Immune Modulation of NYVAC-Based HIV Vaccines by Combined Deletion of Viral Genes that Act on Several Signalling Pathways
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Cells and Viruses
2.3. Construction of the Plasmid Transfer Vector pGem-RG-B6R-B10R-wm
2.4. Construction of NYVAC-Based Deletion Mutants
2.5. PCR Analysis of Deletion Mutants
2.6. Western Blot Detection of HIV-1 Protein gp120 and Gag-Pol-Nef Expression
2.7. Analysis of Virus Growth
2.8. DNA Vectors
2.9. Peptides
2.10. Mouse Immunization Schedule
2.11. Intracellular Cytokine Staining Assay (ICS)
2.12. Antibody Measurement by ELISA
2.13. Plaque Neutralization Assay
2.14. Murine Intranasal Challenge
2.15. Data Analysis and Statistics
3. Results
3.1. Generation and In Vitro Characterization of NYVAC-C Deletion Mutants
3.2. Cellular Immune Profile Induced by NYVAC-C Recombinants after Combined Deletion of VACV Immunomodulatory Genes
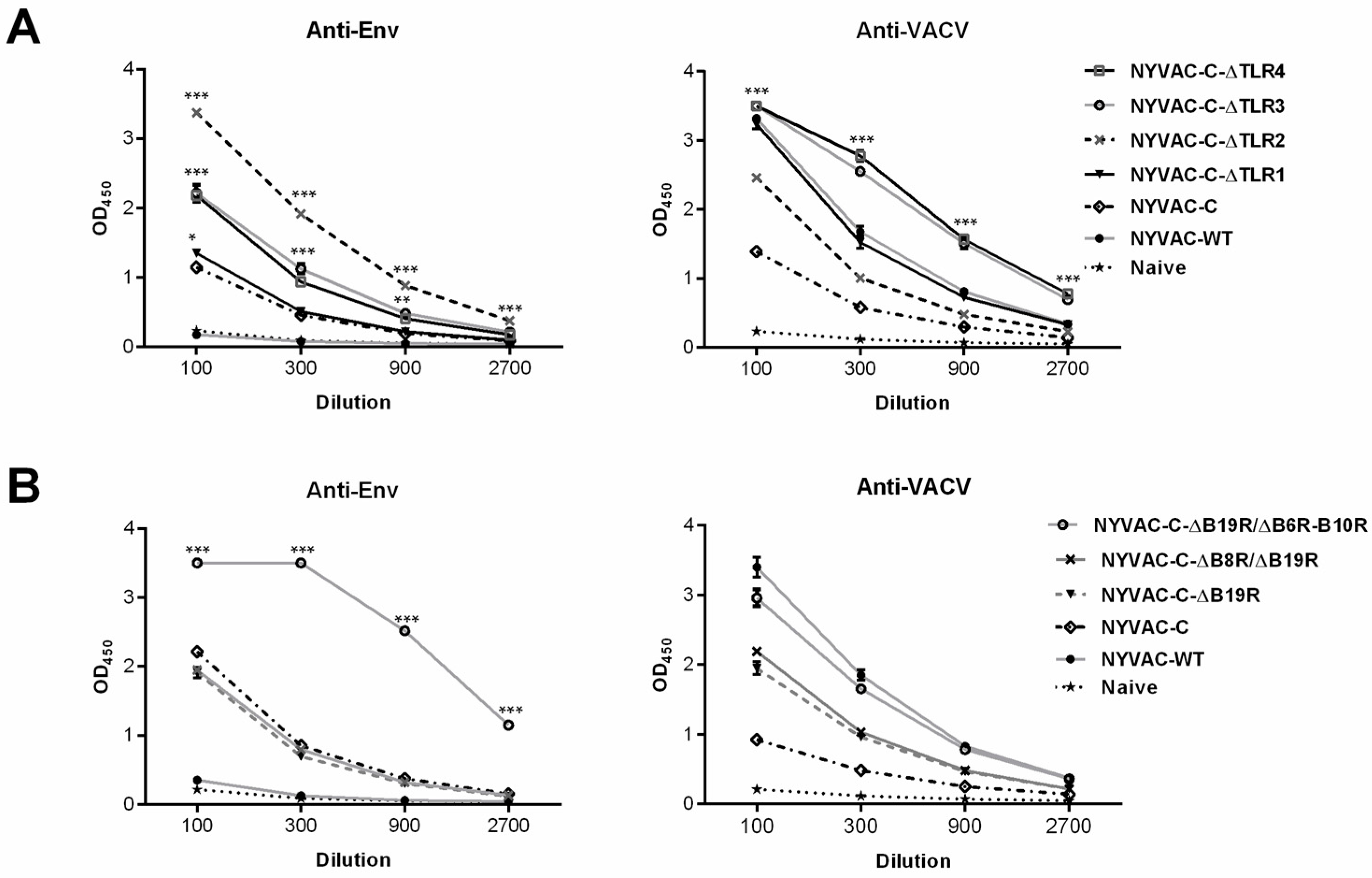
3.2.1. Immunogenicity of NYVAC-C Recombinants Induced by Combined Deletion of VACV-TLR Inhibitors A46R, A52R, K7R and B15R
3.2.2. Effect of Combined Deletion of Several Unknown Non-Essential Genes and Cytokine Inhibitors on NYVAC-C Recombinant Immunogenicity
3.3. Combined Deletion of Immunomodulatory Genes Alters Humoral Responses to Env and VACV Vector
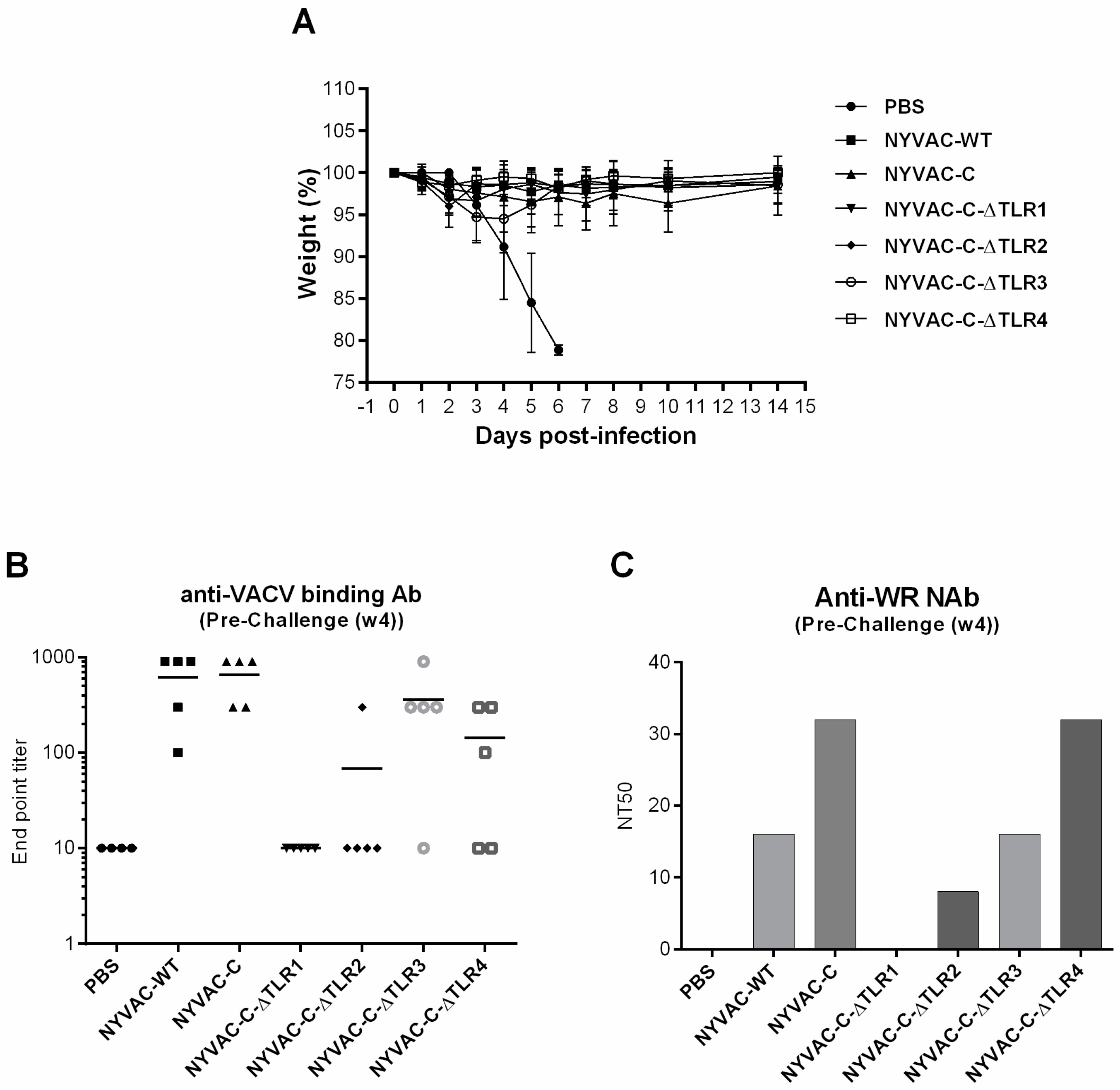
3.4. Single and Combined Deletion of VACV-TLR Inhibitors A46R, A52R, K7R and B15R Induced Similar “In Vivo” Protection against an Intranasal Challenge with a Lethal Dose of Wild-Type VACV WR
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Rerks-Ngarm, S.; Pitisuttithum, P.; Nitayaphan, S.; Kaewkungwal, J.; Chiu, J.; Paris, R.; Premsri, N.; Namwat, C.; de Souza, M.; Adams, E.; et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 2009, 361, 2209–2220. [Google Scholar] [CrossRef] [PubMed]
- Haynes, B.F.; Gilbert, P.B.; McElrath, M.J.; Zolla-Pazner, S.; Tomaras, G.D.; Alam, S.M.; Evans, D.T.; Montefiori, D.C.; Karnasuta, C.; Sutthent, R.; et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 2012, 366, 1275–1286. [Google Scholar] [CrossRef] [PubMed]
