Visualization of HIV-1 RNA Transcription from Integrated HIV-1 DNA in Reactivated Latently Infected Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Virus
2.2. Antibodies and Compounds
2.3. Nucleic Acid Probes
2.4. In Situ vRNA Detection via Microscopy
2.5. Simultaneous In Situ vRNA and vDNA Detection via Microscopy
2.6. Immunostaining for Microscopy
2.7. Imaging and Imaging Quantification
2.8. In Situ vRNA Detection via Flow Cytometry Analysis
2.9. Statisitcal Analyses
3. Results
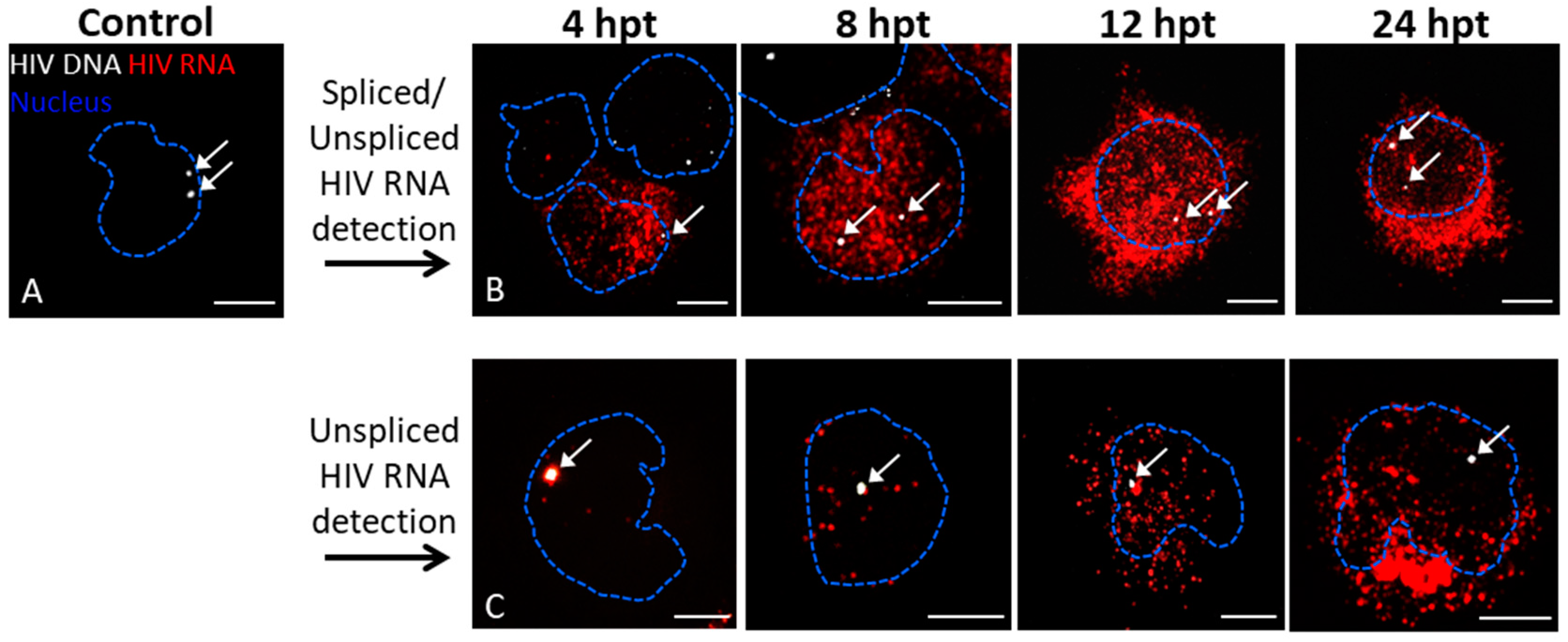
3.1. Detection of Protein and Nucleic Acids in a Model Latent Cell Line
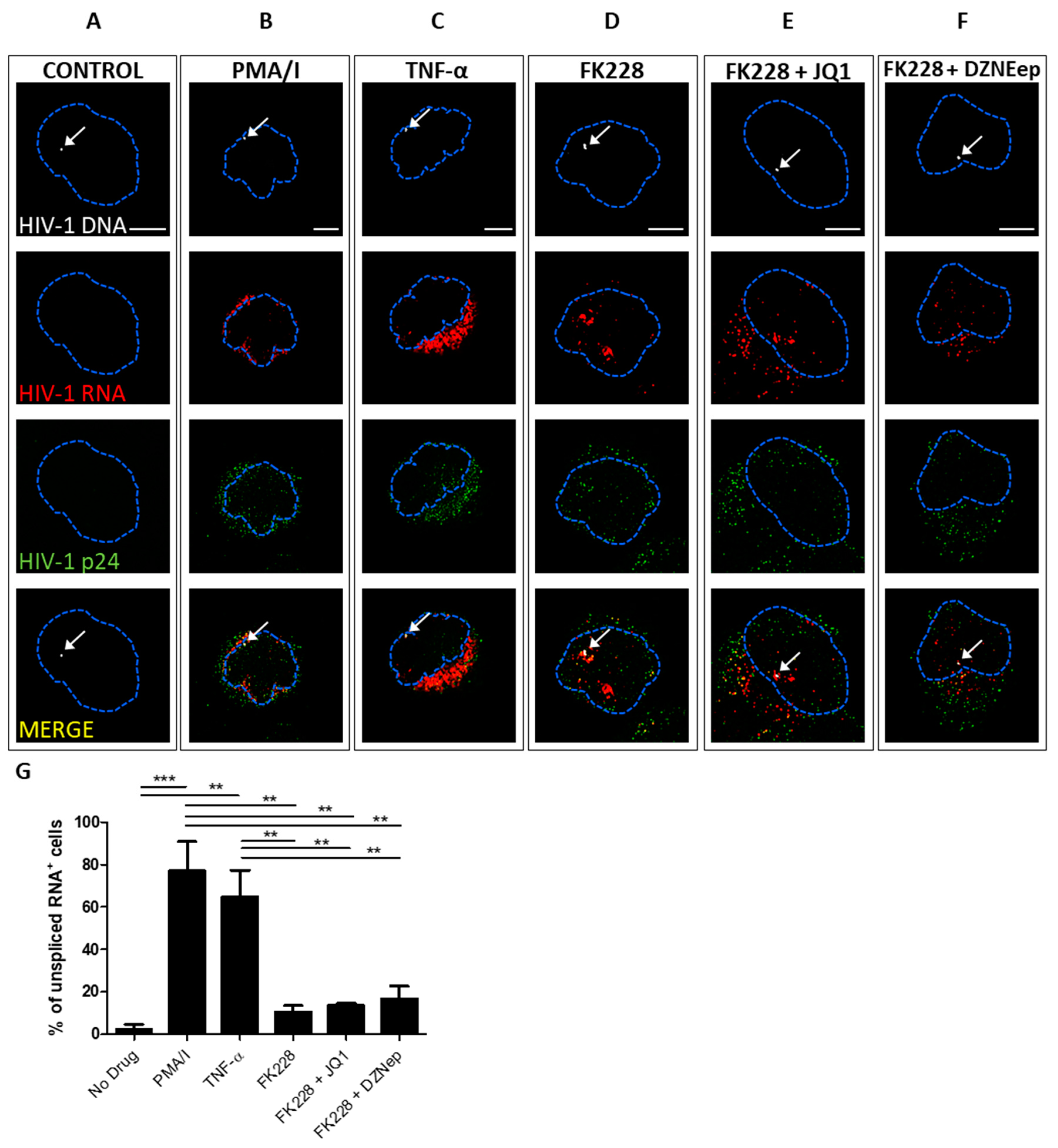
3.2. Quantifying the Efficacy of Latency Reactivating Agents
3.3. Quantification of Latency Reactivation by Flow Cytometry
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bruner, K.M.; Hosmane, N.N.; Siliciano, R.F. Towards an HIV-1 cure: Measuring the latent reservoir. Trends Microbiol. 2015, 23, 192–203. [Google Scholar] [CrossRef] [PubMed]
- Siliciano, J.D.; Siliciano, R.F. Recent developments in the effort to cure HIV infection: Going beyond N = 1. J. Clin. Investig. 2016, 126, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Kimata, J.T.; Rice, A.P.; Wang, J. Challenges and strategies for the eradication of the HIV reservoir. Curr. Opin. Immunol. 2016, 42, 65–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Margolis, D.M.; Garcia, J.V.; Hazuda, D.J.; Haynes, B.F. Latency reversal and viral clearance toc cure HIV-1. Science 2016, 353, aaf6517. [Google Scholar] [CrossRef] [PubMed]
- Finzi, D. Identification of a Reservoir for HIV-1 in Patients on Highly Active Antiretroviral Therapy. Science 1997, 278, 1295–1300. [Google Scholar] [CrossRef] [PubMed]
- Finzi, D.; Blankson, J.; Siliciano, J.D.; Margolick, J.B.; Chadwick, K.; Pierson, T.; Smith, K.; Lisziewicz, J.; Lori, F.; Flexner, C.; et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 1999, 5, 512–517. [Google Scholar] [CrossRef] [PubMed]
- Siliciano, J.D.; Siliciano, R.F. Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol. Biol. 2005, 304, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Laird, G.M.; Eisele, E.E.; Rabi, S.A.; Lai, J.; Chioma, S.; Blankson, J.N.; Siliciano, J.D.; Siliciano, R.F. Rapid Quantification of the Latent Reservoir for HIV-1 Using a Viral Outgrowth Assay. PLoS Pathog. 2013, 9, e1003398. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.C.; Shan, L.; Hosmane, N.N.; Wang, J.; Laskey, S.B.; Rosenbloom, D.I.S.; Lai, J.; Blankson, J.N.; Siliciano, J.D.; Siliciano, R.F. XReplication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 2013, 155, 540–551. [Google Scholar] [CrossRef] [PubMed]
