Towards the Identification of Candidate Genes for Pollen Morphological Traits in Rubus L. Using Association Mapping
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Genotypic Observations
2.3. Statistical Methods
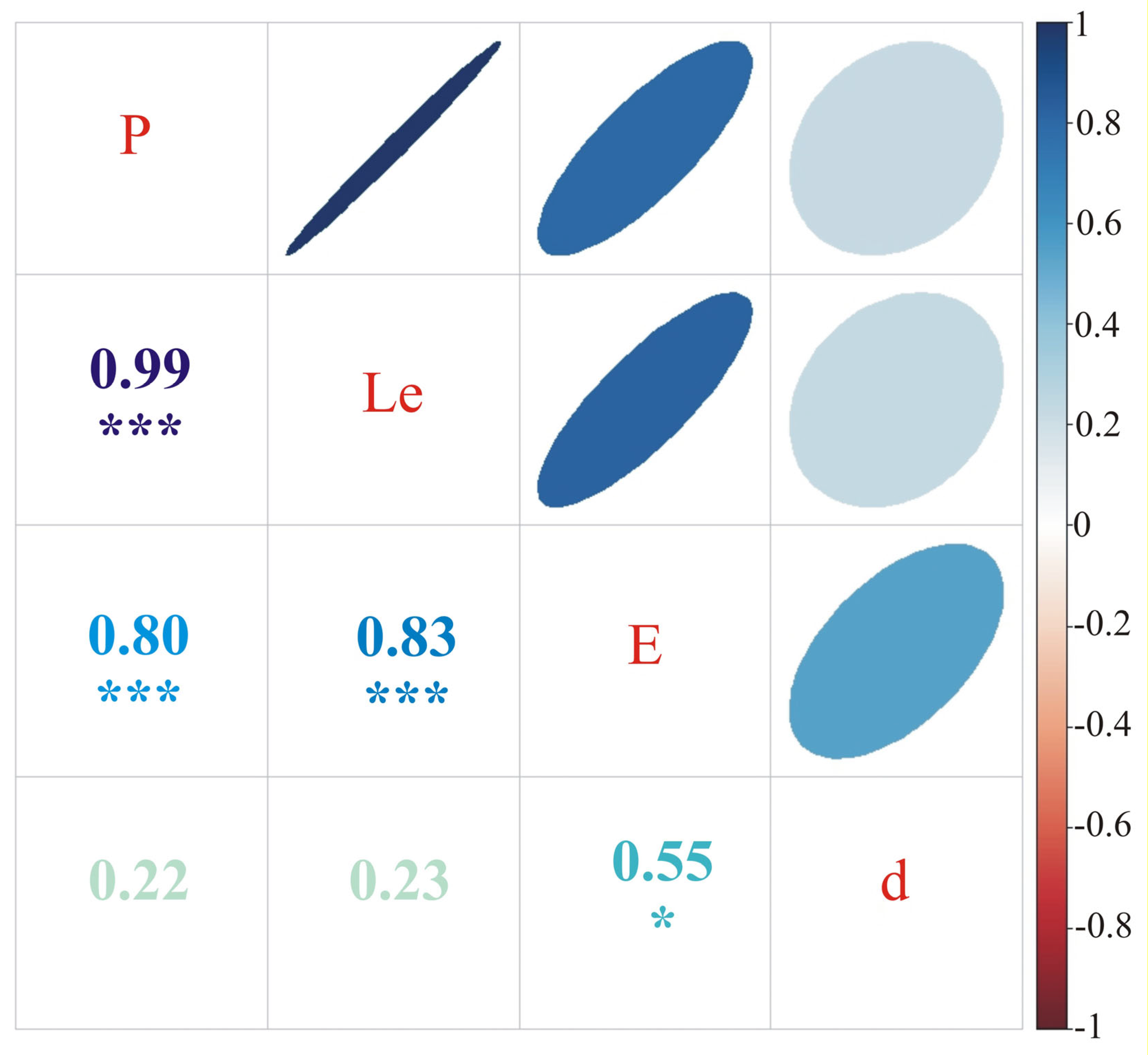
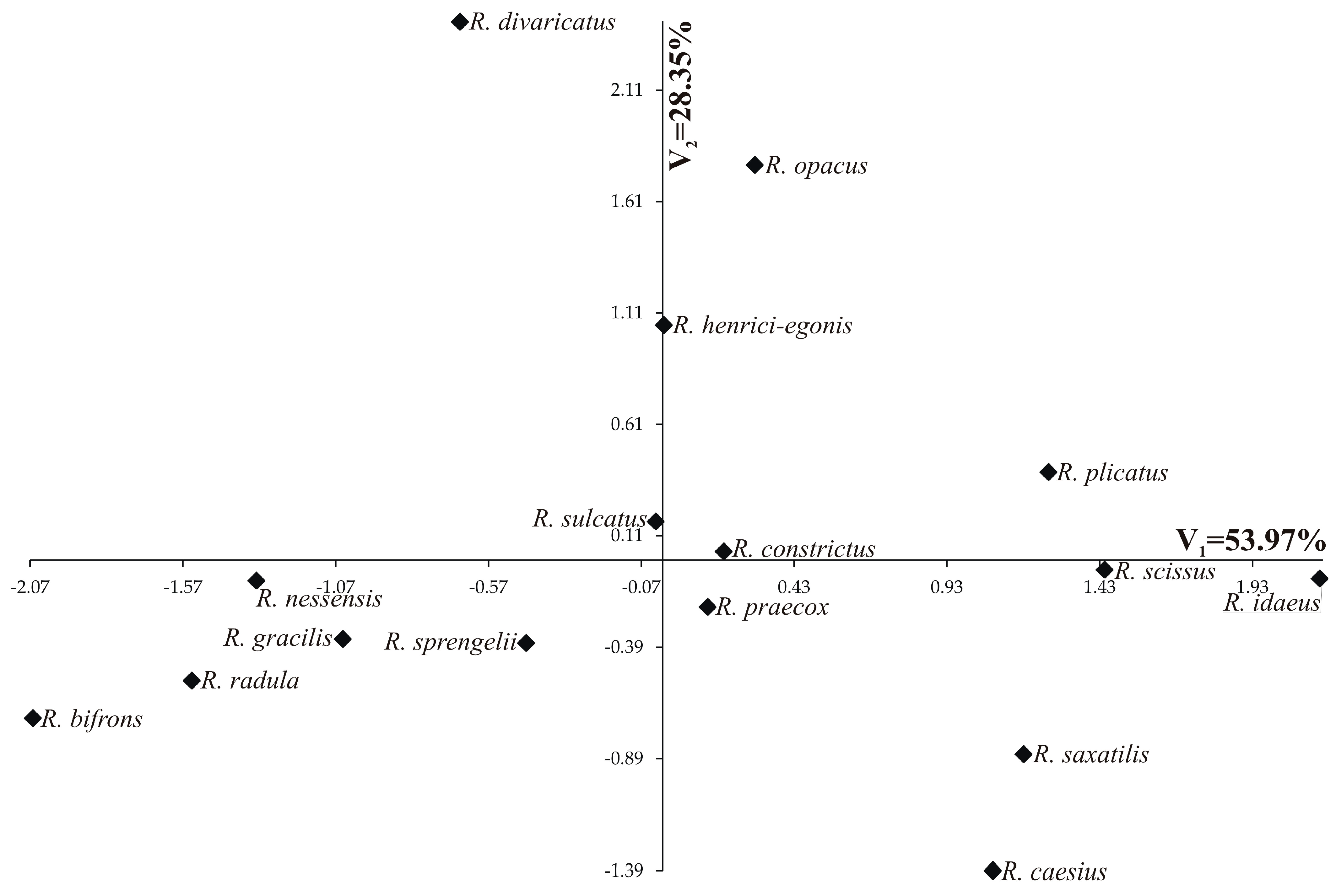
3. Results
3.1. Phenotyping
3.2. Association Mapping
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| ISSR | Inter Simple Sequence Repeats |
| SSR | Simple Sequence Repeats |
| DNA | Deoxyribonucleic acid |
| QTL | Quantitative Trait Loci |
| MANOVA | Multivariate analysis of variance |
| ANOVA | Analysis of variance |
| LSD | Least significant difference |
| s.d. | Standard deviation |
| P | Length of the polar axis |
| Le | Length of the ectoaperture |
| d | The distance between the apices of two ectocolpi |
| E | Equatorial diameter |
| LOD | The limit of detection |
| SNP | Single-nucleotide polymorphism |
| AFLP | Amplified fragment length polymorphism |
Appendix A
| No. | Species | P 1 | Le 2 | d 3 | E 4 | No. | Species | P | Le | d | E |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Rubus bifrons | 25.53 | 21.22 | 2.360 | 21.40 | 9 | Rubus opacus | 20.54 | 17.73 | 2.147 | 16.88 |
| 1 | Rubus bifrons | 24.84 | 20.89 | 2.968 | 21.54 | 9 | Rubus opacus | 22.96 | 18.46 | 3.061 | 18.61 |
| 1 | Rubus bifrons | 26.69 | 22.98 | 2.842 | 22.36 | 10 | Rubus plicatus | 23.11 | 19.75 | 2.638 | 21.30 |
| 1 | Rubus bifrons | 24.78 | 21.09 | 2.580 | 21.35 | 10 | Rubus plicatus | 21.15 | 18.36 | 2.833 | 18.97 |
| 1 | Rubus bifrons | 25.33 | 21.31 | 2.531 | 21.01 | 10 | Rubus plicatus | 24.29 | 19.74 | 3.138 | 21.79 |
| 2 | Rubus caesius | 21.89 | 18.26 | 2.347 | 18.39 | 10 | Rubus plicatus | 23.46 | 19.33 | 3.333 | 21.25 |
| 2 | Rubus caesius | 26.15 | 22.38 | 3.850 | 22.92 | 10 | Rubus plicatus | 22.70 | 19.30 | 3.653 | 21.95 |
| 2 | Rubus caesius | 25.72 | 21.75 | 3.770 | 22.97 | 11 | Rubus praecox | 24.29 | 20.70 | 2.811 | 21.67 |
| 2 | Rubus caesius | 25.58 | 21.03 | 4.553 | 21.77 | 11 | Rubus praecox | 22.27 | 18.87 | 3.384 | 19.53 |
| 2 | Rubus caesius | 23.68 | 19.83 | 4.075 | 22.22 | 11 | Rubus praecox | 23.04 | 19.78 | 2.700 | 20.44 |
| 3 | Rubus constrictus | 23.89 | 20.10 | 3.283 | 21.32 | 11 | Rubus praecox | 25.36 | 21.77 | 2.979 | 21.90 |
| 3 | Rubus constrictus | 23.46 | 19.71 | 2.734 | 20.18 | 12 | Rubus radula | 26.76 | 23.15 | 2.428 | 22.14 |
| 4 | Rubus divaricatus | 22.34 | 19.15 | 1.834 | 18.93 | 12 | Rubus radula | 23.63 | 20.09 | 2.620 | 20.58 |
| 4 | Rubus divaricatus | 21.01 | 17.70 | 2.066 | 16.81 | 12 | Rubus radula | 25.63 | 21.29 | 3.153 | 21.84 |
| 4 | Rubus divaricatus | 21.76 | 18.17 | 2.088 | 19.27 | 12 | Rubus radula | 24.27 | 19.98 | 3.324 | 21.11 |
| 4 | Rubus divaricatus | 21.72 | 18.19 | 2.047 | 19.15 | 12 | Rubus radula | 25.05 | 21.25 | 2.153 | 21.82 |
| 5 | Rubus gracilis | 25.63 | 21.65 | 3.098 | 22.08 | 13 | Rubus saxatilis | 24.48 | 21.57 | 3.669 | 20.31 |
| 5 | Rubus gracilis | 23.70 | 20.33 | 2.581 | 20.70 | 13 | Rubus saxatilis | 24.12 | 20.00 | 3.421 | 20.86 |
| 5 | Rubus gracilis | 24.69 | 21.41 | 3.000 | 21.43 | 13 | Rubus saxatilis | 22.63 | 19.24 | 3.345 | 20.83 |
| 5 | Rubus gracilis | 22.74 | 19.29 | 2.442 | 18.23 | 13 | Rubus saxatilis | 22.22 | 18.80 | 3.544 | 19.77 |
| 5 | Rubus gracilis | 25.46 | 21.23 | 2.720 | 21.30 | 13 | Rubus saxatilis | 22.54 | 19.15 | 3.239 | 19.37 |
| 6 | Rubus henrici-egonis | 22.67 | 18.84 | 2.266 | 18.64 | 14 | Rubus scissus | 22.73 | 19.18 | 2.597 | 20.94 |
| 6 | Rubus henrici-egonis | 24.42 | 20.87 | 3.722 | 22.53 | 14 | Rubus scissus | 23.29 | 19.80 | 3.529 | 21.68 |
| 6 | Rubus henrici-egonis | 22.42 | 18.91 | 2.151 | 19.13 | 14 | Rubus scissus | 24.36 | 20.64 | 3.576 | 21.13 |
| 6 | Rubus henrici-egonis | 21.23 | 17.92 | 2.288 | 18.91 | 15 | Rubus sprengelii | 24.80 | 21.15 | 3.222 | 21.08 |