- Zolla-Pazner, S.; deCamp, A.; Gilbert, P.B.; Williams, C.; Yates, N.L.; Williams, W.T.; Howington, R.; Fong, Y.; Morris, D.E.; Soderberg, K.A.; et al. Vaccine-induced IgG antibodies to V1V2 regions of multiple HIV-1 subtypes correlate with decreased risk of HIV-1 infection. PLoS ONE 2014, 9, e87572. [Google Scholar] [CrossRef] [PubMed]
- Bonsignori, M.; Montefiori, D.C.; Wu, X.; Chen, X.; Hwang, K.K.; Tsao, C.Y.; Kozink, D.M.; Parks, R.J.; Tomaras, G.D.; Crump, J.A.; et al. Two distinct broadly neutralizing antibody specificities of different clonal lineages in a single HIV-1-infected donor: Implications for vaccine design. J. Virol. 2012, 86, 4688–4692. [Google Scholar] [CrossRef] [PubMed]
- Tartaglia, J.; Perkus, M.E.; Taylor, J.; Norton, E.K.; Audonnet, J.C.; Cox, W.I.; Davis, S.W.; van der Hoeven, J.; Meignier, B.; Riviere, M.; et al. NYVAC: A highly attenuated strain of vaccinia virus. Virology 1992, 188, 217–232. [Google Scholar] [CrossRef]
- Patterson, L.J.; Peng, B.; Abimiku, A.G.; Aldrich, K.; Murty, L.; Markham, P.D.; Kalyanaraman, V.S.; Alvord, W.G.; Tartaglia, J.; Franchini, G.; et al. Cross-protection in NYVAC-HIV-1-immunized/HIV-2-challenged but not in NYVAC-HIV-2-immunized/SHIV-challenged rhesus macaques. AIDS 2000, 14, 2445–2455. [Google Scholar] [CrossRef] [PubMed]
- Asbach, B.; Kliche, A.; Kostler, J.; Perdiguero, B.; Esteban, M.; Jacobs, B.L.; Montefiori, D.C.; LaBranche, C.C.; Yates, N.L.; Tomaras, G.D.; et al. Potential to Streamline Heterologous DNA Prime and NYVAC/Protein Boost HIV Vaccine Regimens in Rhesus Macaques by Employing Improved Antigens. J. Virol. 2016, 90, 4133–4149. [Google Scholar] [CrossRef] [PubMed]
- Gomez, C.E.; Najera, J.L.; Jimenez, V.; Bieler, K.; Wild, J.; Kostic, L.; Heidari, S.; Chen, M.; Frachette, M.J.; Pantaleo, G.; et al. Generation and immunogenicity of novel HIV/AIDS vaccine candidates targeting HIV-1 Env/Gag-Pol-Nef antigens of clade C. Vaccine 2007, 25, 1969–1992. [Google Scholar] [CrossRef] [PubMed]
- Mooij, P.; Balla-Jhagjhoorsingh, S.S.; Koopman, G.; Beenhakker, N.; van Haaften, P.; Baak, I.; Nieuwenhuis, I.G.; Kondova, I.; Wagner, R.; Wolf, H.; et al. Differential CD4+ versus CD8+ T-cell responses elicited by different poxvirus-based human immunodeficiency virus type 1 vaccine candidates provide comparable efficacies in primates. J. Virol. 2008, 82, 2975–2988. [Google Scholar] [CrossRef] [PubMed]
