Effect of Soil Layer and Plant–Soil Interaction on Soil Microbial Diversity and Function after Canopy Gap Disturbance
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site Description
2.2. Experimental Design
2.3. Soil Sampling
2.4. Plant Characteristics
2.5. Analysis of Soil Properties
2.6. PLFA Analysis
2.7. Soil Enzyme Activity
2.8. Statistical Analyses
3. Results
3.1. Plant Characteristics
3.2. Soil Chemical Properties.
3.3. Microbial Community Structure
3.4. Enzyme Activities
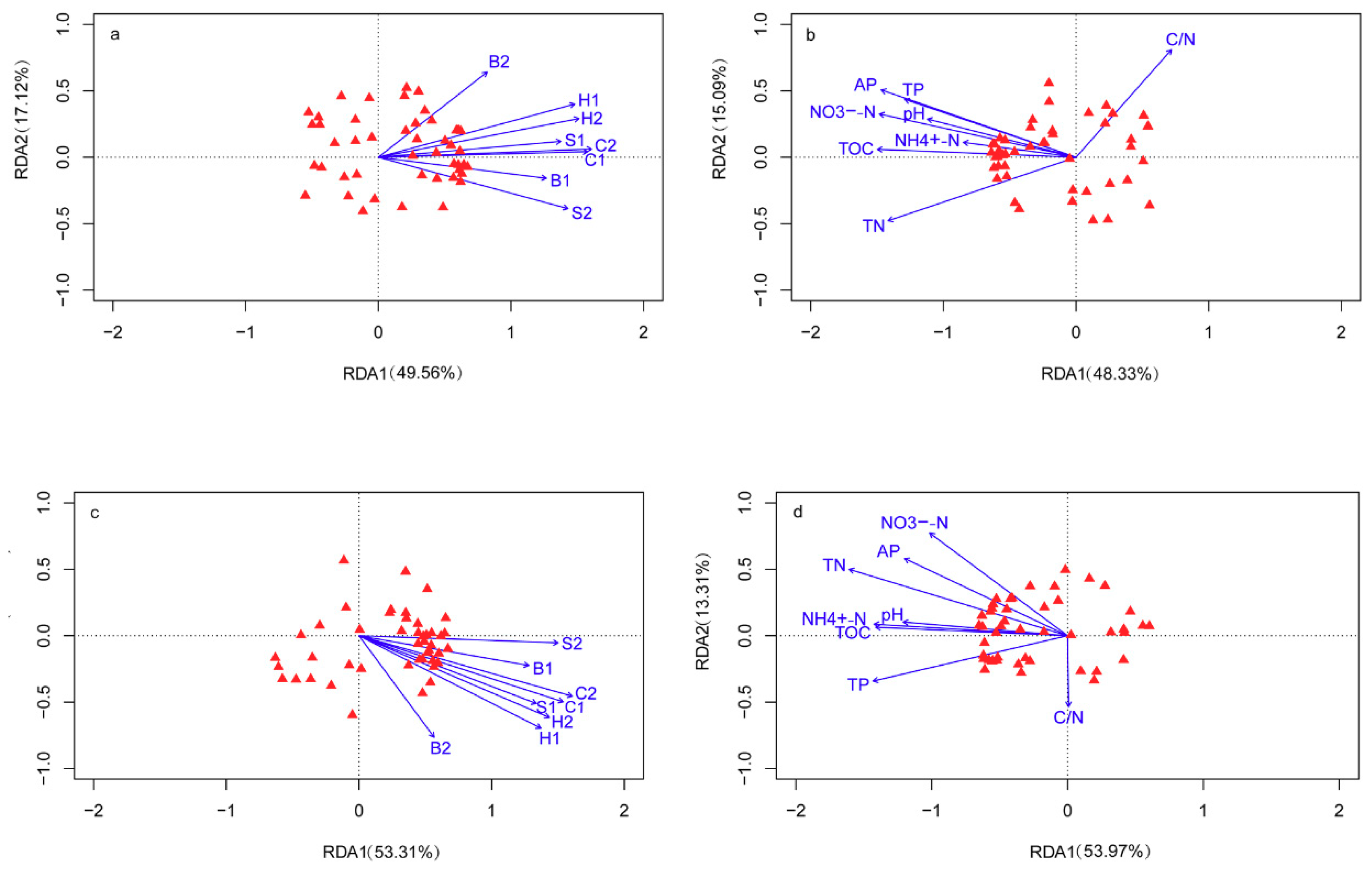
3.5. Correlations among Plant Characteristics, Soil Chemical Properties, and Microbial Communities
3.6. Correlation between Soil Microbial Communities and Enzyme Activities
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Liu, D.; Huang, Y.M.; Sun, H.Y.; An, S.S. The restoration age of Robiniapseudoacacia plantation impacts soil microbial biomass and microbial community structure in the Loess Plateau. Catena 2018, 165, 192–200. [Google Scholar] [CrossRef]
- Parvin, S.; Blagodatskaya, E.; Becker, J.N.; Kuzyakov, Y.; Uddin, S.; Dorodnikov, M. Depth rather than microrelief controls microbial biomass and kinetics of C-, N-, P- and S-cycle enzymes in peatland. Geoderma 2018, 324, 67–76. [Google Scholar] [CrossRef]
- Bolat, I. The effect of thinning on microbial biomass C, N and basal respiration in black pine forest soils in Mudurnu, Turkey. Eur. J. For. Res. 2014, 133, 131–139. [Google Scholar] [CrossRef]
- Scharenbroch, B.C.; Bockheim, J.G. Impacts of forest gaps on soil properties and processes in old growth northern hardwood-hemlock forests. Plant Soil 2007, 294, 219–233. [Google Scholar] [CrossRef]
- Schliemann, S.A.; Bockheim, J.G. Influence of gap size on carbon and nitrogen biogeochemical cycling in Northern hardwood forests of the Upper Peninsula, Michigan. Plant Soil 2014, 377, 323–335. [Google Scholar] [CrossRef]
- Xu, J.X.; Xue, L.; Su, Z.Y. Impacts of Forest Gaps on Soil Properties after a Severe Ice Storm in a Cunninghamialanceolata Stand. Pedosphere 2016, 26, 408–416. [Google Scholar] [CrossRef]
- Muscolo, A.; Sidari, M.; Mercurio, R. Influence of gap size on organic matter decomposition, microbial biomass and nutrient cycle in Calabrian pine (Pinuslaricio, Poiret) stands. For. Ecol. Manag. 2007, 242, 412–418. [Google Scholar] [CrossRef]
- Denslow, J.S.; Ellison, A.M.; Sanford, R.E. Treefall gap size effects on above- and below-ground processes in a tropical wet forest. J. Ecol. 1998, 86, 597–609. [Google Scholar] [CrossRef]
- Hu, W.J.; Wang, P.C.; Zhang, Y.; Teng, M.J.; Zhou, Z.X. Effects of gap disturbances on soil properties and understory plant diversity in a Pinusmassoniana in Hubei, central China. J. Anim. Plant Sci. 2016, 26, 988–1001. [Google Scholar]