| 6 | Rubus henrici-egonis | 22.70 | 19.26 | 2.614 | 19.12 | 15 | Rubus sprengelii | 23.50 | 19.75 | 2.774 | 20.72 |
| 7 | Rubus idaeus | 22.16 | 19.23 | 3.444 | 20.13 | 15 | Rubus sprengelii | 24.74 | 20.91 | 2.707 | 19.92 |
| 7 | Rubus idaeus | 24.62 | 20.32 | 4.053 | 22.46 | 15 | Rubus sprengelii | 24.22 | 20.52 | 2.997 | 21.00 |
| 7 | Rubus idaeus | 22.81 | 19.15 | 3.076 | 19.43 | 15 | Rubus sprengelii | 23.58 | 20.45 | 2.900 | 20.93 |
| 7 | Rubus idaeus | 21.39 | 18.39 | 3.321 | 19.51 | 16 | Rubus sulcatus | 24.34 | 20.41 | 3.011 | 21.65 |
| 7 | Rubus idaeus | 22.52 | 19.12 | 3.542 | 20.77 | 16 | Rubus sulcatus | 22.46 | 19.31 | 2.696 | 18.37 |
| 8 | Rubus nessensis | 23.10 | 19.60 | 2.549 | 19.00 | 16 | Rubus sulcatus | 23.10 | 19.55 | 2.780 | 21.33 |
| 8 | Rubus nessensis | 24.00 | 20.26 | 3.070 | 20.11 | 16 | Rubus sulcatus | 23.09 | 19.38 | 2.927 | 20.66 |
| 8 | Rubus nessensis | 25.63 | 21.34 | 2.597 | 20.80 | 16 | Rubus sulcatus | 24.66 | 21.38 | 2.734 | 21.42 |
| 8 | Rubus nessensis | 24.90 | 20.82 | 2.987 | 20.63 | ||||||
| 8 | Rubus nessensis | 24.31 | 20.34 | 2.440 | 19.79 |
Appendix B
| Integer from Division by 2 | Residual from Division by 2 | Direction of Writing Digits in Binary Number Notation |
|---|---|---|
| 103 | ↑ | |
| 51 | 1 | |
| 25 | 1 | |
| 12 | 1 | |
| 6 | 0 | |
| 3 | 0 | |
| 1 | 1 | |
| 0 | 1 |
References
- Wu, W.; Wang, L.; Huang, W.; Zhang, X.; Li, Y.; Guo, W. A high-quality genome assembly reveals adaptations underlying glossy, wax-coated leaves in the heat-tolerant wild raspberry Rubus leucanthus. DNA Res. 2024, 31, dsae024. [Google Scholar] [CrossRef]
- Zhang, L.; Huang, C.; Zhang, G.; Zhang, C.; Zhao, Y.; Huang, J.; Guo, J.; Cheng, L.; Zhang, T.; Ma, H. Nuclear phylogenomics of angiosperms and evolutionary implications. Diversity 2025, 17, 136. [Google Scholar] [CrossRef]
- Meng, Q.; Manghwar, H.; Hu, W. Study on Supergenus Rubus L.: Edible, Medicinal, and Phylogenetic Characterization. Plants 2022, 11, 1211. [Google Scholar] [CrossRef]
- Shi, Y.; Chen, Z.; Jiang, J.; Wu, W.; Xin, Y.; Zeng, W. Assembly and comparative analysis of the complete mitogenome of Rubus chingii var. suavissimus, an exceptional berry plant possessing sweet leaves. Front. Plant Sci. 2024, 15, 1504687. [Google Scholar] [CrossRef]
- Gaskin, J. Recent contributions of molecular population genetic and phylogenetic studies to classic biological control of weeds. BioControl 2024, 69, 353–360. [Google Scholar] [CrossRef]
- Eathington, S.R.; Crosbie, T.M.; Edwards, M.D.; Reiter, R.S.; Bull, J.K. Molecular markers in a commercial breeding program. Crop Sci. 2007, 47, S-154–S-163. [Google Scholar] [CrossRef]
- Kumar, R.; Das, S.P.; Choudhury, B.U.; Kumar, A.; Prakash, N.R.; Verma, R.; Chakraborti, M.; Devi, A.G.; Bhattacharjee, B.; Das, R.; et al. Advances in genomic tools for plant breeding: Harnessing DNA molecular markers, genomic selection, and genome editing. Biol. Res. 2024, 57, 80. [Google Scholar] [CrossRef]
- da Silva, A.V.; Prado, M.; Campos, G.R.; Borges, K.L.R.; Yassue, R.M.; Husein, G.; Sposito, M.B.; Amorim, L.; Crossa, J.; Fritsche-Neto, R. Comparing strategies for genomic predictions in interspecific biparental populations: A case study with the Rubus genus. Euphytica 2024, 220, 146. [Google Scholar] [CrossRef]
- Abdullah, M.; Ahmad, M.; Ullah, Z.; Khan, A.; Ahmad, S.; Shah, H.; Ullah, A.; Farah, M.A.; Emmanuel, O.; Iqbal, R.; et al. Palynological diversity in the Rosaceae flora of Northern Khyber Pakhtunkhwa, Pakistan. Genet. Resour. Crop Evol. 2024, 72, 4361–4379. [Google Scholar] [CrossRef]
- Hayrapetyan, A.; Asatryan, M.; Sonyan, H.; Balayan, K.; Beketovski, D. Pollen morphological study of some species of the genus Rubus L.(Rosaceae) of the flora of Armenia. Hacquetia 2025, 24, 67–82. [Google Scholar] [CrossRef]