- Bart, P.A.; Goodall, R.; Barber, T.; Harari, A.; Guimaraes-Walker, A.; Khonkarly, M.; Sheppard, N.C.; Bangala, Y.; Frachette, M.J.; Wagner, R.; et al. EV01: A phase I trial in healthy HIV negative volunteers to evaluate a clade C HIV vaccine, NYVAC-C undertaken by the EuroVacc Consortium. Vaccine 2008, 26, 3153–3161. [Google Scholar] [CrossRef] [PubMed]
- Harari, A.; Bart, P.A.; Stohr, W.; Tapia, G.; Garcia, M.; Medjitna-Rais, E.; Burnet, S.; Cellerai, C.; Erlwein, O.; Barber, T.; et al. An HIV-1 clade C DNA prime, NYVAC boost vaccine regimen induces reliable, polyfunctional and long-lasting T cell responses. J. Exp. Med. 2008, 205, 63–77. [Google Scholar] [CrossRef] [PubMed]
- McCormack, S.; Stohr, W.; Barber, T.; Bart, P.A.; Harari, A.; Moog, C.; Ciuffreda, D.; Cellerai, C.; Cowen, M.; Gamboni, R.; et al. EV02: A Phase I trial to compare the safety and immunogenicity of HIV DNA-C prime-NYVAC-C boost to NYVAC-C alone. Vaccine 2008, 26, 3162–3174. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Arriaza, J.; Perdiguero, B.; Heeney, J.; Seaman, M.; Montefiori, D.C.; Labranche, C.; Yates, N.L.; Shen, X.; Tomaras, G.D.; Ferrari, G.; et al. Head-to-Head Comparison of Poxvirus NYVAC and ALVAC Vectors Expressing Identical HIV-1 Clade C Immunogens in Prime-Boost Combination with Env Protein in Nonhuman Primates. J. Virol. 2015, 89, 8525–8539. [Google Scholar] [CrossRef] [PubMed]
- Gomez, C.E.; Perdiguero, B.; Najera, J.L.; Sorzano, C.O.; Jimenez, V.; Gonzalez-Sanz, R.; Esteban, M. Removal of vaccinia virus genes that block interferon type I and II pathways improves adaptive and memory responses of the HIV/AIDS vaccine candidate NYVAC-C in mice. J. Virol. 2012, 86, 5026–5038. [Google Scholar] [CrossRef] [PubMed]
- Perdiguero, B.; Gomez, C.E.; Di Pilato, M.; Sorzano, C.O.; Delaloye, J.; Roger, T.; Calandra, T.; Pantaleo, G.; Esteban, M. Deletion of the vaccinia virus gene A46R, encoding for an inhibitor of TLR signalling, is an effective approach to enhance the immunogenicity in mice of the HIV/AIDS vaccine candidate NYVAC-C. PLoS ONE 2013, 8, e74831. [Google Scholar] [CrossRef] [PubMed]