- Laird, G.M.; Rosenbloom, D.I.S.; Lai, J.; Siliciano, R.F.; Siliciano, J.D. Measuring the Frequency of Latent HIV-1 in Resting CD4+ T Cells Using a Limiting Dilution Coculture Assay. Methods Mol. Biol. 2016, 1354, 239–253. [Google Scholar] [CrossRef] [PubMed]
- Butler, S.L.; Hansen, M.S.; Bushman, F.D. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 2001, 7, 631–634. [Google Scholar] [CrossRef] [PubMed]
- Rouzioux, C.; Mélard, A.; Avéttand-Fénoël, V. Quantification of total HIV1-DNA in peripheral blood mononuclear cells. Methods Mol. Biol. 2014, 1087, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Cillo, A.R.; Vagratian, D.; Bedison, M.A.; Anderson, E.M.; Kearney, M.F.; Fyne, E.; Koontz, D.; Coffin, J.M.; Piatak, M.; Mellors, J.W. Improved single-copy assays for quantification of persistent HIV-1 viremia in patients on suppressive antiretroviral therapy. J. Clin. Microbiol. 2014, 52, 3944–3951. [Google Scholar] [CrossRef] [PubMed]
- O’Doherty, U.; Swiggard, W.J.; Jeyakumar, D.; McGain, D.; Malim, M.H. A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. J. Virol. 2002, 76, 10942–10950. [Google Scholar] [CrossRef] [PubMed]
- Hermankova, M.; Siliciano, J.D.; Zhou, Y.; Monie, D.; Chadwick, K.; Margolick, J.B.; Quinn, T.C.; Siliciano, R.F. Analysis of human immunodeficiency virus type 1 gene expression in latently infected resting CD4+ T lymphocytes in vivo. J. Virol. 2003, 77, 7383–7392. [Google Scholar] [CrossRef] [PubMed]
- Liszewski, M.K.; Yu, J.J.; O’Doherty, U. Detecting HIV-1 integration by repetitive-sampling Alu-gag PCR. Methods 2009, 47, 254–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henrich, T.J.; Gallien, S.; Li, J.Z.; Pereyra, F.; Kuritzkes, D.R. Low-level detection and quantitation of cellular HIV-1 DNA and 2-LTR circles using droplet digital PCR. J. Virol. Methods 2012, 186, 68–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eriksson, S.; Graf, E.H.; Dahl, V.; Strain, M.C.; Yukl, S.A.; Lysenko, E.S.; Bosch, R.J.; Lai, J.; Chioma, S.; Emad, F.; et al. Comparative Analysis of Measures of Viral Reservoirs in HIV-1 Eradication Studies. PLoS Pathog. 2013, 9, e1003174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strain, M.C.; Lada, S.M.; Luong, T.; Rought, S.E.; Gianella, S.; Terry, V.H.; Spina, C.A.; Woelk, C.H.; Richman, D.D. Highly precise measurement of HIV DNA by droplet digital PCR. PLoS ONE 2013, 8, e55943. [Google Scholar] [CrossRef] [PubMed]