- Lechowicz, K.; Wrońska-Pilarek, D.; Bocianowski, J.; Maliński, T. Pollen morphology of Polish species from the genus Rubus L.(Rosaceae) and its systematic importance. PLoS ONE 2020, 15, e0221607. [Google Scholar] [CrossRef]
- Purgina, C.; Ulrich, S.; Weber, M.; Grímsson, F. Morphological and Ultrastructural Features of Selected Epidendroideae Pollen Dispersal Units and New Insights into Their Chemical Nature. Plants 2024, 13, 1114. [Google Scholar] [CrossRef] [PubMed]
- Mimura, M.; Suga, M. Ambiguous species boundaries: Hybridization and morphological variation in two closely related Rubus species along altitudinal gradients. Ecol. Evol. 2020, 10, 7476–7486. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Liu, F.; Xu, Y.; Hu, W. Genetic Diversity Analysis and Core Germplasm Construction of Rubus chingii Hu. Plants 2024, 13, 618. [Google Scholar] [CrossRef]
- Zaman, W.; Ayaz, A.; Park, S. Integrating morphological and molecular data in plant taxonomy. Pak. J. Bot. 2025, 57, 1453–1466. [Google Scholar] [CrossRef]
- Agarwal, M.; Shrivastava, N.; Padh, H. Advances in molecular marker techniques and their applications in plant sciences. Plant Cell Rep. 2008, 27, 617–631. [Google Scholar] [CrossRef]
- Hayward, A.C.; Tollenaere, R.; Dalton-Morgan, J.; Batley, J. Molecular Marker Applications in Plants. In Plant Genotyping. Methods in Molecular Biology; Batley, J., Ed.; Humana Press: New York, NY, USA, 2015; Volume 1245. [Google Scholar] [CrossRef]
- Sharma, S.; Kaur, R.; Kumar, K.; Kumar, D.; Solanke, A.K.U. Genetic variability in Rubus ellipticus collections assessed by morphological traits and EST-SSR markers. J. Plant Biochem. Biotechnol. 2021, 30, 37–55. [Google Scholar] [CrossRef]
- Gao, X.-F.; Xiong, X.-H.; Boufford, D.E.; Gao, Y.-D.; Xu, B.; Zhang, C. Phylogeny of the Diploid Species of Rubus (Rosaceae). Genes 2023, 14, 1152. [Google Scholar] [CrossRef]
- Galeano, E.; Bousquet, J.; Thomas, B.R. SNP-based analysis reveals unexpected features of genetic diversity, parental contributions and pollen contamination in a white spruce breeding program. Sci. Rep. 2021, 11, 4990. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, Y.; Hu, Y.; Zhou, J.; Chen, L.; Lu, X. Systematic review of the characteristic markers in honey of various botanical, geographic, and entomological origins. ACS Food Sci. Technol. 2022, 2, 206–220. [Google Scholar] [CrossRef]
- Zhu, X.; Wu, Y.; Zhou, J.; Lin, J.; Chen, X. Morphological, palynological and transcriptomic-based SSR assessment of peony varieties adaptive in the Jiangnan region. Sci. Hortic. 2023, 310, 111771. [Google Scholar] [CrossRef]
- Luo, J.; Zheng, X.; Xin, Q.; Liu, M.; Zhang, J.; Chen, W.; Chen, C.; Zhang, C.; Wan, G.; Xia, C.; et al. Glycyrrhiza plastid paternal inheritance and a new DNA barcode provide new strategies for molecular identification of three medicinal licorice hybrid complexes. BMC Plant Biol. 2025, 25, 885. [Google Scholar] [CrossRef] [PubMed]
- Karunarathne, P.; Reutemann, A.V.; James, J.E.; Zhou, Q.; Sassone, A.; Rose, L.E.; Hojsgaard, D. Navigating the challenges in apomixis population genetics: Insights from past, present, and future perspectives. Crit. Rev. Plant Sci. 2025, 1–32. [Google Scholar] [CrossRef]
- Šarhanová, P.; Majeský, Ľ.; Sochor, M. A novel strategy to study apomixis, automixis, and autogamy in plants. Plant Reprod. 2024, 37, 379–392. [Google Scholar] [CrossRef]
- Nybom, H.; Lācis, G. Recent large-scale genotyping and phenotyping of plant genetic resources of vegetatively propagated crops. Plants 2021, 10, 415. [Google Scholar] [CrossRef] [PubMed]
- Amiteye, S. Basic concepts and methodologies of DNA marker systems in plant molecular breeding. Heliyon 2021, 7, e08093. [Google Scholar] [CrossRef] [PubMed]
- Korte, A.; Farlow, A. The advantages and limitations of trait analysis with GWAS: A review. Plant Methods 2013, 9, 29. [Google Scholar] [CrossRef]