- Di Pilato, M.; Mejias-Perez, E.; Sorzano, C.O.S.; Esteban, M. Distinct Roles of Vaccinia Virus NF-kappaB Inhibitor Proteins A52, B15 and K7 in the Immune Response. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Di Pilato, M.; Mejias-Perez, E.; Zonca, M.; Perdiguero, B.; Gomez, C.E.; Trakala, M.; Nieto, J.; Najera, J.L.; Sorzano, C.O.; Combadiere, C.; et al. NFkappaB activation by modified vaccinia virus as a novel strategy to enhance neutrophil migration and HIV-specific T-cell responses. Proc. Natl. Acad. Sci. USA 2015, 112, E1333–E1342. [Google Scholar] [CrossRef] [PubMed]
- Kibler, K.V.; Gomez, C.E.; Perdiguero, B.; Wong, S.; Huynh, T.; Holechek, S.; Arndt, W.; Jimenez, V.; Gonzalez-Sanz, R.; Denzler, K.; et al. Improved NYVAC-based vaccine vectors. PLoS ONE 2011, 6, e25674. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, J.C.; Gherardi, M.M.; Esteban, M. Biology of attenuated modified vaccinia virus Ankara recombinant vector in mice: Virus fate and activation of B- and T-cell immune responses in comparison with the Western Reserve strain and advantages as a vaccine. J. Virol. 2000, 74, 923–933. [Google Scholar] [CrossRef] [PubMed]
- Stack, J.; Haga, I.R.; Schroder, M.; Bartlett, N.W.; Maloney, G.; Reading, P.C.; Fitzgerald, K.A.; Smith, G.L.; Bowie, A.G. Vaccinia virus protein A46R targets multiple Toll-like-interleukin-1 receptor adaptors and contributes to virulence. J. Exp. Med. 2005, 201, 1007–1018. [Google Scholar] [CrossRef] [PubMed]
- Harte, M.T.; Haga, I.R.; Maloney, G.; Gray, P.; Reading, P.C.; Bartlett, N.W.; Smith, G.L.; Bowie, A.; O’Neill, L.A. The poxvirus protein A52R targets Toll-like receptor signaling complexes to suppress host defense. J. Exp. Med. 2003, 197, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Schroder, M.; Baran, M.; Bowie, A.G. Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon-mediated IRF activation. EMBO J. 2008, 27, 2147–2157. [Google Scholar] [CrossRef] [PubMed]
- TTeferi, W.M.; Desaulniers, M.A.; Noyce, R.S.; Shenouda, M.; Umer, B.; Evans, D.H. The vaccinia virus K7 protein promotes histone methylation associated with heterochromatin formation. PLoS ONE 2017, 12, e0173056. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.A.; Ryzhakov, G.; Cooray, S.; Randow, F.; Smith, G.L. Inhibition of IkappaB kinase by vaccinia virus virulence factor B14. PLoS Pathog. 2008, 4, e22. [Google Scholar] [CrossRef] [PubMed]