- Cillo, A.R.; Sobolewski, M.D.; Bosch, R.J.; Fyne, E.; Piatak, M.; Coffin, J.M.; Mellors, J.W. Quantification of HIV-1 latency reversal in resting CD4+ T cells from patients on suppressive antiretroviral therapy. Proc. Natl. Acad. Sci. USA 2014, 111, 7078–7083. [Google Scholar] [CrossRef] [PubMed]
- De Spiegelaere, W.; Malatinkova, E.; Lynch, L.; Van Nieuwerburgh, F.; Messiaen, P.; O’Doherty, U.; Vandekerckhove, L. Quantification of integrated HIV DNA by repetitive-sampling Alu-HIV PCR on the basis of poisson statistics. Clin. Chem. 2014, 60, 886–895. [Google Scholar] [CrossRef] [PubMed]
- Hanley, M.B.; Lomas, W.; Mittar, D.; Maino, V.; Park, E. Detection of Low Abundance RNA Molecules in Individual Cells by Flow Cytometry. PLoS ONE 2013, 8, e57002. [Google Scholar] [CrossRef] [PubMed]
- Deleage, C.; Wietgrefe, S.W.; Del Prete, G.; Morcock, D.R.; Hao, X.-P.; Piatak, M., Jr.; Bess, J.; Anderson, J.L.; Perkey, K.; Reilly, C.; et al. Defining HIV and SIV Reservoirs in Lymphoid Tissues. Pathog. Immun. 2016, 1, 68–106. [Google Scholar] [CrossRef] [PubMed]
- Baxter, A.E.; Niessl, J.; Fromentin, R.; Richard, J.; Porichis, F.; Charlebois, R.; Massanella, M.; Brassard, N.; Alsahafi, N.; Delgado, G.G.; et al. Single-Cell Characterization of Viral Translation-Competent Reservoirs in HIV-Infected Individuals. Cell Host Microbe 2016, 20, 368–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baxter, A.E.; Niessl, J.; Fromentin, R.; Richard, J.; Porichis, F.; Massanella, M.; Brassard, N.; Alsahafi, N.; Routy, J.P.; Finzi, A.; et al. Multiparametric characterization of rare HIV-infected cells using an RNA-flow FISH technique. Nat. Protoc. 2017, 12, 2029–2049. [Google Scholar] [CrossRef] [PubMed]
- Grau-Expósito, J.; Serra-Peinado, C.; Miguel, L.; Navarro, J.; Curran, A.; Burgos, J.; Ocaña, I.; Ribera, E.; Torrella, A.; Planas, B.; et al. A novel single-cell FISH-flow assay identifies effector memory CD4+ T cells as a major niche for HIV-1 transcription in HIV-infected patients. MBio 2017, 8, e00876-17. [Google Scholar] [CrossRef]
