Mediterranean Islands Hosting Marginal and Peripheral Forest Tree Populations: The Case of Pinus brutia Ten. in Cyprus
Abstract
:1. Introduction
2. Material and Methods
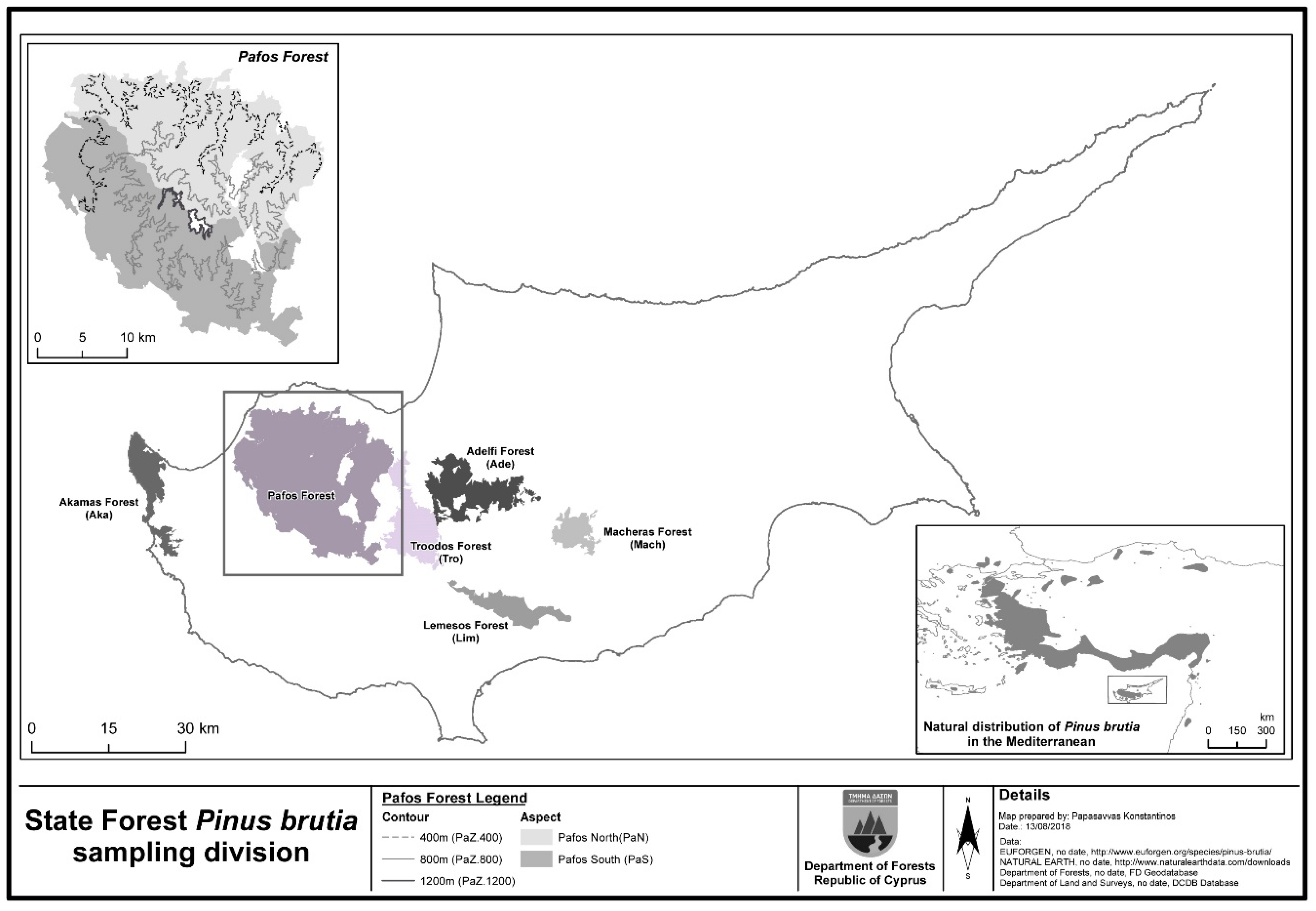
2.1. The P. brutia Forest in Cyprus
2.2. Sampling Design
2.3. Assessment of Genetic Diversity and Structure
2.4. Assessment of Morphoanatomical Diversity and Structure
3. Results
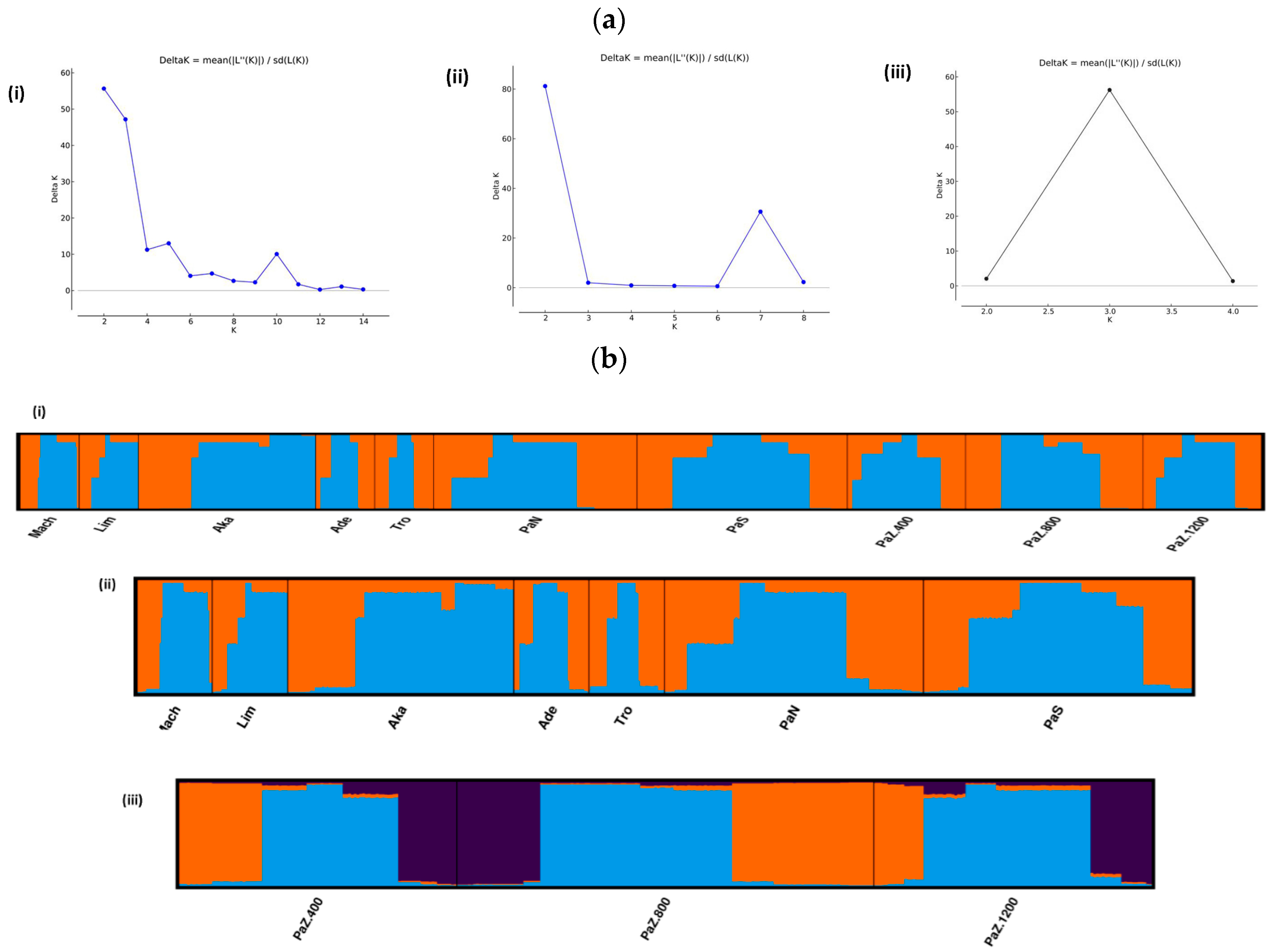
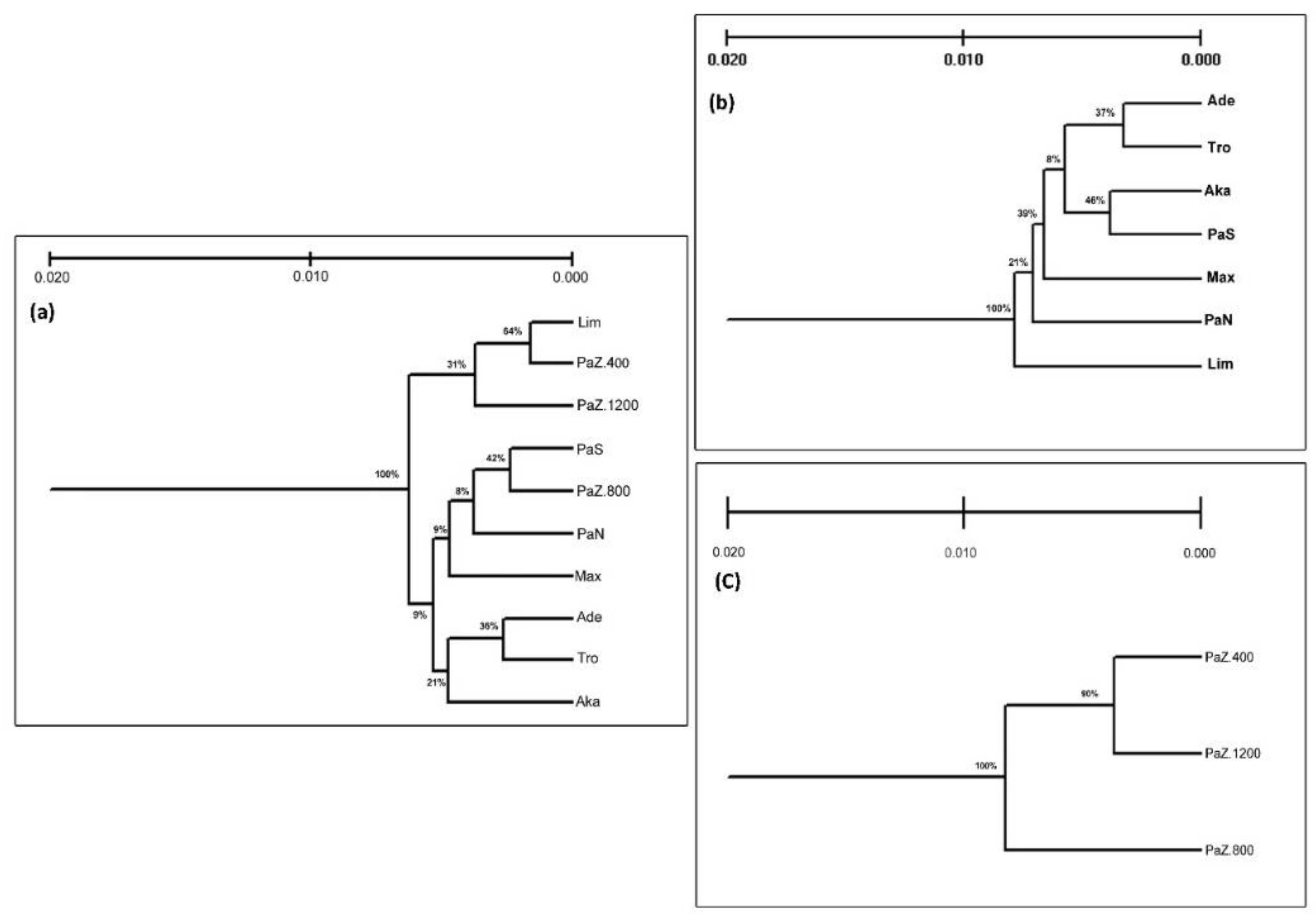
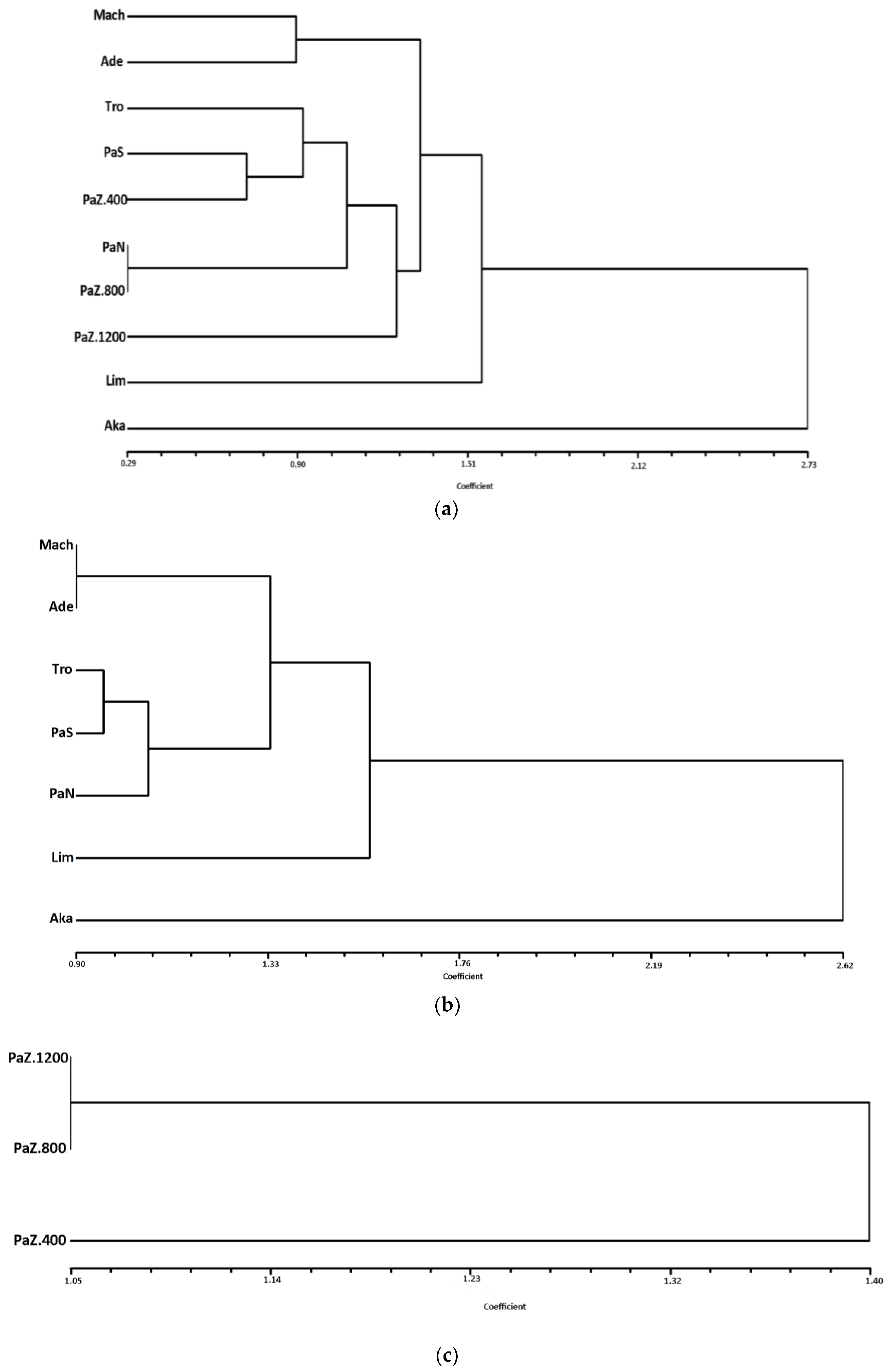
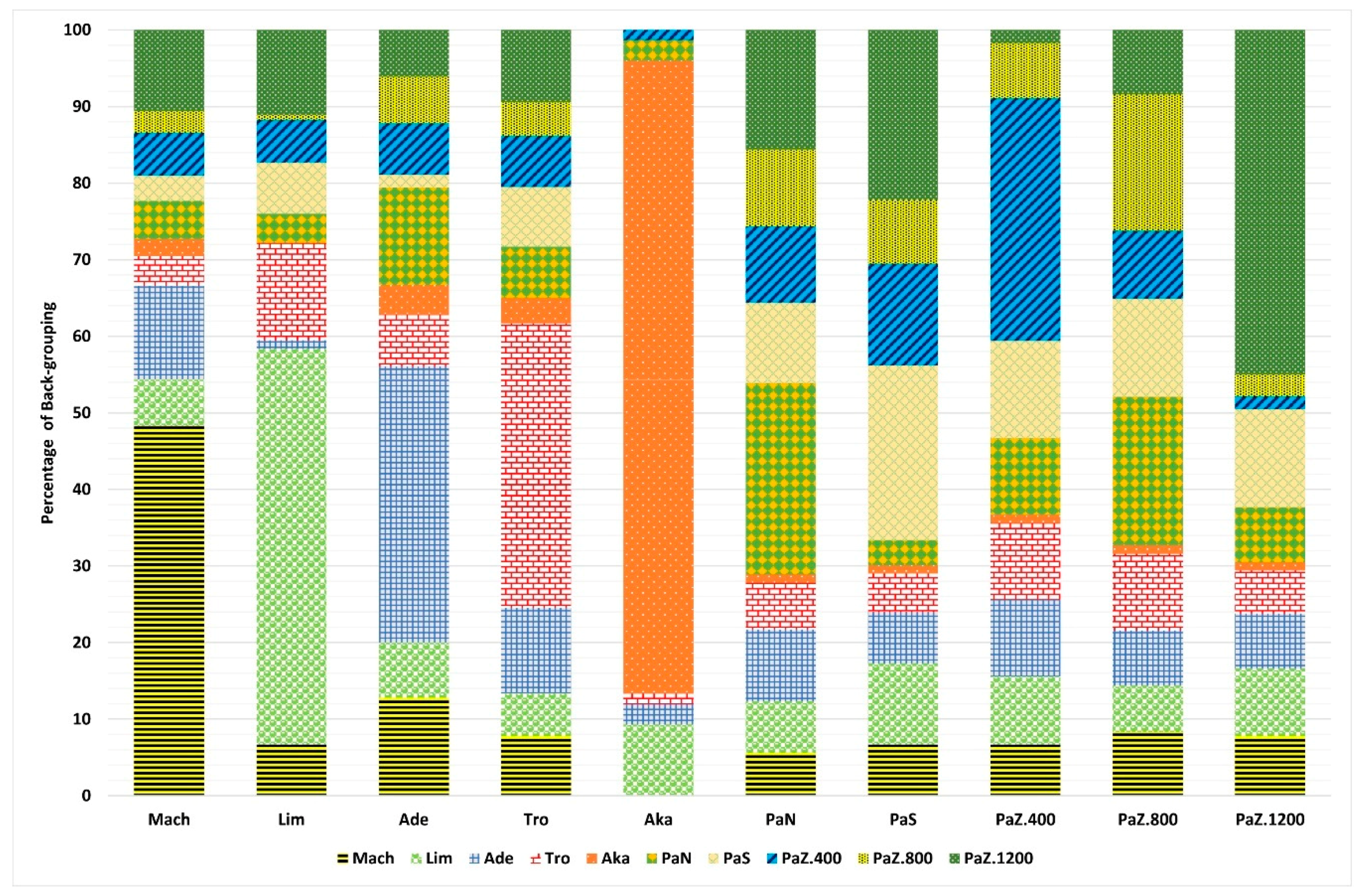
3.1. Genetic Diversity and Subpopulations Structure of P. brutia in Cyprus
3.2. The Patterns of Morphoanatomical Diversity and Structure in P. brutia
4. Discussion
4.1. P. brutia in Cyprus as a Source of Peripheral and Marginalforest Genetic Resources
4.2. Patterns of Genetic Diversity of P. brutia in Cyprus
4.3. Patterns of Morphoanatomical Traits of P. brutia in Cyprus
5. Conclusions and Perspectives of Conservation Action
5.1. Inferences from the Study of a Mediterranean Oceanic Island’s Peripheral Tree Population
5.2. Future Conservation Actions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Médail, F.; Myers, N. Mediterranean Basin. In Hotspots Revisited: Earth’s Biologically Richest and Most Endangered Terrestrial Ecoregions; Mittermeier, R.A., Robles-Gil, P., Hoffmann, M., Pilgrim, J., Brooks, T., Mittermeier, C.G., Lamoreux, J., da Fonseca, G.A.B., Eds.; CEMEX: Monterrey, Mexico; Conservation International: Washington, DC, USA; Agrupación Sierra Madre: Mexico City, Mexico, 2004; pp. 144–147. [Google Scholar]
- Noss, R.F.; Platt, W.J.; Sorrie, B.A.; Weakley, A.S.; Means, D.B.; Costanza, J.; Pee, R.K. How global biodiversity hotspots may go unrecognized: Lessons from the North American Coastal Plain. Divers. Distrib. 2015, 21, 236–244. [Google Scholar] [CrossRef]
- Vogiatzakis, I.N.; Pungetti, G.; Mannion, A.M. Mediterranean Island Landscapes: Natural and Cultural Approaches, Landscape Series; Springer Science & Business Media: New York, NY, USA, 2008; Volume 9. [Google Scholar]
- Médail, F. The specific vulnerability of plant biodiversity and vegetation on Mediterranean islands in the face of global change. Reg. Environ. Chang. 2017, 17, 1775–1790. [Google Scholar] [CrossRef] [Green Version]
- Whittaker, R.J.; Fernández-Palacios, J.M. Island Biogeography, Ecology, Evolution, and Conservation, 1st ed.; Oxford University Press: New York, NY, USA, 2007. [Google Scholar]
- Mansion, G.; Rosenbaum, G.; Schoenenberger, N.; Bacchetta, G.; Rosselló, J.A.; Conti, E. Phylogenetic analysis informed by geological history supports multiple, sequential invasions of the Mediterranean Basin by the angiosperm family Araceae. Syst. Biol. 2008, 57, 269–285. [Google Scholar] [CrossRef] [PubMed]
- Rosselló, J. A perspective of plant microevolution in the Western Mediterranean islands as assessed by molecular markers. In Proceedings of the Islands and Plants: Preservation and Understanding of Flora on Mediterranean Islands, 2nd Botanical conference in Menorca, Maó, Spain, 26–30 April 2011; Cardona Pons, E., Estaún Clarisó, I., Comas Casademont, M., Fraga i Arguimbau, P., Eds.; Institut Menorquı’ d’Estudis: Illes Balears, Spain, 2013; pp. 21–34. [Google Scholar]
- Fady, B. Is there really more biodiversity in Mediterranean forest ecosystems? Taxon 2005, 54, 905–910. [Google Scholar] [CrossRef]
- Fady, B.; Conord, C. Macroecological patterns of species and genetic diversity in vascular plants of the Mediterranean basin. Divers. Distrib. 2010, 16, 53–64. [Google Scholar] [CrossRef]
- Fady, B.; Aravanopoulos, F.A.; Alizoti, P.; Mátyás, C.; von Wühlisch, G.; Westergren, M.; Belletti, P.; Cvjetkovic, B.; Ducci, F.; Huber, G.; et al. Evolution-based approach needed for the conservation and silviculture of peripheral forest tree populations. For. Ecol. Manag. 2016, 375, 66–75. [Google Scholar] [CrossRef]