- Brachi, B.; Faure, N.; Horton, M.; Flahauw, E.; Vazquez, A.; Nordborg, M.; Bergelson, J.; Cuguen, J.; Roux, F. Linkage and association mapping of Arabidopsis thaliana flowering time in nature. PLoS Genet. 2010, 6, e1000940.1. [Google Scholar] [CrossRef]
- Leoni, V.; Panseri, S.; Giupponi, L.; Pavlovic, R.; Gianoncelli, C.; Coatti, G.; Beretta, G.; Giorgi, A. Phytochemical profiling of red raspberry (Rubus idaeus L.) honey and investigation of compounds related to its pollen occurrence. J. Sci. Food Agric. 2024, 104, 5391–5406. [Google Scholar] [CrossRef]
- Kostryco, M.; Chwil, M. Structure of anther epidermis and endothecium, production of pollen, and content of selected nutrients in pollen grains from six Rubus idaeus L. cultivars. Agronomy 2021, 11, 1723. [Google Scholar] [CrossRef]
- Leoni, V.; Giupponi, L.; Pavlovic, R.; Gianoncelli, C.; Cecati, F.; Ranzato, E.; Martinotti, S.; Pedrali, D.; Giorgi, A.; Panseri, S. Multidisciplinary analysis of Italian Alpine wildflower honey reveals criticalities, diversity and value. Sci. Rep. 2021, 11, 19316. [Google Scholar] [CrossRef]
- Lechowicz, K.; Bocianowski, J.; Wrońska-Pilarek, D. Pollen Morphological Inter- and Intraspecific Variability in Selected Species of Rubus L. (Rosaceae). Forests 2022, 13, 1946. [Google Scholar] [CrossRef]
- Shapiro, S.S.; Wilk, M.B. An analysis of variance test for normality (complete samples). Biometrika 1965, 52, 591–611. [Google Scholar] [CrossRef]
- Rencher, A.C. Interpretation of canonical discriminant functions, canonical variates, and principal components. Am. Stat. 1992, 46, 217–225. [Google Scholar] [CrossRef]
- Seidler-Łożykowska, K.; Bocianowski, J. Evaluation of variability of morphological traits of selected caraway (Carum carvi L.) genotypes. Ind. Crops Prod. 2012, 35, 140–145. [Google Scholar] [CrossRef]
- Mahalanobis, P.C. On the generalized distance in statistics. Proc. Natl. Inst. Sci. India 1936, 12, 49–55. [Google Scholar]
- van Eeuwijk, F.A.; Bink, M.C.A.M.; Chenu, K.; Chapman, S.C. Detection and use of QTL for complex traits in multiple environments. Curr. Opin. Plant Biol. 2010, 13, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Malosetti, M.; Ribaut, J.-M.; van Eeuwijk, F.A. The statistical analysis of multi-environment data: Modeling genotype-by-environment interaction and its genetic basis. Front. Physiol. 2013, 4, 44. [Google Scholar] [CrossRef]
- VSN International. VSN International Genstat for Windows, 23rd ed.; VSN International: Hemel Hempstead, UK, 2023. [Google Scholar]
- Patterson, N.; Price, A.L.; Reich, D. Population structure and eigenanalysis. PLoS Genet. 2006, 2, e190. [Google Scholar] [CrossRef] [PubMed]
- Hoban, S.; Archer, F.I.; Bertola, L.D.; Bragg, J.G.; Breed, M.F.; Bruford, M.W.; Coleman, M.A.; Ekblom, R.; Funk, W.C.; Grueber, C.E.; et al. Global genetic diversity status and trends: Towards a suite of Essential Biodiversity Variables (EBVs) for genetic composition. Biol. Rev. 2022, 97, 1511–1538. [Google Scholar] [CrossRef]
- Schneider, H.M. Characterization, costs, cues and future perspectives of phenotypic plasticity. Ann. Bot. 2022, 130, 131–148. [Google Scholar] [CrossRef] [PubMed]
- Eren, B.; Keskin, B.; Demirel, F.; Demirel, S.; Türkoğlu, A.; Yilmaz, A.; Haliloğlu, K. Assessment of genetic diversity and population structure in local alfalfa genotypes using iPBS molecular markers. Genet. Resour. Crop Evol. 2023, 70, 617–628. [Google Scholar] [CrossRef]
- Demirel, S.; Demirel, F. Molecular identification and population structure of emmer and einkorn wheat lines with different ploidy levels using SSR markers. Genet. Resour. Crop Evol. 2024, 71, 363–372. [Google Scholar] [CrossRef]
- Palaz, E.B.; Demirel, F.; Adali, S.; Demirel, S.; Yilmaz, A. Genetic relationships of salep orchid species and gene flow among Serapias vomeracea × Anacamptis morio hybrids. Plant Biotechnol. Rep. 2023, 17, 315–327. [Google Scholar] [CrossRef]