- Niessl, J.; Baxter, A.E.; Kaufmann, D.E. Tools for Visualizing HIV in Cure Research. Curr. HIV/AIDS Rep. 2018, 15, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Puray-Chavez, M.; Tedbury, P.R.; Huber, A.D.; Ukah, O.B.; Yapo, V.; Liu, D.; Ji, J.; Wolf, J.J.; Engelman, A.N.; Sarafianos, S.G. Multiplex single-cell visualization of nucleic acids and protein during HIV infection. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Jordan, A.; Bisgrove, D.; Verdin, E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 2003, 22, 1868–1877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez, V.L.; Rowe, T.; Justement, J.S.; Butera, S.T.; June, C.H.; Folks, T.M. An HIV-1-infected T cell clone defective in IL-2 production and Ca2+ mobilization after CD3 stimulation. J. Immunol. 1991, 147, 3145–3148. [Google Scholar] [PubMed]
- Chavez, L.; Calvanese, V.; Verdin, E. HIV Latency Is Established Directly and Early in Both Resting and Activated Primary CD4 T Cells. PLoS Pathog. 2015, 11, e1004955. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.H.; Fouchier, R.A.; Southerling, T.E.; Guerra, C.B.; Grant, C.K.; Malim, M.H. The Vif and Gag proteins of human immunodeficiency virus type 1 colocalize in infected human T cells. J. Virol. 1997, 71, 5259–5267. [Google Scholar] [PubMed]
- Fouchier, R.A.M.; Meyer, B.E.; Simon, J.H.M.; Fischer, U.; Malim, M.H. HIV-1 infection of non-dividing cells: Evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 1997, 16, 4531–4539. [Google Scholar] [CrossRef] [PubMed]
- Linhoff, M.W.; Garg, S.K.; Mandel, G. A High-Resolution Imaging Approach to Investigate Chromatin Architecture in Complex Tissues. Cell 2015, 163, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Duh, E.J.; Maury, W.J.; Folks, T.M.; Fauci, A.S.; Rabson, A.B. Tumor necrosis factor alpha activates human immunodeficiency virus type 1 through induction of nuclear factor binding to the NF-kappa B sites in the long terminal repeat. Proc. Natl. Acad. Sci. USA 1989, 86, 5974–5978. [Google Scholar] [CrossRef] [PubMed]
- Bagnarelli, P.; Valenza, A.; Menzo, S.; Sampaolesi, R.; Varaldo, P.E.; Butini, L.; Montroni, M.; Perno, C.F.; Aquaro, S.; Mathez, D.; et al. Dynamics and modulation of human immunodeficiency virus type 1 transcripts in vitro and in vivo. J. Virol. 1996, 70, 7603–7613. [Google Scholar] [PubMed]
- Malim, M.H.; Hauber, J.; Le, S.-Y.; Maizel, J.V.; Cullen, B.R. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 1989, 338, 254–257. [Google Scholar] [CrossRef] [PubMed]
