez, A.; Brown, I.H.; et al. Unexpected infection outcomes of China-origin H7N9 low pathogenicity avian influenza virus in turkeys. Sci. Rep. 2018, 8, 7322. [Google Scholar] [CrossRef] [PubMed]
- June Byun, S.; Yuk, S.S.; Jang, Y.J.; Choi, H.; Jeon, M.H.; Erdene-Ochir, T.O.; Kwon, J.H.; Noh, J.Y.; Sun Kim, J.; Gyu Yoo, J.; et al. Transgenic Chickens Expressing the 3D8 Single Chain Variable Fragment Protein Suppress Avian Influenza Transmission. Sci. Rep. 2017, 7, 5938. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.; Smith, N.; Yu, L.; Paton, I.R.; Gutowska, M.W.; Forrest, H.L.; Danner, A.F.; Seiler, J.P.; Digard, P.; Webster, R.G.; et al. A comparative analysis of host responses to avian influenza infection in ducks and chickens highlights a role for the interferon-induced transmembrane proteins in viral resistance. BMC Genom. 2015, 16, 574. [Google Scholar] [CrossRef] [PubMed]
- Barber, M.R.; Aldridge, J.R., Jr.; Webster, R.G.; Magor, K.E. Association of RIG-I with innate immunity of ducks to influenza. Proc. Natl. Acad. Sci. USA 2010, 107, 5913–5918. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Shao, Q.; Zang, Y.; Guo, Q.; Zhang, Y.; Li, Z. Pigeon RIG-I function in innate immunity against H9N2 IAV and IBDV. Viruses 2015, 7, 4131–4151. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Satta, Y. Functional Evolution of Avian RIG-I-Like Receptors. Genes 2018, 9, 456. [Google Scholar] [CrossRef] [PubMed]
- Pasick, J.; Diederich, S.; Berhane, Y.; Embury-Hyatt1, C.; Wanhong, X. Imbalance between innate antiviral and pro-inflammatory immune responses may contribute to different outcomes involving low-and highly pathogenic avian influenza H5N3 infections in chickens. J. Gen. Virol. 2017, 98, 1245–1258. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Okada, H.; Itoh, T.; Tada, T.; Mase, M.; Nakamura, K.; Kubo, M.; Tsukamoto, K. Association of increased pathogenicity of Asian H5N1 highly pathogenic avian influenza viruses in chickens with highly efficient viral replication accompanied by early destruction of innate immune responses. J. Virol. 2009, 83, 7475–7486. [Google Scholar] [CrossRef] [PubMed]
- Liniger, M.; Summerfield, A.; Zimmer, G.; McCullough, K.C.; Ruggli, N. Chicken cells sense influenza A virus infection through MDA5 and CARDIF-signaling involving LGP2. J. Virol. 2012, 86, 705–717. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Meng, S.; Liu, X.; Sun, L.; Liu, W. Chicken cyclophilin A is an inhibitory factor to influenza virus replication. Virol. J. 2010, 7, 372. [Google Scholar] [CrossRef] [PubMed]
- Hsiang, T.-Y.; Zhao, C.; Krug, R.M. Interferon-induced ISG15 conjugation inhibits influenza A virus gene expression and replication in human cells. J. Virol. 2009, 83, 5971–5977. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Hinson, E.R.; Cresswell, P. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe 2007, 2, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Fuse, T.; Asano, I.; Tsukahara, F.; Maru, Y.; Nagata, K.; Kitazato, K.; Kobayashi, N. Identification of Hsc70 as an influenza virus matrix protein (M1) binding factor involved in the virus life cycle. FEBS Lett. 2006, 580, 5785–5790. [Google Scholar] [CrossRef] [PubMed]
- Honda, A.; Okamoto, T.; Ishihama, A. Host factor Ebp1: Selective inhibitor of influenza virus transcriptase. Genes Cells 2007, 12, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Rohaim, M.A.; Santhakumar, D.; Naggar, R.F.E.; Iqbal, M.; Hussein, H.A.; Munir, M. Chickens Expressing IFIT5 Ameliorate Clinical Outcome and Pathology of Highly Pathogenic Avian Influenza and Velogenic Newcastle Disease Viruses. Front. Immunol. 2018, 9, 2025. [Google Scholar] [CrossRef] [PubMed]
- Garas, L.C.; Murray, J.D.; Maga, E.A. Genetically engineered livestock: Ethical use for food and medical models. Annu. Rev. Anim. Biosci. 2015, 3, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Burt, D.W. Chicken genome: Current status and future opportunities. Genome Res. 2005, 15, 1692–1698. [Google Scholar] [CrossRef] [PubMed]
- Fulton, J.E. Selection for avian immune response: A commercial breeding company challenge. Poult. Sci. 2004, 83, 658–661. [Google Scholar] [CrossRef] [PubMed]