- Alleaume-Benharira, M.; Pen, I.R.; Ronce, O. Geographical patterns of adaptation within a species’ range: Interactions between drift and gene flow. J. Evol. Biol. 2006, 19, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Hampe, A.; Petit, R.J. Conserving biodiversity under climate change: The rear edge matters. Ecol. Lett. 2005, 8, 461–467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohsawa, T.; Ide, Y. Global patterns of genetic variation in plant species along vertical and horizontal gradients on mountains. Glob. Ecol. Biogeogr. 2008, 17, 152–163. [Google Scholar] [CrossRef] [Green Version]
- Panetsos, C.P. Monograph of Pinus halepensis (Mill.) and Pinus brutia (Ten.). Annu. For. 1981, 9, 39–77. [Google Scholar]
- Emberger, L.; Gaussen, H.; Kassa, A.; De Phillips, A. Bioclimatic Map of the Mediterranean Zone: Ecological Study of the Mediterranean Zone; UNESCO-FAO: Paris, France, 1963; 58p. [Google Scholar]
- Fady, B.; Semerci, H.; Vendramin, G.G. EUFORGEN Technical Guidelines for Genetic Conservation and Use for Aleppo pine (Pinus halepensis) and Brutia pine (Pinus brutia); Bioversity International: Rome, Italy, 2003. [Google Scholar]
- Selik, M. Botanical investigation on Pinus brutia especially in comparison with P. halepensis, Istanbul Univ. Fac. For. J. 1958, 8, 161–198. [Google Scholar]
- Mirov, N.T. The Genus Pinus; The Roland Press Company: New York, NY, USA, 1967; pp. 252–254. [Google Scholar]
- Oppenheimer, H.R. Mechanisms of Drought Resistance in Conifers of the Mediterranean Zone and the Arid West of the U.S.A. Part 1. Physiological and Anatomical Investigations; Final Report on Project No. A 10-FS 7, Grant No. FG-Is-119; Hebrew University of Jerusalem: Rehovot, Israel, 1967; 73p. [Google Scholar]
- Spencer, D. Conifers in the Dry Countries; A Report for the RIRDC/L&W Australia/FWPRDC Joint Venture Agroforestry Program, RIRDC Publication No. 01/46; Rural Industries Research and Development Corporation: Kingston, Australia, 2001. [Google Scholar]
- Zohary, M. Geobotanical Foundation of the Middle East; Gustav Fischer Verlag: Stuttgart, Germany, 1973; 740p. [Google Scholar]
- Barbéro, M.; Loisel, R.; Quézel, P.; Richardson, D.M.; Romane, F. Pines of the Mediterranean Basin. In Ecology and Biogeography of Pinus; Richardson, D.M., Ed.; Cambridge University Press: Cambridge, UK, 1998; pp. 153–170. [Google Scholar]
- Isik, K. Altitudinal variation in Pinus brutia Ten.: Seed and seedling characteristics. Silvae Genet. 1986, 35, 58–67. [Google Scholar]
- Bariteau, M. Variabilité géographique et adaptation aux contraintes du milieu méditerranéen des pins de la section halepensis: Resultants (provisoires) d’ún essai en plantations comparatives en France. Annu. Sci. For. 1992, 49, 261–276. [Google Scholar] [CrossRef]
- Souvannavong, O.; Malagnoux, M.; Palmberglerche, C. Nations join to conserve forests and woodlands of the Mediterranean region. Diversity 1995, 11, 19–20. [Google Scholar]
- Spencer, D.J. Dry country pines: Provenance evaluation of the Pinus halepensis-P. brutia complex in the semi-arid region of south-east Australia. Aust. For. Res. 1985, 15, 263–279. [Google Scholar]
- Boydak, M. Silvicultural characteristics and natural regeneration of Pinus brutia Ten.—A review. Plant Ecol. 2004, 171, 153–163. [Google Scholar] [CrossRef]
- Kurt, Y.; González-Martínez, S.C.; Alía, R.; Isik, K. Genetic differentiation in Pinus brutia Ten. using molecular markers and quantitative traits: The role of altitude. Annu. For. Sci. 2012, 69, 345–351. [Google Scholar] [CrossRef]
- Department of Forests. Criteria and Indicators—For the Sustainable Forest Management in Cyprus; Ministry of Agriculture, Natural Resources and Environment: Nicosia, Cyprus, 2006.
- Barber i Valles, A. Contribution to the Knowledge of the Bioclimate and Vegetation of the Island of Cyprus; Cyprus Forestry College: Nicosia, Cyprus, 1995. [Google Scholar]
- Pantelas, V. The Bioclima and Phytosociology in Cyprus; Report in Department of Forests, Ministry of Agriculture, Natural Resources and Environment: Nicosia, Cyprus, 1996.
- Shen, H.; Rudin, D.; Lindgren, D. Study of the pollination pattern in a scots pine seed orchard by means of Isozyme Analysis. Silvae Genet. 1981, 30, 7–15. [Google Scholar]