- Hanly, S.M.; Rimsky, L.T.; Malim, M.H.; Kim, J.H.; Hauber, J.; Duc Dodon, M.; Le, S.Y.; Maizel, J.V.; Cullen, B.R.; Greene, W.C. Comparative analysis of the HTLV-I Rex and HIV-1 Rev trans-regulatory proteins and their RNA response elements. Genes Dev. 1989, 3, 1534–1544. [Google Scholar] [CrossRef] [PubMed]
- Karn, J.; Stoltzfus, C.M. Transcriptional and posttranscriptional regulation of HIV-1 gene expression. Cold Spring Harb. Perspect. Med. 2012, 2, a006916. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, J.D.; Valandra, G.; Roderiquez, G.; Bushar, G.; Giri, C.; Norcross, M.A. Phorbol ester enhances human immunodeficiency virus-promoted gene expression and acts on a repeated 10-base-pair functional enhancer element. Mol. Cell. Biol. 1987, 7, 3759–3766. [Google Scholar] [CrossRef] [PubMed]
- Chatila, T.; Silverman, L.; Miller, R.; Geha, R. Mechanisms of T cell activation by the calcium ionophore ionomycin. J. Immunol. 1989, 143, 1283–1289. [Google Scholar] [PubMed]
- Wei, D.G.; Chiang, V.; Fyne, E.; Balakrishnan, M.; Barnes, T.; Graupe, M.; Hesselgesser, J.; Irrinki, A.; Murry, J.P.; Stepan, G.; et al. Histone deacetylase inhibitor romidepsin induces HIV expression in CD4 T cells from patients on suppressive antiretroviral therapy at concentrations achieved by clinical dosing. PLoS Pathog. 2014, 10, e1004071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jonsson, K.L.; Tolstrup, M.; Vad-Nielsen, J.; Kjær, K.; Laustsen, A.; Andersen, M.N.; Rasmussen, T.A.; Søgaard, O.S.; Østergaard, L.; Denton, P.W.; et al. Histone deacetylase inhibitor romidepsin inhibits de novo HIV-1 infections. Antimicrob. Agents Chemother. 2015, 59, 3984–3994. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Guo, J.; Wu, Y.; Zhou, Q. The BET bromodomain inhibitor JQ1 activates HIV latency through antagonizing Brd4 inhibition of Tat-transactivation. Nucleic Acids Res. 2013, 41, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Boehm, D.; Calvanese, V.; Dar, R.D.; Xing, S.; Schroeder, S.; Martins, L.; Aull, K.; Li, P.-C.; Planelles, V.; Bradner, J.E.; et al. BET bromodomain-targeting compounds reactivate HIV from latency via a Tat-independent mechanism. Cell Cycle 2013, 12, 452–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laird, G.M.; Bullen, C.K.; Rosenbloom, D.I.S.; Martin, A.R.; Hill, A.L.; Durand, C.M.; Siliciano, J.D.; Siliciano, R.F. Ex vivo analysis identifies effective HIV-1 latency-reversing drug combinations. J. Clin. Investig. 