- Symons, J.A.; Alcami, A.; Smith, G.L. Vaccinia virus encodes a soluble type I interferon receptor of novel structure and broad species specificity. Cell 1995, 81, 551–560. [Google Scholar] [CrossRef]
- Symons, J.A.; Tscharke, D.C.; Price, N.; Smith, G.L. A study of the vaccinia virus interferon-gamma receptor and its contribution to virus virulence. J. Gen. Virol. 2002, 83, 1953–1964. [Google Scholar] [CrossRef] [PubMed]
- Alejo, A.; Ruiz-Arguello, M.B.; Ho, Y.; Smith, V.P.; Saraiva, M.; Alcami, A. A chemokine-binding domain in the tumor necrosis factor receptor from variola (smallpox) virus. Proc. Natl. Acad. Sci. USA 2006, 103, 5995–6000. [Google Scholar] [CrossRef] [PubMed]
- Price, N.; Tscharke, D.C.; Smith, G.L. The vaccinia virus B9R protein is a 6 kDa intracellular protein that is non-essential for virus replication and virulence. J. Gen. Virol. 2002, 83, 873–878. [Google Scholar] [CrossRef] [PubMed]
- Tscharke, D.C.; Woo, W.P.; Sakala, I.G.; Sidney, J.; Sette, A.; Moss, D.J.; Bennink, J.R.; Karupiah, G.; Yewdell, J.W. Poxvirus CD8+ T-cell determinants and cross-reactivity in BALB/c mice. J. Virol. 2006, 80, 6318–6323. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Arriaza, J.; Najera, J.L.; Gomez, C.E.; Sorzano, C.O.; Esteban, M. Immunogenic profiling in mice of a HIV/AIDS vaccine candidate (MVA-B) expressing four HIV-1 antigens and potentiation by specific gene deletions. PLoS ONE 2010, 5, e12395. [Google Scholar] [CrossRef] [PubMed]
- Najera, J.L.; Gomez, C.E.; Garcia-Arriaza, J.; Sorzano, C.O.; Esteban, M. Insertion of vaccinia virus C7L host range gene into NYVAC-B genome potentiates immune responses against HIV-1 antigens. PLoS ONE 2010, 5, e11406. [Google Scholar] [CrossRef] [PubMed]
- Excler, J.L.; Michael, N.L. Lessons from HIV-1 vaccine efficacy trials. Curr. Opin. HIV AIDS 2016. [Google Scholar] [CrossRef] [PubMed]