- Conkle, M.T.; Hodgkiss, P.D.; Nunnally, L.B.; Hunter, S.C. Starch Gel Electrophoresis of Conifer Seeds, A Laboratory Mannual; Gen. Techn. Rep. PSW-64; Pacific Southwest Forest and Range Experiment Station: Berkeley, CA, USA, 1982. [Google Scholar]
- Cheliak, W.M.; Pitel, J.A. Genetic control of allozyme variants in mature tissues of white spruce trees. J. Hered. 1984, 75, 34–40. [Google Scholar] [CrossRef]
- Nei, M. F-statistics and analysis of gene diversity in subdivided populations. Annu. Hum. Genet. 1977, 41, 225–233. [Google Scholar] [CrossRef]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 89, 583–590. [Google Scholar] [PubMed]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Genetics Society of America Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. CLUMPAK: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Res. 2015, 15, 1179–1191. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Genetic distance between populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Miller, M.P. Tools for Population Genetic Analyses (TFPGA) Version 1.3—A Windows Program for the Analysis of Allozyme and Molecular Population Genetic Data; Northern Arizona University: Flagstaff, AZ, USA, 1997. [Google Scholar]
- Rohlf, F.J. NTSyS-p.c: Numerical Taxonomy and Multivariate Analysis System, version 2.0; Exeter Software Publishers Ltd.: Setauket, NY, USA, 1998. [Google Scholar]
- Breiman, L.; Friedman, J.; Olshen, R.; Stone, C. Classification and Regression Trees; Wadsworth International Group: Belmont, CA, USA, 1984. [Google Scholar]
- De’ath, G.; Fabricius, K.E. Classification and regression trees: A powerful yet simple technique for the analysis of complex ecological data. Ecology 2000, 81, 3178–3192. [Google Scholar] [CrossRef]
- Mantel, N.A. The detection of disease clustering and a generalized regression approach. Can. Res. 1967, 27, 209–220. [Google Scholar]
- Sokal, R. Testing statistical significance of geographic variation patterns. Syst. Zool. 1979, 28, 227–232. [Google Scholar] [CrossRef]
- Pujol, B.; Pannell, J.R. Reduced responses to selection after species range expansion. Science 2008, 321, 96. [Google Scholar] [CrossRef] [PubMed]
- Holt, R.D.; Keitt, T.H.; Lewis, M.A.; Maurer, B.A.; Taper, M.L. Theoretical models of species’ borders: Single species approaches. Oikos 2005, 108, 18–27. [Google Scholar] [CrossRef]
- Adams, W.T.; Joly, R.J. Genetics of allozyme variants in loblolly pine. J. Hered. 1980, 71, 33–40. [Google Scholar] [CrossRef]
- Morgante, M.; Vendramin, G.G.; Giannini, R. Inheritance and linkage relationships of isozymes variants of Pinus leucodermis Ant. Silvae Genet. 1993, 42, 231–237. [Google Scholar]
- Conkle, M.T.; Schiller, G.; Grunwald, C. Electrophoretic analysis of diversity and phylogeny of Pinus brutia and closely related taxa. Syst. Bot. 1988, 13, 411–424. [Google Scholar] [CrossRef]
- Kara, N.; Korol, L.; Isik, K.; Schiller, G. Genetic Diversity in Pinus brutia TEN.: Altitudinal Variation. Silvae Genet. 1997, 46, 2–3. [Google Scholar]
- Schiller, G.; Conkle, M.T.; Grunwald, C. Local differentiation among Mediterranean populations of Aleppo pine in their isoenzymes. Silvae Genet. 1985, 35, 11–19. [Google Scholar]
- Panetsos, K.P.; Aravanopoulos, F.; Scaltsoyiannes, A. Genetic Variation of Pinus brutia from Islands of the Northeastern Aegean Sea. Silvae Genet. 1998, 47, 2–3. [Google Scholar]
- van Staaden, M.J.; Michener, G.R.; Chesser, R.K. Spatial analysis of microgeographic genetic structure in Richardson’s ground squirrels. Can. J. Zool. 1996, 74, 1187–1195. [Google Scholar] [CrossRef]
- Panetsos, K.; Scaltsoyiannes, A.; Aravanopoulos, F.A.; Dounavi, K.; Demetrakopoulos, A. Identification of Pinus brutia TEN., P. halepensis MILL. and Their Putative Hybrids. Silvae Genet. 1997, 46, 253–257. [Google Scholar]
- Dangasuk, O.G.; Panetsos, K.P. Altitudinal and longitudinal variations in Pinus brutia (Ten.) of Crete Island, Greece: Some needle, cone and seed traits under natural habitats. New For. 2004, 27, 269–284. [Google Scholar] [CrossRef]