2015, 125, 1901–1912. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.; Cho, W.-K.; Chu, C.K.; Keedy, K.S.; Archin, N.M.; Margolis, D.M.; Karn, J. Epigenetic silencing of HIV-1 by the histone H3 lysine 27 methyltransferase enhancer of Zeste 2. J. Virol. 2011, 85, 9078–9089. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-S.; Liu, Y.; Wu, T.-C.; Du, G.-Y.; Zhang, F.-J. EZH2 phosphorylation regulates Tat-induced HIV-1 transactivation via ROS/Akt signaling pathway. FEBS Lett. 2015, 589, 4106–4111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vranckx, L.S.; Demeulemeester, J.; Saleh, S.; Boll, A.; Vansant, G.; Schrijvers, R.; Weydert, C.; Battivelli, E.; Verdin, E.; Cereseto, A.; et al. LEDGIN-mediated Inhibition of Integrase-LEDGF/p75 Interaction Reduces Reactivation of Residual Latent HIV. EBioMedicine 2016, 8, 248–264. [Google Scholar] [CrossRef] [PubMed]
- Spina, C.A.; Anderson, J.; Archin, N.M.; Bosque, A.; Chan, J.; Famiglietti, M.; Greene, W.C.; Kashuba, A.; Lewin, S.R.; Margolis, D.M.; et al. An In-Depth Comparison of Latent HIV-1 Reactivation in Multiple Cell Model Systems and Resting CD4+ T Cells from Aviremic Patients. PLoS Pathog. 2013, 9, e1003834. [Google Scholar] [CrossRef] [PubMed]
- McGary, C.S.; Deleage, C.; Harper, J.; Micci, L.; Ribeiro, S.P.; Paganini, S.; Kuri-Cervantes, L.; Benne, C.; Ryan, E.S.; Balderas, R.; et al. CTLA-4+PD-1−Memory CD4+ T Cells Critically Contribute to Viral Persistence in Antiretroviral Therapy-Suppressed, SIV-Infected Rhesus Macaques. Immunity 2017, 47, 776–788. [Google Scholar] [CrossRef] [PubMed]
- Popovic, M.; Sarngadharan, M.G.; Read, E.; Gallo, R.C. Detection, isolation, and continuous production of cytopathic retroviruses (HTLV-III) from patients with AIDS and pre-AIDS. Science 1984, 224, 497–500. [Google Scholar] [CrossRef] [PubMed]
- Popovic, M.; Read-Connole, E.; Gallo, R.C. T4 positive human neoplastic cell lines susceptible to and permissive for HTLV-III. Lancet 1984, 2, 1472–1473. [Google Scholar] [CrossRef]
- Mann, D.L.; O’Brien, S.J.; Gilbert, D.A.; Reid, Y.; Popovic, M.; Read-Connole, E.; Gallo, R.C.; Gazdar, A.F. Origin of the HIV-susceptible human CD4+ cell line H9. AIDS Res. Hum. Retroviruses 1989, 5, 253–255. [Google Scholar] [CrossRef] [PubMed]