- Gomez, C.E.; Perdiguero, B.; Garcia-Arriaza, J.; Esteban, M. Poxvirus vectors as HIV/AIDS vaccines in humans. Hum. Vaccin. Immunother. 2012, 8, 1192–1207. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, K.E.; D’Couto, H.T.; Barouch, D.H. New concepts in HIV-1 vaccine development. Curr. Opin. Immunol. 2016, 41, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Falivene, J.; del Medico Zajac, M.P.; Pascutti, M.F.; Rodriguez, A.M.; Maeto, C.; Perdiguero, B.; Gomez, C.E.; Esteban, M.; Calamante, G.; Gherardi, M.M. Improving the MVA vaccine potential by deleting the viral gene coding for the IL-18 binding protein. PLoS ONE 2012, 7, e32220. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Arriaza, J.; Arnaez, P.; Gomez, C.E.; Sorzano, C.O.; Esteban, M. Improving Adaptive and Memory Immune Responses of an HIV/AIDS Vaccine Candidate MVA-B by Deletion of Vaccinia Virus Genes (C6L and K7R) Blocking Interferon Signaling Pathways. PLoS ONE 2013, 8, e66894. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Arriaza, J.; Najera, J.L.; Gomez, C.E.; Tewabe, N.; Sorzano, C.O.; Calandra, T.; Roger, T.; Esteban, M. A candidate HIV/AIDS vaccine (MVA-B) lacking vaccinia virus gene C6L enhances memory HIV-1-specific T-cell responses. PLoS ONE 2011, 6, e24244. [Google Scholar] [CrossRef] [PubMed]
- Perdiguero, B.; Gomez, C.E.; Najera, J.L.; Sorzano, C.O.; Delaloye, J.; Gonzalez-Sanz, R.; Jimenez, V.; Roger, T.; Calandra, T.; Pantaleo, G.; et al. Deletion of the viral anti-apoptotic gene F1L in the HIV/AIDS vaccine candidate MVA-C enhances immune responses against HIV-1 antigens. PLoS ONE 2012, 7, e48524. [Google Scholar] [CrossRef] [PubMed]
- Garber, D.A.; O’Mara, L.A.; Gangadhara, S.; McQuoid, M.; Zhang, X.; Zheng, R.; Gill, K.; Verma, M.; Yu, T.; Johnson, B.; et al. Deletion of specific immune-modulatory genes from modified vaccinia virus Ankara-based HIV vaccines engenders improved immunogenicity in rhesus macaques. J. Virol. 2012, 86, 12605–12615. [Google Scholar] [CrossRef] [PubMed]
- Holgado, M.P.; Falivene, J.; Maeto, C.; Amigo, M.; Pascutti, M.F.; Vecchione, M.B.; Bruttomesso, A.; Calamante, G.; del Medico-Zajac, M.P.; Gherardi, M.M. Deletion of A44L, A46R and C12L Vaccinia Virus Genes from the MVA Genome Improved the Vector Immunogenicity by Modifying the Innate Immune Response Generating Enhanced and Optimized Specific T-Cell Responses. Viruses 2016, 8, 139. [Google Scholar] [CrossRef] [PubMed]