- Wang, Z.M.; Pan, L.; Xue, L. Effects of forest gaps on soil microbial diversity in a Cunninghamialanceolata stand after a serve storm. Int. J. Agric. Biol. 2018, 20, 1161–1166. [Google Scholar]
- He, R.Y.; Yang, K.J.; Li, Z.J.; Schaedler, M.; Yang, W.Q.; Wu, F.Z.; Tan, B.; Zhang, L.; Xu, Z.F. Effects of forest conversion on soil microbial communities depend on soil layer on the eastern Tibetan Plateau of China. PLoS ONE 2017, 12, e0186053. [Google Scholar] [CrossRef] [PubMed]
- Han, W.J.; He, J.F.; Zhang, W.H.; Li, J.X. Effects of gap size on root growth of seedlings and soil physical and chemical properties in Pinustabulaeformis plantation in the Huanglong Forest. Sci. Silvae Sin. 2013, 49, 16–23. [Google Scholar]
- Wang, Z.B.; Yang, H.J.; Dong, B.Q.; Zhou, M.M.; Ma, L.Y.; Jia, Z.K.; Duan, J. Effects of canopy gap size on growth and spatial patterns of Chinese pine (Pinustabulaeformis) regeneration. For. Ecol. Manag. 2017, 385, 46–56. [Google Scholar] [CrossRef]
- Wang, Z.B.; Yang, H.J.; Dong, B.Q.; Zhou, M.M.; Ma, L.Y.; Jia, Z.K.; Duan, J. Regeneration response to canopy gap size in a Chinese pine plantation: Species diversity patterns, size structures and spatial distributions. For. Ecol. Manag. 2017, 397, 97–107. [Google Scholar] [CrossRef]
- Wang, Z.B.; Zhao, K.J.; Yang, H.J.; Ma, L.Y.; Jia, Z.K. Various response of Pinustabulaeformis Carr. regeneration in artifical gaps. Sci. Rep. 2017, 7, 1–12. [Google Scholar]
- Brokaw, N.V.L. The definition of treefall gap and its effect on measures of forest dynamics. Biotropica 1982, 14, 158–160. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, G.B.; Xue, S.; Wang, G.L. Soil bacterial community dynamics reflect changes in plant community and soil properties during the secondary succession of abandoned farmland in the Loess Plateau. Soil Biol. Biochem. 2016, 97, 40–49. [Google Scholar] [CrossRef]
- Tscherko, D.; Hammesfahr, U.; Marx, M.C.; Kandeler, E. Shifts in rhizosphere microbial communities and enzyme activity of Poaalpina across an alpine chronosequence. Soil Biol. Biochem. 2004, 36, 1685–1698. [Google Scholar] [CrossRef]
- Liu, G.S. Soil Physical and Chemical Analysis and Description of Soil Profiles, 1st ed.; Standards Press of China: Beijing, China, 1996; pp. 5–42. ISBN 7-5066-1218-6. [Google Scholar]
- White, D.C.; Davis, W.M.; Nickels, J.S.; King, J.D.; Bobbie, R.J. Determination of the sedimentary microbial biomass by extractible lipid phosphate. Oecologia 1979, 40, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Yang, D.; Yu, X. Soil microbial communities and enzyme activities in sea-buckthorn (Hippophae rhamnoides) plantation at different ages. PLoS ONE 2018. [Google Scholar] [CrossRef] [PubMed]