- Ratner, L.; Haseltine, W.; Patarca, R.; Livak, K.J.; Starcich, B.; Josephs, S.F.; Doran, E.R.; Rafalski, J.A.; Whitehorn, E.A.; Baumeister, K. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature 1985, 313, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Symons, J.; Chopra, A.; Malatinkova, E.; De Spiegelaere, W.; Leary, S.; Cooper, D.; Abana, C.O.; Rhodes, A.; Rezaei, S.D.; Vandekerckhove, L.; et al. HIV integration sites in latently infected cell lines: Evidence of ongoing replication. Retrovirology 2017, 14, 2. [Google Scholar] [CrossRef] [PubMed]
- Imamichi, H.; Dewar, R.L.; Adelsberger, J.W.; Rehm, C.A.; O’Doherty, U.; Paxinos, E.E.; Fauci, A.S.; Lane, H.C. Defective HIV-1 proviruses produce novel protein-coding RNA species in HIV-infected patients on combination antiretroviral therapy. Proc. Natl. Acad. Sci. USA 2016, 113, 8783–8788. [Google Scholar] [CrossRef] [PubMed]
- Pollack, R.A.; Jones, R.B.; Pertea, M.; Bruner, K.M.; Martin, A.R.; Thomas, A.S.; Capoferri, A.A.; Beg, S.A.; Huang, S.H.; Karandish, S.; et al. Defective HIV-1 Proviruses Are Expressed and Can Be Recognized by Cytotoxic T Lymphocytes, which Shape the Proviral Landscape. Cell Host Microbe 2017, 21, 494–506.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ukah, O.B.; Puray-Chavez, M.; Tedbury, P.R.; Herschhorn, A.; Sodroski, J.G.; Sarafianos, S.G. Visualization of HIV-1 RNA Transcription from Integrated HIV-1 DNA in Reactivated Latently Infected Cells. Viruses 2018, 10, 534. https://doi.org/10.3390/v10100534
Ukah OB, Puray-Chavez M, Tedbury PR, Herschhorn A, Sodroski JG, Sarafianos SG. Visualization of HIV-1 RNA Transcription from Integrated HIV-1 DNA in Reactivated Latently Infected Cells. Viruses. 2018; 10(10):534. https://doi.org/10.3390/v10100534
Chicago/Turabian StyleUkah, Obiaara B., Maritza Puray-Chavez, Philip R. Tedbury, Alon Herschhorn, Joseph G. Sodroski, and Stefan G. Sarafianos. 2018. "Visualization of HIV-1 RNA Transcription from Integrated HIV-1 DNA in Reactivated Latently Infected Cells" Viruses 10, no. 10: 534. https://doi.org/10.3390/v10100534
APA StyleUkah, O. B., Puray-Chavez, M., Tedbury, P. R., Herschhorn, A., Sodroski, J. G., & Sarafianos, S. G. (2018). Visualization of HIV-1 RNA Transcription from Integrated HIV-1 DNA in Reactivated Latently Infected Cells. Viruses, 10(10), 534. https://doi.org/10.3390/v10100534