- Demirel, S.; Pehluvan, M.; Aslantaş, R. Evaluation of genetic diversity and population structure of peach (Prunus persica L.) genotypes using inter-simple sequence repeat (ISSR) markers. Genet. Resour. Crop Evol. 2024, 71, 1301–1312. [Google Scholar] [CrossRef]
- Orman, E.; Çakar, D.; Alkan, M.; Özer, G.; Güler, E.; Gündoğdu, M. Genetic diversity and population structure of Turkish European chestnut (Castanea sativa) genotypes assessed using start codon targeted polymorphism (SCoT) markers. Genet. Resour. Crop Evol. 2025, 72, 6507–6519. [Google Scholar] [CrossRef]
- Paudel, D.; Parrish, S.B.; Peng, Z.; Parajuli, S.; Deng, Z. A chromosome-scale and haplotype-resolved genome assembly of tetraploid blackberry (Rubus L. subgenus Rubus Watson). Hortic. Res. 2025, 12, uhaf052. [Google Scholar] [CrossRef]
- De Mori, G.; Cipriani, G. Marker-assisted selection in breeding for fruit trait improvement: A review. Int. J. Mol. Sci. 2023, 24, 8984. [Google Scholar] [CrossRef]
- Sun, L.; Lai, M.; Ghouri, F.; Nawaz, M.A.; Ali, F.; Baloch, F.S.; Nadeem, M.A.; Aasim, M.; Shahid, M.Q. Modern Plant Breeding Techniques in Crop Improvement and Genetic Diversity: From Molecular Markers and Gene Editing to Artificial Intelligence—A Critical Review. Plants 2024, 13, 2676. [Google Scholar] [CrossRef]
- Song, L.; Wang, R.; Yang, X.; Zhang, A.; Liu, D. Molecular Markers and Their Applications in Marker-Assisted Selection (MAS) in Bread Wheat (Triticum aestivum L.). Agriculture 2023, 13, 642. [Google Scholar] [CrossRef]
- Moreno–Medina, B.L.; Casierra–Posada, F. Molecular characterization of a species in the genus Rubus in Boyacá, Colombia. Rev. Bras. Frutic. 2021, 43, e-713. [Google Scholar] [CrossRef]
- Huang, T.-R.; Chen, J.-H.; Hummer, K.E.; Alice, L.A.; Wang, W.-H.; He, Y.; Yu, S.-X.; Yang, M.-F.; Chai, T.-Y.; Zhu, X.-Y.; et al. Phylogeny of Rubus (Rosaceae): Integrating molecular and morphological evidence into an infrageneric revision. Taxon 2023, 72, 278–306. [Google Scholar] [CrossRef]
- Sochor, M.; Manning, J.C. Evolutionary patterns in South African brambles (Rubus L.)—new insights from molecular markers. Bothalia 2023, 53, a8. [Google Scholar] [CrossRef]
- Han, S.K.; Shin, H.; Lee, J.W.; Hong, K.N.; Ahn, J.Y. Genetic diversity and population relationships in wild Korean black raspberry (Rubus coreanus Miq.) based on microsatellite markers: Establishing a fruit tree breeding strategy. Hortic. Environ. Biotechnol. 2024, 65, 293–302. [Google Scholar] [CrossRef]
- Ward, J.A.; Bhangoo, J.; Fernández-Fernández, F.; Moore, P.; Swanson, J.D.; Viola, R.; Velasco, R.; Bassil, N.; Weber, C.A.; Sargent, D.J. Saturated linkage map construction in Rubus idaeus using genotyping by sequencing and genome-independent imputation. BMC Genom. 2013, 14, 2. [Google Scholar] [CrossRef]
- Pinczinger, D.; von Reth, M.; Hanke, M.V.; Flachowsky, H. Self-incompatibility of raspberry cultivars assessed by SSR markers. Sci. Hortic. 2021, 288, 110384. [Google Scholar] [CrossRef]
- Ryu, J.; Kim, W.J.; Im, J.; Kim, S.H.; Lee, K.S.; Jo, H.J.; Kim, E.Y.; Kang, S.Y.; Lee, J.H.; Ha, B.K. Genotyping-by-sequencing based single nucleotide polymorphisms enabled Kompetitive Allele Specific PCR marker development in mutant Rubus genotypes. Electron. J. Biotechnol. 2018, 35, 57–62. [Google Scholar] [CrossRef]
- Bushakra, J.M.; Bryant, D.W.; Dossett, M.; Vining, K.J.; VanBuren, R.; Gilmore, B.S.; Lee, J.; Mockler, T.C.; Finn, C.E.; Bassil, N.V. A genetic linkage map of black raspberry (Rubus occidentalis) and the mapping of Ag4 conferring resistance to the aphid Amphorophora agathonica. Theor. Appl. Genet. 2015, 128, 1631–1646. [Google Scholar] [CrossRef]
- Kollmann, J.; Steinger, T.; Roy, B.A. Evidence of sexuality in European Rubus (Rosaceae) species based on AFLP and allozyme analysis. Am. J. Bot. 2000, 87, 1592–1598. [Google Scholar] [CrossRef]