- Alharbi, N.K.; Spencer, A.J.; Hill, A.V.; Gilbert, S.C. Deletion of Fifteen Open Reading Frames from Modified Vaccinia Virus Ankara Fails to Improve Immunogenicity. PLoS ONE 2015, 10, e0128626. [Google Scholar] [CrossRef] [PubMed]
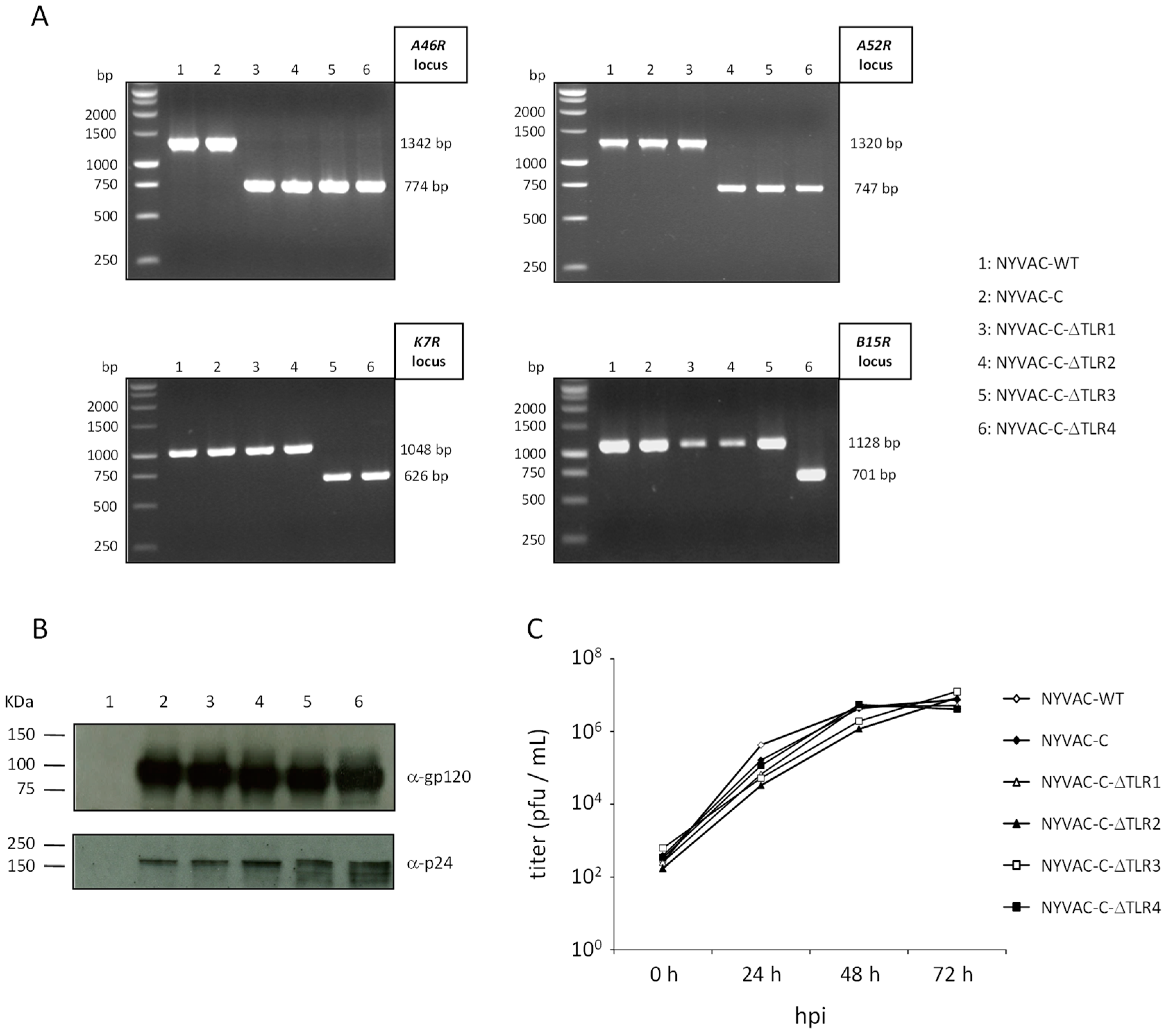
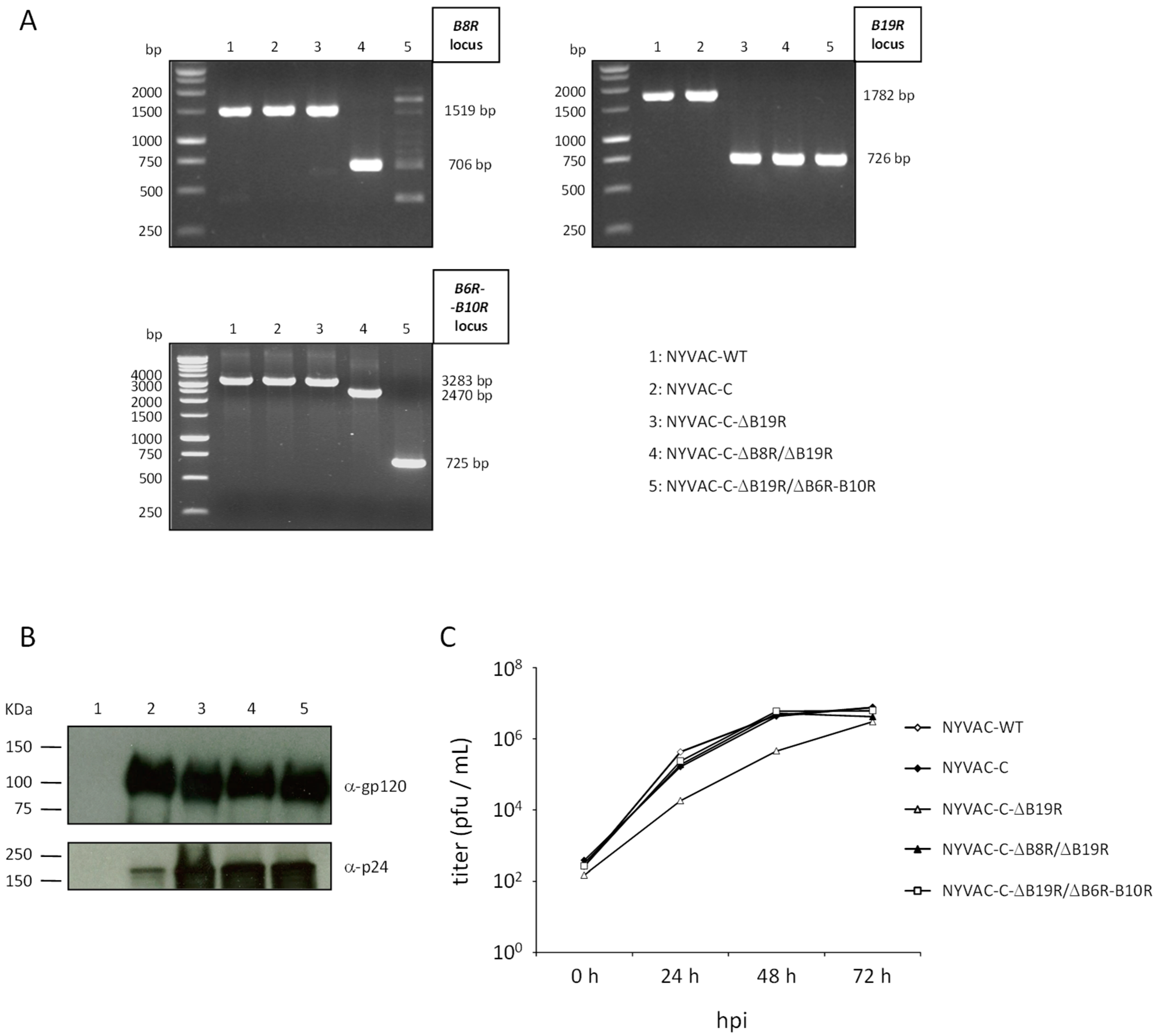



| NYVAC Viruses | Deleted Gene/Function | Generation (Infection/Transfection Protocol) | |
|---|---|---|---|
| Parental Virus | Plasmid Transfer Vector | ||
| NYVAC-WT | - | Not recombinant virus. Used as a control | |
| NYVAC-C | - | NYVAC recombinant virus expressing HIV-1 Env and Gag-Pol-Nef (clade C) antigens from the TK locus [8]. Used as a control for HIV responses | |
| NYVAC-C-∆TLR1 | A46R/TLR signalling (NF-κB/IRF3) inhibitor [20] | [15] | |
| NYVAC-C-∆TLR2 | A46R; A52R/TLR signalling (NF-κB) inhibitor [21] | NYVAC-C-∆TLR1 | pGem-RG-A52R-wm |
| NYVAC-C-∆TLR3 | A46R; A52R; K7R/TLR signalling (NF-κB/IRF3) inhibitor [22], promotes histone methylation [23] | NYVAC-C-∆TLR2 | pGem-RG-K7R-wm |
| NYVAC-C-∆TLR4 | A46R; A52R; K7R; B15R/TLR signalling (IkappaB kinase) inhibitor [24] | NYVAC-C-∆TLR3 | pGem-RG-B15R-wm |
| NYVAC-C-∆B19R | B19R/IFN-α/β soluble receptor [25] | [18] | |
| NYVAC-C-∆TLR4/∆B19R | A46R; A52R; K7R; B15R; B19R | NYVAC-C-∆TLR4 | pGem-RG-B19R-wm [18] |
| NYVAC-C-∆B8R/∆B19R | B8R/IFN-γ soluble receptor [26]; B19R | [14] | |
| NYVAC-C-∆B19R/∆B6R-B10R | B6R/unknown; B7R/TNF-α soluble receptor and chemokine binding protein [27]; B8R; B9R/intracellular protein [28]; B10R/unknown; B19R | NYVAC-C-∆B19R | pGem-RG-B6R-B10R-wm |
| Locus | Sequence of Primers (5′→3′) | Size of Amplified Product (bp) | |
|---|---|---|---|
| Parental Virus | Deletion Mutant | ||
| A46R | LFA46R-Aat: CACGATGACGTCAGAGGAGTTAT RFA46R-Bam: ATTTAAGGATCCAGAACGGCAAC | 1342 | 774 |
| A52R | LF′A52R-Eco: ATTAGAGAATTCTACGATTAACGA RFA52R-Bam: TCTGCCGGATCCAATGTAGTAATG | 1320 | 747 |
| K7R | K7R-F: TATGATCATGTGAGAATACTAAAATTCC K7R-R: CCGAATTGGGTAGACGATGTATGAATCC | 1048 | 626 |
| B15R | LF′B15R-Aat: TTCTTTGACGTCTGTTTTCCTGAAG RFB15R-Bam: GTGTCGGGATCCGAATTAGCATATT | 1128 | 701 |
| B8R | LFB8R-AatII-F: TTTTTTGACGTCATTGACTCGTCTACTATTC RFB8R-BamHI-R: TTTTTTGGATCCAAACAGCGGACACATTGC | 1519 | 706 |