- Zelles, L. Fatty acid patterns of phospholipids and lipopolysaccharides in the characterization of microbial communities in soil: A review. Biol. Fertil. Soils 1999, 29, 111–129. [Google Scholar] [CrossRef]
- Frostegård, A.; Bååth, E. The use of phospholipid fatty acid analysis to estimate bacterial and fungal biomass in soils. Biol. Fertil. Soils 1996, 22, 59–65. [Google Scholar] [CrossRef]
- Bossio, D.A.; Fleck, J.A.; Scow, K.M.; Fujii, R. Alteration of soil microbial communities and water quality in restored wetlands. Soil Biol. Biochem. 2006, 38, 1223–1233. [Google Scholar] [CrossRef]
- Frostegård, A.; Tunlid, A.; Bååth, E. Use and misuse of PLFA measurements in soils. Soil Biol. Biochem. 2011, 43, 1621–1625. [Google Scholar] [CrossRef]
- Myers, R.T.; Zak, D.R.; White, D.C.; Peacock, A. Landscape-level patterns of microbial community composition and substrate use in upland forest ecosystems. Soil. Sci. Soc. Am. J. 2001, 65, 359–367. [Google Scholar] [CrossRef]
- Yarwood, S.A.; Myrold, D.D.; Hogberg, M.N. Termination of belowground C allocation by trees alters soil fungal and bacterial communities in a boreal forest. Fems. Microbiol. Ecol. 2009, 70, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Guan, S.Y. Soil Enzyme and Its Study Methods, 1st ed.; Agriculture Press: Beijing, China, 1986; pp. 274–339. ISBN 161443123. [Google Scholar]
- Yang, Y.; Geng, Y.Q.; Zhou, H.J.; Zhao, G.L.; Wang, L. Effects of gaps in the forest canopy on soil microbial communities and enzyme activity in a Chinese pine forest. Pedobiologia 2017, 61, 51–60. [Google Scholar] [CrossRef]
- Pang, X.Y.; Hu, B.; Bao, W.K.; Vargas, T.D.; Tian, G.L. Effect of thinning-induced gap size on soil CO2 efflux in a reforested spruce forest in the eastern Tibetan Plateau. Agric. For. Meteorol. 2016, 220, 1–9. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, J.; Yang, W.Q.; Wu, F.Z.; Xu, Z.F.; Tan, B.; Zhang, L.; He, X.H.; Guo, L. Canopy gaps accelerate soil organic carbon retention by soil microbial biomass in the organic horizon in a subalpine fir forest. Appl. Soil. Ecol. 2018, 125, 169–176. [Google Scholar] [CrossRef]
- Hu, B.; Yang, B.; Pang, X.Y.; Bao, W.K.; Tian, G.L. Reponses of soil phosphorus fractions to gap size in a reforested spruce forest. Geoderma 2016, 279, 61–69. [Google Scholar] [CrossRef]
- Özcan, M.; Gökbulak, F. Effect of size and surrounding forest vegetation on chemical properties of soil in forest gaps. iForest 2015, 8, 67–72. [Google Scholar] [CrossRef]