- Graham, J.; Smith, K.; MacKenzie, K.; Jorgenson, L.; Hackett, C.; Powell, W. The construction of a genetic linkage map of red raspberry (Rubus idaeus subsp. idaeus) based on AFLPs, genomic-SSR and EST-SSR markers. Theor. Appl. Genet. 2004, 109, 740–749. [Google Scholar] [CrossRef]
- Bushakra, J.M.; Stephens, M.J.; Atmadjaja, A.N.; Lewers, K.S.; Symonds, V.V.; Udall, J.A.; Chagné, D.; Buck, E.J.; Gardiner, S.E. Construction of black (Rubus occidentalis) and red (R. idaeus) raspberry linkage maps and their comparison to the genomes of strawberry, apple, and peach. Theor. Appl. Genet. 2012, 125, 311–327. [Google Scholar] [CrossRef]


| Species | Repl | P 1 | Le 2 | d 3 | E 4 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | s.d. | Mean | s.d. | Mean | s.d. | Mean | s.d. | ||||||
| Rubus bifrons | 5 | 25.43 | a | 0.771 | 21.5 | a | 0.8434 | 2.656 | de | 0.2454 | 21.53 | ab | 0.502 |
| Rubus caesius | 5 | 24.6 | abc | 1.789 | 20.65 | abcd | 1.638 | 3.719 | a | 0.8253 | 21.65 | a | 1.892 |
| Rubus constrictus | 2 | 23.68 | abcd | 0.304 | 19.91 | abcde | 0.2758 | 3.008 | bcd | 0.3882 | 20.75 | ab | 0.806 |
| Rubus divaricatus | 4 | 21.71 | e | 0.545 | 18.3 | ef | 0.6087 | 2.009 | e | 0.1177 | 18.54 | cd | 1.162 |
| Rubus gracilis | 5 | 24.44 | abcd | 1.22 | 20.78 | abc | 0.972 | 2.768 | cd | 0.2767 | 20.75 | ab | 1.491 |
| Rubus henrici-egonis | 5 | 22.69 | de | 1.14 | 19.16 | def | 1.0766 | 2.608 | de | 0.646 | 19.67 | bc | 1.613 |
| Rubus idaeus | 5 | 22.7 | de | 1.198 | 19.24 | cdef | 0.6913 | 3.487 | ab | 0.3611 | 20.46 | ab | 1.242 |
| Rubus nessensis | 5 | 24.39 | abcd | 0.951 | 20.47 | abcd | 0.6514 | 2.729 | d | 0.2812 | 20.07 | abc | 0.72 |
| Rubus opacus | 2 | 21.75 | e | 1.711 | 18.09 | f | 0.5162 | 2.604 | de | 0.6463 | 17.74 | d | 1.223 |
| Rubus plicatus | 5 | 22.94 | cde | 1.16 | 19.3 | cdef | 0.5658 | 3.119 | abcd | 0.4014 | 21.05 | ab | 1.203 |
| Rubus praecox | 4 | 23.74 | abcd | 1.364 | 20.28 | abcd | 1.2429 | 2.969 | bcd | 0.2998 | 20.88 | ab | 1.108 |
| Rubus radula | 5 | 25.07 | ab | 1.213 | 21.15 | ab | 1.2769 | 2.736 | d | 0.4919 | 21.5 | ab | 0.638 |
| Rubus saxatilis | 5 | 23.2 | cde | 1.025 | 19.75 | bcde | 1.1065 | 3.444 | abc | 0.1681 | 20.23 | abc | 0.655 |
| Rubus scissus | 3 | 23.46 | bcde | 0.828 | 19.87 | bcde | 0.7328 | 3.234 | abcd | 0.5522 | 21.25 | ab | 0.384 |
| Rubus sprengelii | 5 | 24.17 | abcd | 0.617 | 20.56 | abcd | 0.5339 | 2.92 | bcd | 0.2026 | 20.73 | ab | 0.472 |
| Rubus sulcatus | 5 | 23.53 | bcde | 0.93 | 20.01 | abcd | 0.8853 | 2.83 | bcd | 0.134 | 20.69 | ab | 1.346 |
| LSD0.05 | 1.869 | 1.61 | 0.6935 | 1.914 | |||||||||
| F-ANOVA | 3.91 *** | 3.9 *** | 4.49 *** | 2.9 ** | |||||||||
| Shapiro–Wilk test for normality | W-test | 0.987 | 0.982 | 0.986 | 0.969 | ||||||||
| p-value | 0.658 | 0.41 | 0.626 | 0.066 | |||||||||
| Bartlett test | χ2 | 9.51 | 12.02 | 16.99 | 17.7 | ||||||||
| p-value | 0.849 | 0.677 | 0.329 | 0.279 | |||||||||
| Characteristics | P 1 | Le | d | E | |
|---|---|---|---|---|---|
| The number of significant markers | 3346 | 3340 | 3342 | 2882 | |
| Effect (absolute value) | Min | 1.040 | 0.916 | 0.400 | 1.025 |
| Max | 2.131 | 1.989 | 0.980 | 2.905 | |
| The percentage of variation explained | Min | 19.4 | 19.4 | 19.4 | 19.4 |
| Max | 63.6 | 65.1 | 60.4 | 70.8 | |
| LOD | Min | 1.30 | 1.30 | 1.30 | 1.30 |
| Max | 3.88 | 4.01 | 3.62 | 4.57 | |
| Marker | P | Le | d | E | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Effect | Perc 1 | p-Value | LOD | Effect | Perc | p-Value | LOD | Effect | Perc | p-Value | LOD | Effect | Perc | p-Value | LOD | |
| m [4224]=0001000010000000 | −2.131 | 41.6 | 0.00416 | 2.38 | −1.989 | 47.7 | 0.00184 | 2.74 | −0.71 | 29.5 | 0.01742 | 1.76 | −2.658 | 70.8 | 0.00003 | 4.57 |
| m [4225] = 0001000010000001 | −1.556 | 29.1 | 0.01821 | 1.74 | −1.4 | 30.7 | 0.01518 | 1.82 | −0.55 | 23.4 | 0.03302 | 1.48 | −1.819 | 43.7 | 0.00317 | 2.50 |