- Calamassi, R.; Puglisi, S.R.; Vendramin, G.G. Genetic variation in morphological and anatomical needle characteristics in Pinus brutia Ten. Silvae Genet. 1988, 37, 5–6. [Google Scholar]
- Tian, Μ.; Yu, G.; He, N.; Hou, J. Leaf morphological and anatomical traits from tropical to temperate coniferous forests: Mechanisms and influencing factors. Sci. Rep. 2016, 6, 19703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niinemets, Ü.; Cescatti, A.; Lukjanova, A.; Tobias, M.; Truus, L. Modification of light-acclimation of Pinus sylvestris shoot architecture by site fertility. Agric. For Meteorol. 2002, 111, 121–140. [Google Scholar] [CrossRef]
- Smolander, S.; Stenberg, P. A method to account for shoot scale clumping in coniferous canopy reflectance models. Remote Sens. Environ. 2003, 88, 363–373. [Google Scholar] [CrossRef] [Green Version]
- Niinemets, Ü. A review of light interception in plant stands from leaf to canopy in different plant functional types and in species with varying shade tolerance. Ecol. Res. 2010, 25, 693–714. [Google Scholar] [CrossRef]
- Gebauer, R.; Volařík, D.; Urban, J.; Børja, I.; Nagy, N.E.; Eldhuset, T.D.; Krokene, P. Effect of thinning on the anatomical adaptation of Norway spruce needles. Tree Physiol. 2011, 31, 1103–1113. [Google Scholar] [CrossRef] [PubMed]
- Knapp, B. Cyprus’s earliest prehistory: Seafarers, Forgers and Settlers. J. World Prehist. 2010, 23, 79–120. [Google Scholar] [CrossRef]
- Vigne, J.-D.; Briois, F.; Zazzo, A.; Willcox, G.; Cucchi, T.; Thiébault, S.; Carrère, I.; Franel, Y.; Touquet, R.; Martin, C.; et al. First wave of cultivators spread to Cyprus at least 10,600 y ago. Proc. Natl. Acad. Sci. USA 2012, 109, 8445–8449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thirgood, J.V. Cyprus, a Chronicle of Its Forest, Land, and People; University of British Columbia Press: Vancouver, BC, Canada, 1987. [Google Scholar]
- Geological Survey Department. The Geology of Cyprus; Article 10; Ministry of Agriculture, Natural Resources and Environment: Nicosia, Cyprus, 2002.
- Hampe, A.; Arroyo, J.; Jordano, P.; Petit, R.J. Range wide phylogeography of a bird-dispersed Eurasian shrub: Contrasting Mediterranean and temperate glacial refugia. Mol. Ecol. 2003, 12, 3415–3426. [Google Scholar] [CrossRef] [PubMed]
- Petit, R.J.; Aguinagalde, I.; de Beaulieu, J.L.; Bittkau, C.; Brewer, S.; Cheddadi, R.; Ennos, R.; Fineschi, S.; Grivet, D.; Lascoux, M.; et al. Glacial refugia: Hotspots but not melting pots of genetic diversity. Science 2003, 300, 1563–1565. [Google Scholar] [CrossRef] [PubMed]
- Martin, P.R.; McKay, J.K. Latitudinal variation in genetic divergence of populations and the potential for future speciation. Evolution 2004, 58, 938–945. [Google Scholar] [CrossRef] [PubMed]
- Eliades, N.-G.; Gailing, O.; Leinemann, L.; Fady, B.; Finkeldey, R. High genetic diversity and significant population structure in Cedrus brevifolia Henry, a narrow Mediterranean endemic tree from Cyprus. Plant Syst. Evol. 2011, 294, 185–198. [Google Scholar] [CrossRef]
- Gonzalo-Turpin, H.; Hazard, L. Local adaptation occurs along altitudinal gradient despite the existence of gene flow in the alpine plant species Festuca eskia. J. Ecol. 2009, 97, 742–751. [Google Scholar] [CrossRef]
- Sáenz-Romero, C.; Guzmán-Reyna, R.R.; Rehfeldt, G.E. Altitudinal genetic variation among Pinus oocarpa populations in Michoacán, Mexico Implications for seed zoning, conservation, tree breeding and global warming. For. Ecol. Manag. 2006, 229, 340–350. [Google Scholar] [CrossRef]
- Byars, S.G.; Papst, W.; Hoffmann, A.A. Local adaptation and cogradient selection in the Alpine plant, Poa hiemata, along a narrow altitudinal gradient. Evolution 2007, 61, 2925–2941. [Google Scholar] [CrossRef] [PubMed]
- Brousseau, L.; Postolache, D.; Lascoux, M.; Drouzas, A.D.; Källman, T.; Leonarduzzi, C.; Liepelt, S.; Piotti, A.; Popescu, F.; Roschanski, A.M.; et al. Local Adaptation in European Firs Assessed through Extensive Sampling across Altitudinal Gradients in Southern Europe. PLoS ONE 2016, 11, e0158216. [Google Scholar] [CrossRef] [PubMed]