- Coulombe, D.; Sirois, L.; Paré, D. Effect of harvest gap formation and thinning on soil nitrogen cycling at the boreal-temperate interface. Can. J. For. Res. 2017, 47, 308–318. [Google Scholar] [CrossRef]
- Han, Z.M.; Deng, M.W.; Yuan, A.Q.; Wang, J.H.; Li, H.; Ma, J.C. Vertical variation of a black soil’s properties in response to freeze-thaw cycles and its links to shift of microbial community structure. Sci. Total Environ. 2018, 625, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.J.; Zhao, F.Z.; Kang, D.; Yang, G.H.; Han, X.H.; Tong, X.G.; Feng, Y.Z.; Ren, G.X. Linkages of C:N:P stoichiometry and bacterial community in soil following afforestation of former farmland. For. Ecol. Manag. 2016, 376, 59–66. [Google Scholar] [CrossRef]
- Wang, Y.; Li, C.Y.; Tu, C.; Hoyt, G.D.; DeForest, J.L.; Hu, S.J. Long-term no-tillage and organic input management enhanced the diversity and stability of soil microbial community. Sci. Total Environ. 2017, 609, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Knelman, J.E.; Graham, E.B.; Trahan, N.A.; Schmidt, S.K.; Nemergut, D.R. Fire severity shapes plant colonization effects on bacterial community structure, microbial biomass, and soil enzyme activity in secondary succession of a burned forest. Soil Biol. Biochem. 2015, 90, 161–168. [Google Scholar] [CrossRef]
- Knelman, J.E.; Graham, E.B.; Prevéy, J.S.; Robeson, M.S.; Kelly, P.; Hood, E.; Schmldt, S.K. Interspecific plant interactions reflected in soil bacterial community structure and nitrogen cycling in primary succession. Front. Microbiol. 2018, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.Z.; Wang, J.Y.; Zhang, L.; Ren, C.J.; Han, X.H.; Yang, G.H.; Doughty, R.; Deng, J. Understory plants regulate soil respiration through changes in soil enzyme activity and microbial C, N, and P stoichiometry following afforestation. Forests 2018, 9, 436. [Google Scholar] [CrossRef]
- Zhou, Z.C.; Shangguan, Z.P. Vertical distribution of fine roots in relation to soil factors in Pinustabulaeformis Carr. forest of the Loess Plaetau of China. Plant Soil 2007, 291, 119–129. [Google Scholar] [CrossRef]
- Reverchon, F.; Bai, S.H.; Liu, X.; Blumfield, T.J. Tree plantation systems influence nitrogen retention and the abundance of nitrogen functional genes in the solomon islands. Front. Microbiol. 2015, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.E.; Smith, F.A. Roles of arbuscularmycorrhizas in plant nutrition and growth: New paradigms from cellular to ecosystem scales. Annu. Rev. Plant Biol. 2011, 62, 227–250. [Google Scholar] [CrossRef] [PubMed]