- Drobik-Czwarno, W.; Wolc, A.; Fulton, J.E.; Jankowski, T.; Arango, J.; O’Sullivan, N.P.; Dekkers, J.C.M. Identifying the genetic basis for resistance to avian influenza in commercial egg layer chickens. Animal 2018, 12, 1363–1371. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Hernandez, R.; Mwangi, W.; Peroval, M.; Sadeyen, J.R.; Ascough, S.; Balkissoon, D.; Staines, K.; Boyd, A.; McCauley, J.; Smith, A.; et al. Host genetics determine susceptibility to avian influenza infection and transmission dynamics. Sci. Rep. 2016, 6, 26787. [Google Scholar] [CrossRef] [PubMed]
- Matsuu, A.; Kobayashi, T.; Patchimasiri, T.; Shiina, T.; Suzuki, S.; Chaichoune, K.; Ratanakorn, P.; Hiromoto, Y.; Abe, H.; Parchariyanon, S.; et al. Pathogenicity of Genetically Similar, H5N1 Highly Pathogenic Avian Influenza Virus Strains in Chicken and the Differences in Sensitivity among Different Chicken Breeds. PLoS ONE 2016, 11, e0153649. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lupiani, B.; Reddy, S.M.; Lamont, S.J.; Zhou, H. RNA-seq analysis revealed novel genes and signaling pathway associated with disease resistance to avian influenza virus infection in chickens. Poult. Sci. 2014, 93, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Bernasconi, D.; Schultz, U.; Staeheli, P. The interferon-induced Mx protein of chickens lacks antiviral activity. J. Interferon Cytokine Res. 1995, 15, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.H.; Jin, H.K.; Asano, A.; Takada, A.; Ninomiya, A.; Kida, H.; Hokiyama, H.; Ohara, M.; Tsuzuki, M.; Nishibori, M.; et al. Polymorphisms and the differential antiviral activity of the chicken Mx gene. Genome Res. 2002, 12, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Qu, L.J.; Yao, J.F.; Yang, N. Skewed allele frequencies of an Mx gene mutation with potential resistance to avian influenza virus in different chicken populations. Poult. Sci. 2006, 85, 1327–1329. [Google Scholar] [CrossRef] [PubMed]
- Ewald, S.J.; Kapczynski, D.R.; Livant, E.J.; Suarez, D.L.; Ralph, J.; McLeod, S.; Miller, C. Association of Mx1 Asn631 variant alleles with reductions in morbidity, early mortality, viral shedding, and cytokine responses in chickens infected with a highly pathogenic avian influenza virus. Immunogenetics 2011, 63, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Benfield, C.T.; Lyall, J.W.; Kochs, G.; Tiley, L.S. Asparagine 631 variants of the chicken Mx protein do not inhibit influenza virus replication in primary chicken embryo fibroblasts or in vitro surrogate assays. J. Virol. 2008, 82, 7533–7539. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Brahmakshatriya, V.; Lupiani, B.; Reddy, S.; Okimoto, R.; Li, X.; Chiang, H.; Zhou, H. Associations of chicken Mx1 polymorphism with antiviral responses in avian influenza virus infected embryos and broilers. Poult. Sci. 2012, 91, 3019–3024. [Google Scholar] [CrossRef] [PubMed]
- Sironi, L.; Williams, J.L.; Moreno-Martin, A.M.; Ramelli, P.; Stella, A.; Jianlin, H.; Weigend, S.; Lombardi, G.; Cordioli, P.; Mariani, P. Susceptibility of different chicken lines to H7N1 highly pathogenic avian influenza virus and the role of Mx gene polymorphism coding amino acid position 631. Virology 2008, 380, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Steinman, L. Elaborate interactions between the immune and nervous systems. Nat. Immunol. 2004, 5, 575–581. [Google Scholar] [CrossRef]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Looi, F.Y.; Baker, M.L.; Townson, T.; Richard, M.; Novak, B.; Doran, T.J.; Short, K.R. Creating Disease Resistant Chickens: A Viable Solution to Avian Influenza? Viruses 2018, 10, 561. https://doi.org/10.3390/v10100561
Looi FY, Baker ML, Townson T, Richard M, Novak B, Doran TJ, Short KR. Creating Disease Resistant Chickens: A Viable Solution to Avian Influenza? Viruses. 2018; 10(10):561. https://doi.org/10.3390/v10100561
Chicago/Turabian StyleLooi, Fong Yang, Michelle L. Baker, Thomas Townson, Mathilde Richard, Ben Novak, Tim J. Doran, and Kirsty R. Short. 2018. "Creating Disease Resistant Chickens: A Viable Solution to Avian Influenza?" Viruses 10, no. 10: 561. https://doi.org/10.3390/v10100561
APA StyleLooi, F. Y., Baker, M. L., Townson, T., Richard, M., Novak, B., Doran, T. J., & Short, K. R. (2018). Creating Disease Resistant Chickens: A Viable Solution to Avian Influenza? Viruses, 10(10), 561. https://doi.org/10.3390/v10100561