- Forsman, A. Rethinking phenotypic plasticity and its consequences for individuals, populations and species. Heredity 2015, 115, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Matyas, C.; Yeatman, C.W. Effect of geographical transfer on growth and survival of jack pine (Pinus banksiana Lamb.) populations. Silvae Genet. 1992, 41, 370–376. [Google Scholar]
- Körner, C. The use of “altitude” in ecological research. Trends Ecol. Evol. 2007, 22, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Corcuera, L.; Cochard, H.; Gil-Pelegrin, E.; Notivol, E. Phenotypic plasticity in mesic populations of Pinus pinaster improves resistance to xylem embolism (P50) under severe drought. Trees 2011, 25, 1033–1042. [Google Scholar] [CrossRef]
- Papageorgiou, A.C.; Kostoudi, C.; Sorotos, I.; Varsamis, G.; Korakis, G.; Drouzas, A.D. Diversity in needle morphology and genetic markers in a marginal Abies cephalonica (Pinaceae) population. Annu. For. Res. 2015, 58, 217–234. [Google Scholar] [CrossRef]
- Denk, T.; Grimm, G.; Stögerer, K.; Langer, M.; Hemleben, V. The evolutionary history of Fagus in western Eurasia: Evidence from genes, morphology and the fossil record. Plant Syst. Evol. 2002, 232, 213–236. [Google Scholar] [CrossRef]
- Chevin, L.-M.; Collins, S.; Lefèvre, F. Phenotypic plasticity and evolutionary demographic responses to climate change: Taking theory out to the field. Funct. Ecol. 2013, 27, 966–979. [Google Scholar] [CrossRef]
- Chevin, L.M.; Lande, R. Adaptation to marginal habitats by evolution of increased phenotypic plasticity. J. Evol. Biol. 2011, 24, 1462–1476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiwari, S.P.; Kumar, P.; Yadav, D.; Chauhan, D.K. Comparative morphological, epidermal, and anatomical studies of Pinus roxburghii needles at different altitudes in the North-West Indian Himalayas. Turk. J. Bot. 2013, 37, 65–73. [Google Scholar]
- Sheue, C.R.; Yang, Y.P.; Huang, L.L.K. Altitudinal variation of resin ducts in Pinus taiwanensis Hayata (Pinaceae) needles. Bot. Bull. Acad. Sin. 2003, 44, 305–313. [Google Scholar]
- Brown, J.H.; Stevens, G.C.; Kaufman, D.M. The geographic range: Size, shape, boundaries, and internal structure. Annu. Rev. Ecol. Syst. 1996, 27, 597–623. [Google Scholar] [CrossRef]
- Sagarin, R.D.; Gaines, S.D.; Gaylord, B. Moving beyond assumptions to understand abundance distributions across the ranges of species. Trends Ecol. Evol. 2006, 21, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Eckert, C.G.; Samis, K.E.; Lougheed, S.C. Genetic variation across species’ geographical ranges: The centralmarginal hypothesis and beyond. Mol. Ecol. 2008, 17, 1170–1188. [Google Scholar] [CrossRef] [PubMed]
- Rehfeldt, G.E.; Tchebakova, N.M.; Parfenova, Y.I.; Wykoff, W.R.; Kuzmina, N.A.; Milyutin, L.I. Intraspecific responses to climate in Pinus sylvestris. Glob. Chang. Biol. 2002, 8, 912–929. [Google Scholar] [CrossRef]
- Schlichting, C.; Pigliucci, M. Phenotypic Evolution: A Reaction Norm Perspective; Sinauer Associates Incorporated: Sunderland, MA, USA, 1998. [Google Scholar]
- Sultan, S.E. Phenotypic plasticity for plant development, function and life-history. Trends Plant Sci. 2000, 5, 537–542. [Google Scholar] [CrossRef]
- Lefèvre, F.; Boivin, T.; Bontemps, A.; Courbet, F.; Davi, H.; Durand-Gillmann, M.; Fady, B.; Gaüzere, J.; Gidoin, C.; Karam, M.J.; et al. Considering evolutionary processes in adaptive forestry. Annu. For. Sci. 2014, 71, 723–739. [Google Scholar] [CrossRef]
- Aravanopoulos, F.A. Do silviculture and forest management affect the genetic diversity and structure of long-impacted forest tree populations? Forests 2018, 9, 355. [Google Scholar] [CrossRef]
- Hadjinicolaou, P.; Giannakopoulos, C.; Zerefos, C.; Lange, M.A.; Pashiardis, S.; Lelieveld, J. Mid-21st century climate and weather extremes in Cyprus as projected by six regional climate models. Reg. Environ. Chang. 2011, 11, 441–457. [Google Scholar] [CrossRef]

| Location (Abbr.) | Distribution Area ǂ (ha/km) | Elevation Range (m) | Sample Size | |||||