- Herman, D.J.; Firestone, M.K.; Nuccio, E.; Hodge, A. Interactions between an arbuscularmycorrhizal fungus and a soil microbial community mediating litter decomposition. FEMS Microbiol. Ecol. 2012, 80, 236–247. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.S.; Jia, Y.Y.; Song, F.Q.; Tian, K.; Lin, H.; Bei, Z.L.; Jia, X.Q.; Yao, B.; Guo, P.; Tian, X.J. Insight into litter decomposition driven by demands of symbiosis system through the hypha bridge of arbuscularmycorrhizal fungi. Environ. Sci. Pollut. Res. 2018, 25, 5369–5378. [Google Scholar] [CrossRef] [PubMed]
- Cavagnaro, T.R.; Bender, S.F.; Asghari, H.R.; van der Heijden, M.G.A. The role of arbuscularmycorrhizas in reducing soil nutrient loss. Trends Plant Sci. 2015, 20, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.F.; Yuan, S.H.; Song, H.C.; Su, X.L.; Mao, H.Q.; Shen, W.Y.; Qu, X.L.; Dong, J.Y. Arbuscularmycorrhizal and dark septate fungal associations in riparian plants of the three Gorges Reservoir Region, Southwest China. Aquat. Bot. 2016, 133, 28–37. [Google Scholar] [CrossRef]
- Guo, X.H.; Gong, J. Differential effects of abiotic factors and host plant traits on diversity and community composition of root-colonizing arbuscularmycorrhizal fungi in a salt-stressed ecosystem. Mycorrhiza 2014, 24, 79–94. [Google Scholar] [CrossRef] [PubMed]
- Marosz, A. Effect of green waste compost and mycorrhizal fungi on calcium, potassium, and sodium uptake of woody plants growth under salt stress. Water Air Soil Pollut. 2012, 223, 223–787. [Google Scholar] [CrossRef]
- Bongard, C.L.; Navaranjan, G.; Yan, W.; Fulthorpe, R.R. Fungal colonization of the invasive vine Vincetoxiumrossicumand native plants. Plant Ecol. Evol. 2013, 146, 45–52. [Google Scholar] [CrossRef]
- Zhang, W.; Qiao, W.J.; Gao, D.X.; Dai, Y.Y.; Deng, J.; Yang, G.H.; Han, X.H.; Ren, G.X. Relationship between soil nutrient properties and biological activities along a restoration chronosequence of Pinustabulaeformis plantation forests in the Ziwuling Mountains, China. Catena 2018, 161, 85–95. [Google Scholar] [CrossRef]
- Yuan, B.C.; Yue, D.X. Soil Microbial and Enzymatic Activities across a Chronosequence of Chinese Pine Plantation Development on the Loess Plateau of China. Pedosphere 2012, 22, 1–12. [Google Scholar] [CrossRef]

| Undergrowth Plant Species | Gap | Coverage (%) | Biomass (g cm−3) | Hplant | Splant |
|---|---|---|---|---|---|
| Shrub species | G1 | 17.13 ± 2.68b | 1.03 ± 0.32ab | 1.20 ± 0.03b | 7.33 ± 0.58b |
| G2 | 25.70 ± 1.40a | 1.53 ± 0.18a | 1.69 ± 0.08a | 9.33 ± 0.58a | |
| G3 | 11.57 ± 1.10c | 0.72 ± 0.21b | 1.08 ± 0.06b | 6.33 ± 0.58b | |