| m [5248] = 0001010010000000 | −1.902 | 46.9 | 0.00205 | 2.69 | −1.747 | 51.8 | 0.00101 | 3.00 | −0.641 | 34.4 | 0.01001 | 2.00 | −2.237 | 69.8 | 0.00003 | 4.46 |
| m [5249] = 0001010010000001 | −1.566 | 38.0 | 0.00653 | 2.19 | −1.397 | 39.2 | 0.00559 | 2.25 | −0.553 | 31.0 | 0.01477 | 1.83 | −1.745 | 50.5 | 0.00123 | 2.91 |
| m [5250] = 0001010010000010 | −1.354 | 26.6 | 0.02381 | 1.62 | −1.214 | 27.9 | 0.02069 | 1.68 | −0.523 | 26.9 | 0.02292 | 1.64 | −1.731 | 49.5 | 0.00142 | 2.85 |
| m [5251] = 0001010010000011 | −1.2 | 23.2 | 0.03384 | 1.47 | −1.04 | 22.3 | 0.03712 | 1.43 | −0.485 | 26.4 | 0.02421 | 1.62 | −1.447 | 38.2 | 0.00634 | 2.20 |
| m [5280] = 0001010010100000 | −1.496 | 34.0 | 0.01041 | 1.98 | −1.306 | 33.4 | 0.01123 | 1.95 | −0.507 | 24.8 | 0.02854 | 1.54 | −1.679 | 46.2 | 0.00227 | 2.64 |
| m [5281] = 0001010010100001 | −1.324 | 29.8 | 0.01673 | 1.78 | −1.12 | 27.0 | 0.02267 | 1.64 | −0.471 | 24.5 | 0.02964 | 1.53 | −1.402 | 35.4 | 0.00884 | 2.05 |
| m [5504] = 0001010110000000 | −1.28 | 23.0 | 0.03452 | 1.46 | −1.242 | 29.5 | 0.01733 | 1.76 | −0.587 | 35.7 | 0.00854 | 2.07 | −1.952 | 64.9 | 0.00010 | 4.00 |
| m [5505] = 0001010110000001 | −1.136 | 20.0 | 0.04664 | 1.33 | −1.064 | 23.7 | 0.03214 | 1.49 | −0.541 | 34.6 | 0.00981 | 2.01 | −1.64 | 51.1 | 0.00111 | 2.95 |
| m [5509] = 0001010110000101 | −1.077 | 19.5 | 0.04920 | 1.31 | −0.993 | 22.2 | 0.03769 | 1.42 | −0.414 | 19.5 | 0.04925 | 1.31 | −1.295 | 32.5 | 0.01242 | 1.91 |
| m [5568] = 0001010111000000 | −1.307 | 28.9 | 0.01860 | 1.73 | −1.271 | 36.8 | 0.00750 | 2.12 | −0.456 | 22.6 | 0.03609 | 1.44 | −1.534 | 43.8 | 0.00311 | 2.51 |
| m [5569] = 0001010111000001 | −1.215 | 26.8 | 0.02322 | 1.63 | −1.147 | 31.9 | 0.01323 | 1.88 | −0.444 | 23.6 | 0.03246 | 1.49 | −1.348 | 35.8 | 0.00848 | 2.07 |
| m [5601] = 0001010111100001 | −1.12 | 23.1 | 0.03407 | 1.47 | −1.006 | 24.4 | 0.02986 | 1.52 | −0.413 | 20.7 | 0.04362 | 1.36 | −1.178 | 27.3 | 0.02205 | 1.66 |
| m [7296] = 0001110010000000 | −1.262 | 22.1 | 0.03775 | 1.42 | −1.139 | 23.7 | 0.03227 | 1.49 | −0.574 | 33.8 | 0.01067 | 1.97 | −1.725 | 49.1 | 0.00150 | 2.82 |
| m [7360] = 0001110011000000 | −1.291 | 28.0 | 0.02049 | 1.69 | −1.181 | 30.8 | 0.01500 | 1.82 | −0.445 | 21.1 | 0.04197 | 1.38 | −1.335 | 31.5 | 0.01391 | 1.86 |
| m [7361] = 0001110011000001 | −1.2 | 26.0 | 0.02536 | 1.60 | −1.064 | 26.5 | 0.02394 | 1.62 | −0.434 | 22.2 | 0.03766 | 1.42 | −1.166 | 25.0 | 0.02810 | 1.55 |
| m [13440] = 0011010010000000 | −1.518 | 35.2 | 0.00905 | 2.04 | −1.431 | 41.5 | 0.00421 | 2.38 | −0.493 | 23.2 | 0.03391 | 1.47 | −1.724 | 49.1 | 0.00151 | 2.82 |
| m [13441] = 0011010010000001 | −1.343 | 30.9 | 0.01488 | 1.83 | −1.229 | 34.0 | 0.01046 | 1.98 | −0.459 | 22.9 | 0.03477 | 1.46 | −1.441 | 37.9 | 0.00662 | 2.18 |
| m [13442] = 0011010010000010 | −1.158 | 21.1 | 0.04193 | 1.38 | −1.069 | 24.0 | 0.03120 | 1.51 | −0.433 | 19.6 | 0.04884 | 1.31 | −1.428 | 37.1 | 0.00729 | 2.14 |
| m [13443] = 0011010010000011 | −1.078 | 19.6 | 0.04887 | 1.31 | −0.962 | 20.4 | 0.04520 | 1.34 | −0.423 | 20.7 | 0.04372 | 1.36 | −1.251 | 29.9 | 0.01666 | 1.78 |
| m [13761] = 0011010111000001 | −1.136 | 24.0 | 0.03105 | 1.51 | −1.101 | 30.7 | 0.01525 | 1.82 | −0.403 | 19.4 | 0.04998 | 1.30 | −1.212 | 29.3 | 0.01769 | 1.75 |
| m [51774] = 1100101000111110 | 1.136 | 24.0 | 0.03105 | 1.51 | 1.101 | 30.7 | 0.01525 | 1.82 | 0.403 | 19.4 | 0.04998 | 1.30 | 1.212 | 29.3 | 0.01769 | 1.75 |
| m [52092] = 1100101101111100 | 1.078 | 19.6 | 0.04887 | 1.31 | 0.962 | 20.4 | 0.04520 | 1.34 | 0.423 | 20.7 | 0.04372 | 1.36 | 1.251 | 29.9 | 0.01666 | 1.78 |
| m [52093] = 1100101101111101 | 1.158 | 21.1 | 0.04193 | 1.38 | 1.069 | 24.0 | 0.03120 | 1.51 | 0.433 | 19.6 | 0.04884 | 1.31 | 1.428 | 37.1 | 0.00729 | 2.14 |
| m [52094] = 1100101101111110 | 1.343 | 30.9 | 0.01488 | 1.83 | 1.229 | 34.0 | 0.01046 | 1.98 | 0.459 | 22.9 | 0.03477 | 1.46 | 1.441 | 37.9 | 0.00662 | 2.18 |