|---|---|---|---|---|---|---|---|---|
| Genetic Analysis | Morphoanatomical Analysis | |||||||
| No. Trees | No. Mega-Gametophytes | No. Trees | No. Cones | No. Seeds | No. Needles | |||
| Macheras (Mach) | 5583 ha | 400–1200 | 60 | 50 | 60 | 180 | 540 | 180 |
| Limassol (Lim) | 7896 ha | 800–1200 | 60 | 50 | 60 | 180 | 540 | 180 |
| Akamas (Aka) | 5743 ha | 400 | 25 | 150 * | 25 | 75 | 225 | 180 |
| Adelphi (Ade) | 12,826 ha | 400–1200 | 60 | 50 | 60 | 180 | 540 | 180 |
| Troodos (Tro) | 8843 ha | 400–1200 | 60 | 50 | 60 | 180 | 540 | 180 |
| Pafos North (PaN) | 37,794 ha | 400–1200 | 60 | 172 | 60 | 180 | 540 | 180 |
| Pafos South (PaS) | 22,365 ha | 400–1200 | 60 | 178 | 60 | 180 | 540 | 180 |
| Pafos z.400 (PaZ.400) | 320 km | 400 | 60 | 100 | 60 | 180 | 540 | 180 |
| Pafos z.800 (PaZ.800) | 438 km | 800 | 25 | 150 * | 60 | 180 | 540 | 180 |
| Pafos z.1200 (PaZ.1200) | 35 km | 1200 | 60 | 100 | 60 | 180 | 540 | 180 |
| Total | 580 | 1050 | 565 | 1695 | 5085 | 1695 | ||
| Subpopulation | Genetic Parameters | ||||
|---|---|---|---|---|---|
| PPL | Na | Ne | I | HE | |
| Mach | 60% | 1.800 | 1.435 | 0.331 | 0.203 |
| Lim | 60% | 1.700 | 1.300 | 0.269 | 0.170 |
| Aka | 70% | 2.000 | 1.442 | 0.371 | 0.217 |
| Ade | 80% | 2.100 | 1.473 | 0.393 | 0.242 |
| Tro | 60% | 1.800 | 1.430 | 0.335 | 0.214 |
| PaN | 90% | 2.300 | 1.415 | 0.393 | 0.228 |
| PaS | 80% | 2.100 | 1.415 | 0.375 | 0.220 |
| PaZ.400 | 80% | 2.000 | 1.375 | 0.350 | 0.209 |
| PaZ.800 | 80% | 2.200 | 1.464 | 0.418 | 0.244 |
| PaZ.1200 | 90% | 2.200 | 1.315 | 0.327 | 0.186 |
| Overall | 90% | 2.300 | 1.421 | 0.381 | 0.220 |
| (a) | ||||||||
| Source of Variation | All Sampled Subpopulations§ | Range-Wide Subpopulations† | ||||||
| d.f. | Variance Components | Percentage Variation | Fixation Indicesǂ | d.f. | Variance Components | Percentage Variation | Fixation Indicesǂ | |
| Among Groups | 5 | 0.137 | 12% | PhiRT = 0.118 *** | 3 | 0.005 | 0% | PhiRT = 0.004 n.s |
| Among subpopulations within Groups | 4 | 0.013 | 1% | PhiPR = 0.012 *** | 3 | 0.011 | 1% | PhiPR = 0.010 * |
| Within subpopulations within Groups | 1040 | 1.017 | 87% | PhiPT = 0.129 *** | 693 | 1.088 | 99% | PhiPT = 0.014 *** |
| Total | 1049 | 1.167 | 100% | 693 | 1.104 | 100% | ||
| (b) | ||||||||
| Source of Variation | Altitudinal Subpopulations | Aspect Subpopulations | ||||||
| d.f. | Variance Components | Percentage Variation | Fixation Indicesǂ | d.f. | Variance Components | Percentage Variation | Fixation Indicesǂ | |
| Among subpopulations | 2 | 0.020 | 2% | 1 | 0.014 | 1% | ||
| Within subpopulations | 347 | 1.087 | 98% | PhiPT = 0.018 ** | 348 | 1.118 | 99% | PhiPT = 0.012 ** |
| Total | 349 | 1.107 | 100% | 349 | 1.132 | 100% | ||
| (a) | ||||||||||||||||
| Traits | Mach | Lim | Aka | Ade | Tro | |||||||||||
| μ | SD | CV | μ | SD | CV | μ | SD | CV | μ | SD | CV | μ | SD | CV | ||
| Clen (mm) | 69.47 | 8.50 | 0.12 | 68.85 | 7.80 | 0.11 | 63.27 | 7.16 | 0.11 | 68.23 | 9.61 | 0.14 | 67.39 | 8.38 | 0.12 | |
| Cwid (mm) | 40.40 | 3.17 | 0.08 | 37.77 | 3.31 | 0.08 | 35.335 | 3.60 | 0.10 | 38.56 | 3.65 | 0.34 | 38.53 | 3.11 | 0.08 | |
| Clen/Cwid | 1.72 | 0.18 | 0.10 | 1.77 | 0.12 | 0.06 | 1.79 | 0.13 | 0.07 | 1.77 | 0.17 | 0.09 | 1.75 | 0.17 | 0.09 | |
| Slen (mm) | 8.15 | 0.61 | 0.07 | 7.76 | 0.57 | 0.07 | 7.33 | 0.55 | 0.07 | 8.09 | 0.72 | 0.09 | 7.88 | 0.57 | 0.06 | |
| SWid (mm) | 5.01 | 0.45 | 0.09 | 4.63 | 0.43 | 0.09 | 4.44 | 0.35 | 0.08 | 4.88 | 0.46 | 0.09 | 4.71 | 0.39 | 0.08 | |
| SWing (mm) | 18.18 | 1.90 | 0.10 | 17.02 | 2.17 | 0.12 | 15.97 | 2.52 | 0.16 | 17.69 | 2.29 | 0.13 | 16.98 | 1.86 | 0.11 | |
| SLenWing (mm) | 26.33 | 4.80 | 0.08 | 24.78 | 2.45 | 0.09 | 23.30 | 2.74 | 0.12 | 25.78 | 2.68 | 0.10 | 24.86 | 2.12 | 0.08 | |
| NShLen (mm) | 6.82 | 1.48 | 0.21 | 6.40 | 0.99 | 0.15 | 6.81 | 0.87 | 0.13 | 6.57 | 0.96 | 0.14 | 6.46 | 1.03 | 0.16 | |
| NLen (mm) | 126.76 | 15.62 | 0.12 | 106.10 | 13.01 | 0.12 | 115.72 | 12.83 | 0.11 | 131.05 | 15.63 | 0.12 | 119.33 | 14.91 | 0.12 | |