| G0 | 15.37 ± 2.10b | 1.01 ± 0.21ab | 1.15 ± 0.09b | 7.00 ± 1.00b | |
| Herb species | G1 | 18.47 ± 0.75b | 0.32 ± 0.13b | 0.91 ± 0.04b | 5.67 ± 0.58a |
| G2 | 27.43 ± 0.91a | 0.62 ± 0.09a | 1.44 ± 0.06a | 6.33 ± 0.58a | |
| G3 | 12.13 ± 1.05d | 0.41 ± 0.22ab | 0.74 ± 0.07c | 4.33 ± 0.58b | |
| G0 | 16.47 ± 1.07c | 0.30 ± 0.07b | 0.81 ± 0.05bc | 5.33 ± 0.58ab |
| Layer | Gap | pH | TOC | TN | TP | NH4+–N | NO3−–N | AP |
|---|---|---|---|---|---|---|---|---|
| OL | G1 | 7.51 ± 0.15 abA | 42.61 ± 2.50 abA | 7.18 ± 0.17 bA | 0.29 ± 0.13 bA | 13.56 ± 0.65 abA | 8.25 ± 1.32 bA | 9.11 ± 0.28 bA |
| G2 | 7.73 ± 0.12 aA | 47.09 ± 3.25 aA | 7.89 ± 0.35 aA | 0.55 ± 0.21 aA | 16.02 ± 2.34 aA | 11.13 ± 1.60 aA | 13.12 ± 1.23 aA | |
| G3 | 7.41 ± 0.12 bA | 38.41 ± 1.92 bA | 5.46 ± 0.35 cA | 0.24 ± 0.11 bA | 12.61 ± 0.57 bA | 7.02 ± 0.93 bA | 8.33 ± 0.70 bcA | |
| G0 | 7.50 ± 0.12 abA | 40.62 ± 2.30 bA | 7.35 ± 0.05 abA | 0.26 ± 0.11 bA | 15.36 ± 1.06 abA | 8.05 ± 0.53 bA | 8.85 ± 0.42 bcA | |
| ML | G1 | 7.14 ± 0.17 abB | 13.65 ± 1.24 abB | 1.20 ± 0.19 bB | 0.20 ± 0.01 abA | 5.26 ± 1.06 abB | 3.95±1.37 bB | 4.06 ± 2.57 bB |
| G2 | 7.36 ± 0.05 aB | 16.15 ± 1.65 aB | 1.58 ± 0.19 aB | 0.23 ± 0.12 aA | 6.45 ± 0.63 aB | 7.17 ± 1.51 aB | 8.41 ± 1.75 aB | |
| G3 | 7.06 ± 0.13 bB | 11.36 ± 1.81 bB | 1.09 ± 0.08 bB | 0.11 ± 0.03 abA | 4.09 ± 1.12 bB | 4.45 ± 0.73 bB | 3.48 ± 2.18 bB | |
| G0 | 7.10 ± 0.19 abB | 12.45 ± 1.79 abB | 1.12 ± 0.10 bB | 0.10 ± 0.03 bA | 4.56 ± 0.68 abB | 4.08 ± 1.03 bB | 3.88 ± 2.23 bB | |
| L | *** | *** | *** | ** | *** | *** | *** | |
| G | * | ** | *** | * | ** | *** | *** | |
| L × G | ns | ns | *** | ns | ns | ns | ns |
| Layer | Gap | Total Lipid | G+ | G− | Bacteria | Fungi | AMF |
|---|---|---|---|---|---|---|---|
| OL | G1 | 59.21 ± 2.40 bA | 16.86 ± 1.98 aA | 9.83 ± 1.94 aA | 35.33 ± 4.61 abA | 6.14 ± 1.60 abA | 2.15 ± 0.23 bA |
| G2 | 66.90 ± 4.80 aA | 18.78 ± 1.30 aA | 11.13 ± 0.95 aA | 39.21 ± 3.12 aA | 7.46 ± 0.53 aA | 2.65 ± 0.24 aA | |
| G3 | 43.35 ± 2.44 dA | 9.59 ± 0.58 bA | 5.60 ± 0.56 bA | 19.31 ± 0.40 cA | 4.66 ± 0.59 bA | 1.90 ± 0.27 bA | |
| G0 | 54.74 ± 1.78 cA | 10.62 ± 2.46 bA | 6.17 ± 1.36 bA | 29.76 ± 4.05 bA | 5.80 ± 0.75 abA | 2.04 ± 0.30 bA | |
| ML | G1 | 46.70 ± 3.05 bB | 13.74 ±1.64 aB | 5.11 ± 0.23 aB | 28.09 ± 2.35 bB | 3.93 ± 0.52 bB | 1.06 ± 0.48 aB |
| G2 | 57.88 ± 3.17 aB | 15.32 ±1.77 aB | 6.36 ± 0.28 aB | 31.58 ± 1.64 aB | 5.02 ± 0.43 aB | 1.35 ± 0.38 aB | |
| G3 | 31.61 ± 1.23 dB | 7.36 ±0.02 bB | 3.24 ± 0.67 bB | 14.33 ± 0.43 dB | 2.14 ± 0.21 cB | 0.82 ± 0.33 aB | |
| G0 | 42.19 ± 1.01 cB | 9.88 ± 2.2 bB | 3.77 ± 0.47 bB | 21.29 ± 2.03 cB | 3.25 ± 0.37 bcB | 0.79 ± 0.18 aB | |
