Genetic Variation Associated with Leaf Phenology in Pedunculate Oak (Quercus robur L.) Implicates Pathogens, Herbivores, and Heat Stress as Selective Drivers
Abstract
1. Introduction
1.1. Background
1.2. Aims and Hypotheses
2. Materials and Methods
2.1. Study Species, Sampling Locations, and Data Collection
2.1.1. Leaf Phenology Scoring Method
2.1.2. Temperature Data
2.2. Statistical Testing for Associations of Temperature and Latitude with Phenology
2.3. Acquisition of RADseq Data
2.3.1. DNA Extraction and Sequencing
2.3.2. Bioinformatics
2.4. Analysis of Genetic Variation Associated with Phenology
2.4.1. Investigating Population Structure
2.4.2. Linkage Decay
2.4.3. Genotype–Phenotype Association
2.5. Regional Genetic Variation in Loci Putatively Associated with Leaf Phenology and the Neutral Dataset
3. Results
3.1. Patterns and Associations of Budburst and Senescence
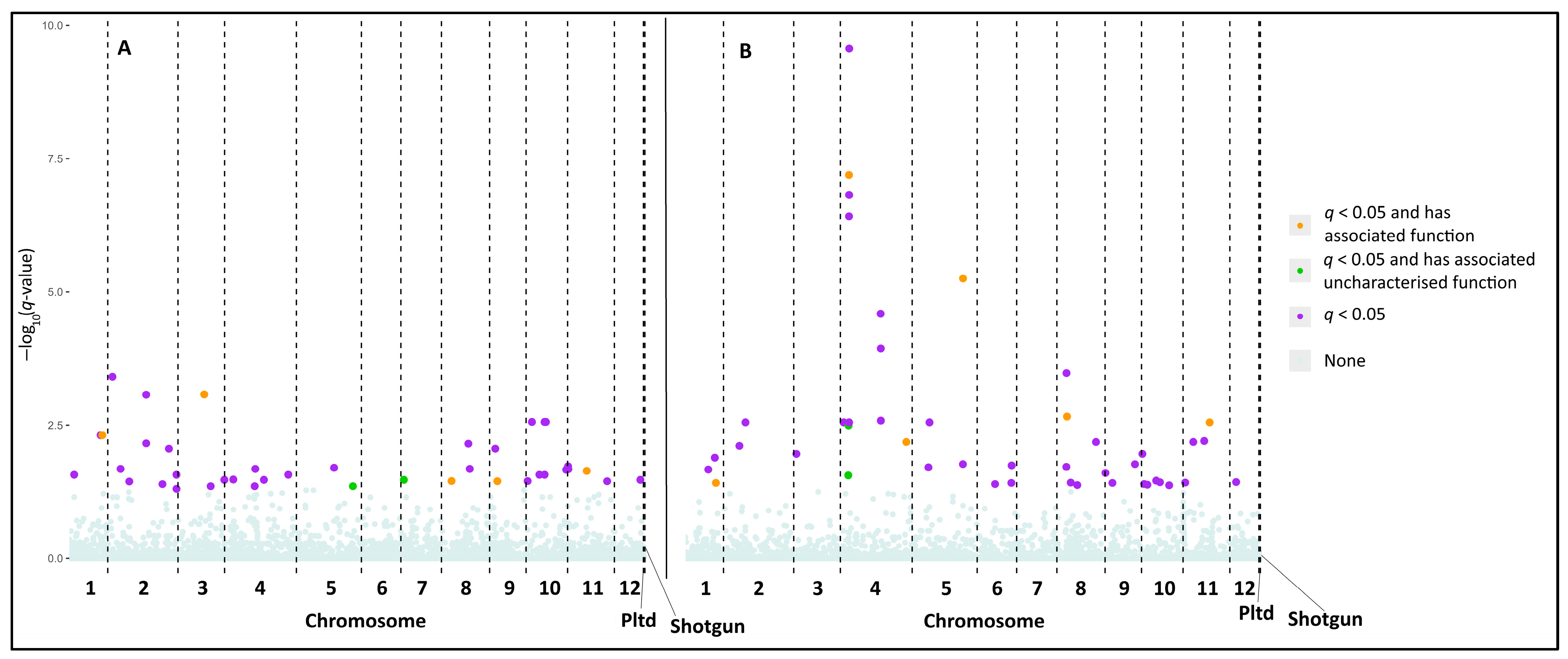
3.2. Loci Putatively Associated with Leaf Phenology
3.2.1. Population Structure
3.2.2. Genotype–Phenotype Associations
3.3. Variation in the Full SNP Dataset and in Loci Putatively Associated with Phenology
3.3.1. Genetic Diversity Indices