| m [52095] = 1100101101111111 | 1.518 | 35.2 | 0.00905 | 2.04 | 1.431 | 41.5 | 0.00421 | 2.38 | 0.493 | 23.2 | 0.03391 | 1.47 | 1.724 | 49.1 | 0.00151 | 2.82 |
| m [58174] = 1110001100111110 | 1.2 | 26.0 | 0.02536 | 1.60 | 1.064 | 26.5 | 0.02394 | 1.62 | 0.434 | 22.2 | 0.03766 | 1.42 | 1.166 | 25.0 | 0.02810 | 1.55 |
| m [58175] = 1110001100111111 | 1.291 | 28.0 | 0.02049 | 1.69 | 1.181 | 30.8 | 0.01500 | 1.82 | 0.445 | 21.1 | 0.04197 | 1.38 | 1.335 | 31.5 | 0.01391 | 1.86 |
| m [58239] = 1110001101111111 | 1.262 | 22.1 | 0.03775 | 1.42 | 1.139 | 23.7 | 0.03227 | 1.49 | 0.574 | 33.8 | 0.01067 | 1.97 | 1.725 | 49.1 | 0.00150 | 2.82 |
| m [59934] = 1110101000011110 | 1.12 | 23.1 | 0.03407 | 1.47 | 1.006 | 24.4 | 0.02986 | 1.52 | 0.413 | 20.7 | 0.04362 | 1.36 | 1.178 | 27.3 | 0.02205 | 1.66 |
| m [59966] = 1110101000111110 | 1.215 | 26.8 | 0.02322 | 1.63 | 1.147 | 31.9 | 0.01323 | 1.88 | 0.444 | 23.6 | 0.03246 | 1.49 | 1.348 | 35.8 | 0.00848 | 2.07 |
| m [59967] = 1110101000111111 | 1.307 | 28.9 | 0.01860 | 1.73 | 1.271 | 36.8 | 0.00750 | 2.12 | 0.456 | 22.6 | 0.03609 | 1.44 | 1.534 | 43.8 | 0.00311 | 2.51 |
| m [60026] = 1110101001111010 | 1.077 | 19.5 | 0.04920 | 1.31 | 0.993 | 22.2 | 0.03769 | 1.42 | 0.414 | 19.5 | 0.04925 | 1.31 | 1.295 | 32.5 | 0.01242 | 1.91 |
| m [60030] = 1110101001111110 | 1.136 | 20.0 | 0.04664 | 1.33 | 1.064 | 23.7 | 0.03214 | 1.49 | 0.541 | 34.6 | 0.00981 | 2.01 | 1.64 | 51.1 | 0.00111 | 2.95 |
| m [60031] = 1110101001111111 | 1.28 | 23.0 | 0.03452 | 1.46 | 1.242 | 29.5 | 0.01733 | 1.76 | 0.587 | 35.7 | 0.00854 | 2.07 | 1.952 | 64.9 | 0.00010 | 4.00 |
| m [60254] = 1110101101011110 | 1.324 | 29.8 | 0.01673 | 1.78 | 1.12 | 27.0 | 0.02267 | 1.64 | 0.471 | 24.5 | 0.02964 | 1.53 | 1.402 | 35.4 | 0.00884 | 2.05 |
| m [60255] = 1110101101011111 | 1.496 | 34.0 | 0.01041 | 1.98 | 1.306 | 33.4 | 0.01123 | 1.95 | 0.507 | 24.8 | 0.02854 | 1.54 | 1.679 | 46.2 | 0.00227 | 2.64 |
| m [60284] = 1110101101111100 | 1.2 | 23.2 | 0.03384 | 1.47 | 1.04 | 22.3 | 0.03712 | 1.43 | 0.485 | 26.4 | 0.02421 | 1.62 | 1.447 | 38.2 | 0.00634 | 2.20 |
| m [60285] = 1110101101111101 | 1.354 | 26.6 | 0.02381 | 1.62 | 1.214 | 27.9 | 0.02069 | 1.68 | 0.523 | 26.9 | 0.02292 | 1.64 | 1.731 | 49.5 | 0.00142 | 2.85 |
| m [60286] = 1110101101111110 | 1.566 | 38.0 | 0.00653 | 2.19 | 1.397 | 39.2 | 0.00559 | 2.25 | 0.553 | 31.0 | 0.01477 | 1.83 | 1.745 | 50.5 | 0.00123 | 2.91 |
| m [60287] = 1110101101111111 | 1.902 | 46.9 | 0.00205 | 2.69 | 1.747 | 51.8 | 0.00101 | 3.00 | 0.641 | 34.4 | 0.01001 | 2.00 | 2.237 | 69.8 | 0.00003 | 4.46 |
| m [61310] = 1110111101111110 | 1.556 | 29.1 | 0.01821 | 1.74 | 1.4 | 30.7 | 0.01518 | 1.82 | 0.55 | 23.4 | 0.03302 | 1.48 | 1.819 | 43.7 | 0.00317 | 2.50 |
| m [61311] = 1110111101111111 | 2.131 | 41.6 | 0.00416 | 2.38 | 1.989 | 47.7 | 0.00184 | 2.74 | 0.71 | 29.5 | 0.01742 | 1.76 | 2.658 | 70.8 | 0.00003 | 4.57 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bocianowski, J.; Leśniewska-Bocianowska, A. Towards the Identification of Candidate Genes for Pollen Morphological Traits in Rubus L. Using Association Mapping. Forests 2025, 16, 1395. https://doi.org/10.3390/f16091395
Bocianowski J, Leśniewska-Bocianowska A. Towards the Identification of Candidate Genes for Pollen Morphological Traits in Rubus L. Using Association Mapping. Forests. 2025; 16(9):1395. https://doi.org/10.3390/f16091395
Chicago/Turabian StyleBocianowski, Jan, and Agnieszka Leśniewska-Bocianowska. 2025. "Towards the Identification of Candidate Genes for Pollen Morphological Traits in Rubus L. Using Association Mapping" Forests 16, no. 9: 1395. https://doi.org/10.3390/f16091395
APA StyleBocianowski, J., & Leśniewska-Bocianowska, A. (2025). Towards the Identification of Candidate Genes for Pollen Morphological Traits in Rubus L. Using Association Mapping. Forests, 16(9), 1395. https://doi.org/10.3390/f16091395