| L | *** | ** | *** | *** | *** | *** | |
| G | *** | *** | *** | *** | *** | * | |
| L × G | ns | ns | ns | ns | ns | ns |
| Layer | Gap | β-glucosidase (μg g−1dw h−1) | Cellulase (mg g−1dw h−1) | Urease (mg g−1dw d−1) | Phosphatase (mg g−1dw d−1) |
|---|---|---|---|---|---|
| OL | G1 | 27.64 ± 0.95 bA | 0.63 ± 0.21 abA | 1.84 ± 0.42 abA | 3.18 ± 0.34 aA |
| G2 | 32.72 ± 0.88 aA | 0.92 ± 0.20 aA | 2.10 ± 0.26 aA | 3.24 ± 0.12 aA | |
| G3 | 11.48 ± 0.61 dA | 0.51 ± 0.18 bA | 1.57 ± 0.48 abA | 3.08 ± 0.33 aA | |
| G0 | 25.45 ± 0.80 cA | 0.70 ± 0.25 abA | 1.32 ± 0.28 bA | 3.51 ± 0.43 aA | |
| ML | G1 | 9.35 ± 1.00 bB | 0.15 ± 0.12 bB | 0.84 ± 0.22 acB | 1.30 ± 0.30 aB |
| G2 | 12.26 ± 0.87 aB | 0.41 ± 0.10 aB | 1.06 ± 0.23 aB | 1.46 ± 0.42 aB | |
| G3 | 7.66 ± 0.19 cB | 0.11 ± 0.04 bB | 0.31 ± 0.21 bcB | 1.26 ± 0.18 aB | |
| G0 | 8.45 ± 0.24 bcB | 0.12 ± 0.04 bB | 0.48 ± 0.15 cB | 1.55 ± 0.32 aB | |
| layer | *** | *** | *** | *** | |
| gap | *** | ** | ** | ns | |
| Layer × gap | *** | ns | ns | ns |
| Total Lipids | Bacteria | Fungi | G+ | G− | |
|---|---|---|---|---|---|
| GLU | 0.937 ** | 0.931 ** | 0.622 * | 0.785 ** | 0.762 ** |
| CEL | 0.546 | 0.529 | 0.464 | 0.411 | 0.462 |
| URE | 0.500 | 0.555 | 0.355 | 0.717 ** | 0.732 ** |
| PHO | 0.696 * | 0.628 * | 0.637 * | 0.573 | 0.608 * |
| Total Lipids | Bacteria | Fungi | G+ | G− | |
|---|---|---|---|---|---|
| GLU | 0.673 * | 0.612 * | 0.424 | 0.496 | 0.692 ** |
| CEL | 0.783 ** | 0.791 ** | 0.411 | 0.710 ** | 0.805 ** |
| URE | 0.885 ** | 0.855 ** | 0.529 | 0.789 ** | 0.784 ** |
| PHO | 0.701 * | 0.646 * | 0.530 | 0.546 | 0.706 ** |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, X.; Yang, L.; Fei, S.; Ma, Z.; Hao, R.; Zhao, Z. Effect of Soil Layer and Plant–Soil Interaction on Soil Microbial Diversity and Function after Canopy Gap Disturbance. Forests 2018, 9, 680. https://doi.org/10.3390/f9110680
Yu X, Yang L, Fei S, Ma Z, Hao R, Zhao Z. Effect of Soil Layer and Plant–Soil Interaction on Soil Microbial Diversity and Function after Canopy Gap Disturbance. Forests. 2018; 9(11):680. https://doi.org/10.3390/f9110680
Chicago/Turabian StyleYu, Xuan, Lin Yang, Shixuan Fei, Zitong Ma, Ruqian Hao, and Zhong Zhao. 2018. "Effect of Soil Layer and Plant–Soil Interaction on Soil Microbial Diversity and Function after Canopy Gap Disturbance" Forests 9, no. 11: 680. https://doi.org/10.3390/f9110680
APA StyleYu, X., Yang, L., Fei, S., Ma, Z., Hao, R., & Zhao, Z. (2018). Effect of Soil Layer and Plant–Soil Interaction on Soil Microbial Diversity and Function after Canopy Gap Disturbance. Forests, 9(11), 680. https://doi.org/10.3390/f9110680