| NWid (mm) | 1.33 | 0.09 | 0.06 | 1.31 | 0.06 | 0.05 | 1.43 | 0.09 | 0.06 | 1.32 | 0.08 | 0.06 | 1.28 | 0.08 | 0.06 | |
| NThic (mm) | 0.82 | 0.05 | 0.06 | 0.79 | 0.04 | 0.05 | 0.88 | 0.06 | 0.07 | 0.82 | 0.06 | 0.07 | 0.78 | 0.06 | 0.07 | |
| NResIn | 3.73 | 0.89 | 0.23 | 3.43 | 0.98 | 0.28 | 4.23 | 0.48 | 0.11 | 3.64 | 0.79 | 0.21 | 3.34 | 0.88 | 0.26 | |
| NResDo | 6.05 | 0.98 | 0.16 | 5.78 | 0.99 | 0.17 | 6.72 | 0.95 | 0.14 | 6.16 | 1.06 | 0.17 | 5.64 | 1.24 | 0.22 | |
| NResTot | 9.78 | 1.48 | 0.15 | 9.21 | 1.69 | 0.18 | 10.95 | 1.10 | 0.10 | 9.80 | 1.60 | 0.16 | 8.99 | 1.85 | 0.20 | |
| NStomDo | 8.98 | 1.17 | 0.13 | 9.00 | 0.83 | 0.09 | 8.92 | 0.87 | 0.10 | 8.90 | 1.07 | 0.12 | 9.06 | 1.21 | 0.13 | |
| NStomIn | 3.75 | 0.76 | 0.21 | 3.87 | 0.65 | 0.16 | 3.49 | 0.74 | 0.21 | 3.66 | 0.60 | 0.16 | 3.73 | 0.68 | 0.18 | |
| NStoRow | 12.74 | 1.63 | 0.12 | 12.71 | 1.64 | 0.13 | 12.41 | 1.26 | 0.10 | 12.56 | 1.39 | 0.11 | 12.78 | 1.57 | 0.12 | |
| NSto/Row | 83.37 | 8.69 | 0.10 | 83.37 | 9.75 | 0.11 | 84.25 | 10.68 | 0.13 | 87.47 | 8.14 | 0.09 | 81.65 | 9.57 | 0.11 | |
| Nstom | 13,471 | 2917 | 0.21 | 11,268 | 2472 | 0.21 | 12,062 | 2171.60 | 0.18 | 14411 | 2730.80 | 0.19 | 12,465 | 2654.40 | 0.21 | |
| NTeh | 70.23 | 9.43 | 0.13 | 71.43 | 12.08 | 0.17 | 75.07 | 8.24 | 0.11 | 70.73 | 10.61 | 0.15 | 67.21 | 13.50 | 0.20 | |
| Traits | PaN | PaS | PaZ.400 | PaZ.800 | PaZ.1200 | |||||||||||
| μ | SD | CV | μ | SD | CV | μ | SD | CV | μ | SD | CV | μ | SD | CV | ||
| CLen (mm) | 69.96 | 10.26 | 0.14 | 67.69 | 10.05 | 0.14 | 68.75 | 9.08 | 0.13 | 68.75 | 10.09 | 0.14 | 67.02 | 8.79 | 0.13 | |
| CWid (mm) | 39.04 | 3.84 | 0.09 | 38.64 | 4.15 | 0.10 | 38.26 | 3.58 | 0.09 | 39.4 | 4.09 | 0.1 | 39.49 | 3.59 | 0.09 | |
| CLen/CWid | 1.74 | 0.16 | 0.09 | 1.74 | 0.17 | 0.09 | 1.79 | 0.17 | 0.09 | 1.75 | 0.16 | 0.09 | 1.69 | 0.15 | 0.08 | |
| SLen (mm) | 8.12 | 0.61 | 0.07 | 8.05 | 0.66 | 0.08 | 7.95 | 0.61 | 0.07 | 8.17 | 0.67 | 0.08 | 8.23 | 0.62 | 0.07 | |
| SWid (mm) | 4.83 | 0.40 | 0.08 | 4.81 | 0.42 | 0.08 | 4.73 | 0.37 | 0.08 | 4.86 | 0.41 | 0.08 | 4.96 | 0.43 | 0.08 | |
| SWing (mm) | 17.71 | 2.55 | 0.14 | 17.51 | 2.51 | 0.14 | 17.44 | 2.32 | 0.13 | 17.89 | 2.51 | 0.14 | 17.66 | 2.08 | 0.11 | |
| SLenWing (mm) | 25.83 | 2.95 | 0.11 | 25.56 | 2.98 | 0.11 | 25.39 | 2.72 | 0.1 | 26.06 | 2.96 | 0.11 | 25.89 | 2.42 | 0.09 | |
| NShLen (mm) | 6.54 | 1.07 | 0.16 | 6.33 | 0.89 | 0.14 | 6.11 | 0.77 | 0.12 | 6.39 | 0.91 | 0.14 | 7.04 | 1.1 | 0.16 | |
| NLen (mm) | 128.47 | 19.12 | 0.15 | 114.50 | 14.14 | 0.12 | 121.7 | 21.62 | 0.17 | 127.4 | 15.86 | 0.12 | 116.49 | 15.55 | 0.13 | |
| NWid (mm) | 1.27 | 0.09 | 0.07 | 1.28 | 0.07 | 0.05 | 1.29 | 0.09 | 0.07 | 1.25 | 0.08 | 0.06 | 1.29 | 0.07 | 0.05 | |
| NThic (mm) | 0.78 | 0.05 | 0.06 | 0.77 | 0.05 | 0.06 | 0.79 | 0.06 | 0.07 | 0.77 | 0.05 | 0.06 | 0.78 | 0.05 | 0.06 | |
| NResIn | 3.55 | 0.90 | 0.25 | 3.15 | 0.87 | 0.27 | 3.24 | 1.08 | 0.33 | 3.37 | 0.8 | 0.23 | 3.46 | 0.79 | 0.22 | |
| NResDo | 5.84 | 1.20 | 0.20 | 5.21 | 1.21 | 0.23 | 5.21 | 1.55 | 0.3 | 5.58 | 1.16 | 0.2 | 5.8 | 1.11 | 0.19 | |
| NResTot | 9.39 | 1.90 | 0.20 | 8.36 | 1.84 | 0.22 | 8.46 | 2.34 | 0.27 | 8.94 | 1.79 | 0.2 | 9.26 | 1.66 | 0.18 | |
| NStomDo | 8.92 | 1.06 | 0.12 | 9.18 | 1.02 | 0.11 | 8.91 | 1.06 | 0.12 | 8.58 | 1.01 | 0.11 | 9.48 | 1.1 | 0.11 | |
| NStomIn | 3.67 | 0.61 | 0.16 | 3.69 | 0.62 | 0.16 | 3.69 | 0.6 | 0.16 | 3.56 | 0.59 | 0.16 | 3.79 | 0.69 | 0.18 | |
| NStoRow | 12.59 | 1.38 | 0.11 | 12.88 | 1.32 | 0.10 | 12.61 | 1.41 | 0.11 | 12.14 | 1.22 | 0.1 | 13.27 | 1.54 | 0.11 | |
| NSto/Row | 84.44 | 7.33 | 0.87 | 86.64 | 8.31 | 0.09 | 85.81 | 8.32 | 0.09 | 85.09 | 8.07 | 0.09 | 85.88 | 8.82 | 0.1 | |
| Nstom | 13,678 | 2837.13 | 0.20 | 12,777 | 2391.70 | 0.18 | 13,138 | 2854.60 | 0.21 | 13,163 | 2430.70 | 0.18 | 13,270.18 | 2676.90 | 0.2 | |
| NTeh | 69.97 | 9.98 | 0.14 | 71.06 | 10.22 | 0.14 | 71.45 | 9.85 | 0.13 | 70.79 | 10.09 | 0.14 | 71.98 | 10.82 | 0.15 | |
| (b) | ||||||||||||||||
| CLen | CWid | CLen/CWid | SLen | SWid | SWing | SLenWing | ||||||||||
| μ | 67.60 | 38.74 | 1.75 | 8.02 | 4.81 | 17.50 | 25.51 | |||||||||