| B19R | LFB19R-AatII-F: TTTTTTGACGTCGAGAAAGTTAAGAAGATAC RFB19R-BamHI-R: TTTTTTGGATCCAGTTCTATCATAATCATC | 1782 | 726 |
| B6R-B10R | LFB6R-AatII-F: GGAATGACGTCCTCCCAATATGTG RFB10R-BamHI-R: CGGGATCCAGTAGATATGATCTATATTC | 3283 | 725 |
| NYVAC Deletion Mutant | HIV-Specific Response | VACV-Specific Response | |||||||
|---|---|---|---|---|---|---|---|---|---|
| CD4 T Cells | CD8 T Cells | Memory Phenotype | BAbs | CD8 T Cells | Memory CD8 T Cells Phenotype | BAbs | NAbs | ||
| CD4 T Cells | CD8 T Cells | ||||||||
| NYVAC-C-∆TLR1 | + | +++ | EM | EM > TEM | + | + | EM ≥ TEM | ++ | + |
| NYVAC-C-∆TLR2 | +++ | ++ | EM | EM < TEM | +++ | + | EM < TEM | + | = |
| NYVAC-C-∆TLR3 | ++ | − | EM | EM < TEM | ++ | ++ | EM < TEM | +++ | +++ |
| NYVAC-C-∆TLR4 | − | ++ | EM | EM ≥ TEM | ++ | ++ | EM ≤ TEM | +++ | ++ |
| NYVAC-C-∆B19R | + | ++ | EM | EM < TEM | = | = | EM < TEM | ++ | + |
| NYVAC-C-∆B8R/∆B19R | = | ++++ | EM | EM < TEM | = | +++ | EM ≤ TEM | ++ | + |
| NYVAC-C-∆B19R/∆B6R-B10R | + | + | EM | EM < TEM | +++ | ++++ | EM < TEM | +++ | + |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gómez, C.E.; Perdiguero, B.; Sánchez-Corzo, C.; Sorzano, C.O.S.; Esteban, M. Immune Modulation of NYVAC-Based HIV Vaccines by Combined Deletion of Viral Genes that Act on Several Signalling Pathways. Viruses 2018, 10, 7. https://doi.org/10.3390/v10010007
Gómez CE, Perdiguero B, Sánchez-Corzo C, Sorzano COS, Esteban M. Immune Modulation of NYVAC-Based HIV Vaccines by Combined Deletion of Viral Genes that Act on Several Signalling Pathways. Viruses. 2018; 10(1):7. https://doi.org/10.3390/v10010007
Chicago/Turabian StyleGómez, Carmen Elena, Beatriz Perdiguero, Cristina Sánchez-Corzo, Carlos Oscar S. Sorzano, and Mariano Esteban. 2018. "Immune Modulation of NYVAC-Based HIV Vaccines by Combined Deletion of Viral Genes that Act on Several Signalling Pathways" Viruses 10, no. 1: 7. https://doi.org/10.3390/v10010007
APA StyleGómez, C. E., Perdiguero, B., Sánchez-Corzo, C., Sorzano, C. O. S., & Esteban, M. (2018). Immune Modulation of NYVAC-Based HIV Vaccines by Combined Deletion of Viral Genes that Act on Several Signalling Pathways. Viruses, 10(1), 7. https://doi.org/10.3390/v10010007