| SD | 0.22 | 0.09 | 0.004 | 0.01 | 0.01 | 0.05 | 0.06 | |||||||||
| CV | 0.14 | 0.10 | 0.10 | 0.08 | 0.09 | 0.13 | 0.11 | |||||||||
| NShLen | NLen | NWid | NThic | NResIn | NResDo | NResTot | NStomDo | NStomIn | NStoRow | NSto/Row | Nstom | NTeh | ||||
| μ | 6.53 | 121.07 | 1.29 | 0.79 | 3.47 | 5.74 | 9.21 | 9.00 | 3.71 | 12.69 | 84.83 | 13026 | 70.74 | |||
| SD | 1.06 | 17.83 | 0.09 | 0.06 | 0.91 | 1.23 | 1.88 | 1.08 | 0.66 | 1.48 | 8.83 | 2769.6 | 10.79 | |||
| CV | 0.16 | 0.15 | 0.07 | 0.66 | 0.26 | 0.21 | 0.20 | 0.12 | 0.18 | 0.12 | 0.10 | 0.21 | 0.15 | |||
| CLen | CWid | CLen/CWid | SLen | SWid | SWing | SLenWing | |||||||
| Altitude zone | −0.79 n.s. | 0.13 ** | −0.24 ** | 0.20 ** | 0.23 ** | 0.04 n.s. | 0.09 ** | ||||||
| NShLen | NLen | NWid | NThic | NResIn | NResDo | NResTot | NStomDo | NStomIn | NStoRow | NSto/Row | Nstom | NTeh | |
| Altitude zone | 0.36 ** | −0.10 * | −0.03 n.s. | −0.05 n.s. | 0.09 * | 0.16 ** | 0.14 ** | 0.20 ** | 0.05 n.s. | 0.16 n.s. | −0.001 n.s. | 0.01 n.s. | 0.03 |
| Variable | Axis 1 | Axis 2 | Axis 3 | Axis 4 | Axis 5 | Axis 6 | Axis 7 | Axis 8 | Axis 9 |
|---|---|---|---|---|---|---|---|---|---|
| % of variance explained | 40.6 | 27.9 | 14.0 | 7.2 | 4.4 | 3.0 | 2.0 | 0.6 | 0.2 |
| NWid | −0.628 * | 0.277 | 0.216 | 0.191 | 0.199 | −0.126 | −0.185 | −0.273 | 0.136 |
| NThic | −0.578 * | 0.557 | 0.159 | 0.103 | 0.296 | −0.074 | −0.268 | −0.126 | 0.086 |
| SLen | 0.446 * | 0.145 | 0.281 | 0.149 | 0.185 | 0.166 | −0.055 | 0.033 | 0.188 |
| NLen | 0.328 | 0.742 * | −0.273 | 0.023 | −0.052 | −0.181 | −0.033 | −0.214 | 0.258 |
| NStoRow | 0.250 | 0.439 * | 0.011 | 0.409 | 0.060 | −0.154 | −0.408 | −0.269 | 0.224 |
| NShLen | −0.047 | 0.171 | 0.579 * | 0.009 | −0.288 | −0.218 | 0.046 | −0.063 | 0.293 |
| SWid | 0.348 | 0.259 | 0.475 * | 0.109 | 0.419 | 0.108 | −0.039 | 0.113 | −0.019 |
| NStom | 0.068 | 0.060 | −0.028 | 0.639 * | 0.012 | 0.334 | −0.193 | 0.262 | 0.018 |
| NResIn | −0.263 | 0.375 | 0.282 | −0.182 | −0.553 * | 0.128 | −0.287 | 0.011 | 0.001 |
| NResTot | −0.296 | 0.415 | 0.278 | −0.168 | −0.530 * | 0.118 | −0.131 | −0.216 | −0.078 |
| CWid | 0.337 | 0.136 | 0.325 | −0.218 | 0.432 * | 0.073 | 0.156 | −0.019 | −0.023 |
| CLen | 0.153 | 0.076 | −0.040 | −0.093 | 0.363 * | 0.166 | −0.018 | −0.060 | 0.231 |
| NStomDo | −0.019 | −0.159 | 0.394 | 0.246 | 0.087 | −0.452 * | −0.388 | −0.424 | −0.002 |
| NTeh | −0.110 | 0.002 | 0.063 | 0.341 | −0.102 | 0.361 * | 0.354 | −0.277 | 0.263 |
| SWing | 0.253 | 0.154 | 0.148 | 0.008 | 0.329 | 0.344 * | 0.079 | −0.122 | −0.167 |
| SLenWing | 0.324 | 0.167 | 0.195 | 0.042 | 0.328 | 0.337 * | 0.055 | −0.098 | −0.099 |
| NStomIn | −0.005 | −0.136 | 0.190 | −0.154 | 0.213 | 0.139 | −0.454 * | −0.411 | 0.376 |
| NStomDo | 0.004 | −0.141 | 0.364 | 0.158 | 0.168 | −0.387 | −0.428 | −0.472 * | 0.134 |
| NResDo | −0.250 | 0.343 | 0.189 | −0.097 | −0.339 | 0.069 | 0.117 | −0.454 * | −0.160 |
| CLen/CWid | −0.128 | −0.022 | −0.380 | 0.083 | 0.105 | 0.159 | −0.136 | −0.007 | 0.424 * |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eliades, N.-G.H.; Aravanopoulos, F.A.; Christou, A.K. Mediterranean Islands Hosting Marginal and Peripheral Forest Tree Populations: The Case of Pinus brutia Ten. in Cyprus. Forests 2018, 9, 514. https://doi.org/10.3390/f9090514
Eliades N-GH, Aravanopoulos FA, Christou AK. Mediterranean Islands Hosting Marginal and Peripheral Forest Tree Populations: The Case of Pinus brutia Ten. in Cyprus. Forests. 2018; 9(9):514. https://doi.org/10.3390/f9090514
Chicago/Turabian StyleEliades, Nicolas-George H., Filippos (Phil) A. Aravanopoulos, and Andreas K. Christou. 2018. "Mediterranean Islands Hosting Marginal and Peripheral Forest Tree Populations: The Case of Pinus brutia Ten. in Cyprus" Forests 9, no. 9: 514. https://doi.org/10.3390/f9090514
APA StyleEliades, N.-G. H., Aravanopoulos, F. A., & Christou, A. K. (2018). Mediterranean Islands Hosting Marginal and Peripheral Forest Tree Populations: The Case of Pinus brutia Ten. in Cyprus. Forests, 9(9), 514. https://doi.org/10.3390/f9090514