3.3.2. Population Structuring in the Putatively Adaptive Candidate Locus Datasets
4. Discussion
4.1. Associations Between Timings and Cues
4.2. Genotype–Phenotype Associations and Signatures of Selection
4.2.1. Functions Identified Among the Candidate Loci
4.2.2. On the Relative Roles of Selection and Neutral Processes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| RADseq | Restriction-site associated sequencing |
| PCoA | Principal coordinates analysis |
| dbRDA | Distance-based redundancy analysis |
| PAs | Private alleles |
| P | Proportion of polymorphic loci |
| RMSE | Root mean square error |
| MAE | Mean absolute error |
| MAPE | Mean absolute percent error |
Appendix A
Appendix A.1. Model Estimation on Timing of Budburst and the Onset of Leaf Senescence

Appendix A.2. Accuracy Assessment of SMHI Data
| Variable | Period | No. Stands | No. Obs. | RMSE | MAE | MAPE (%) | ME | β0 | β1 | R2 |
|---|---|---|---|---|---|---|---|---|---|---|
| April mean | April | 2023 | 21 | 630 | 1.47 | 1.18 | 0.33 | 1.00 | 0.64 | 1.06 |
| August mean | August | 2023 | 21 | 651 | 0.51 | 0.41 | 0.03 | 0.01 | −0.43 | 1.03 |
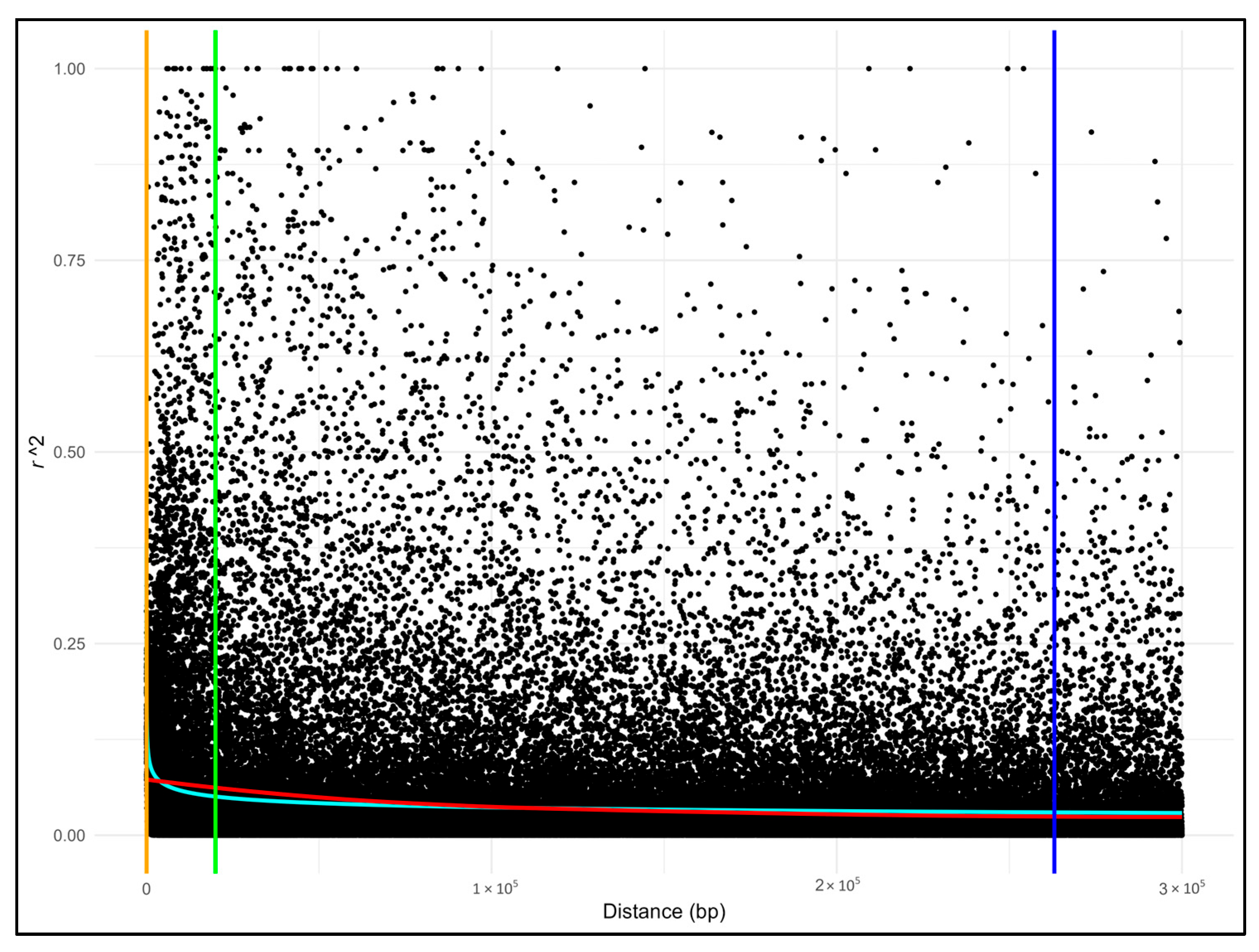
Appendix A.3. Linkage Decay
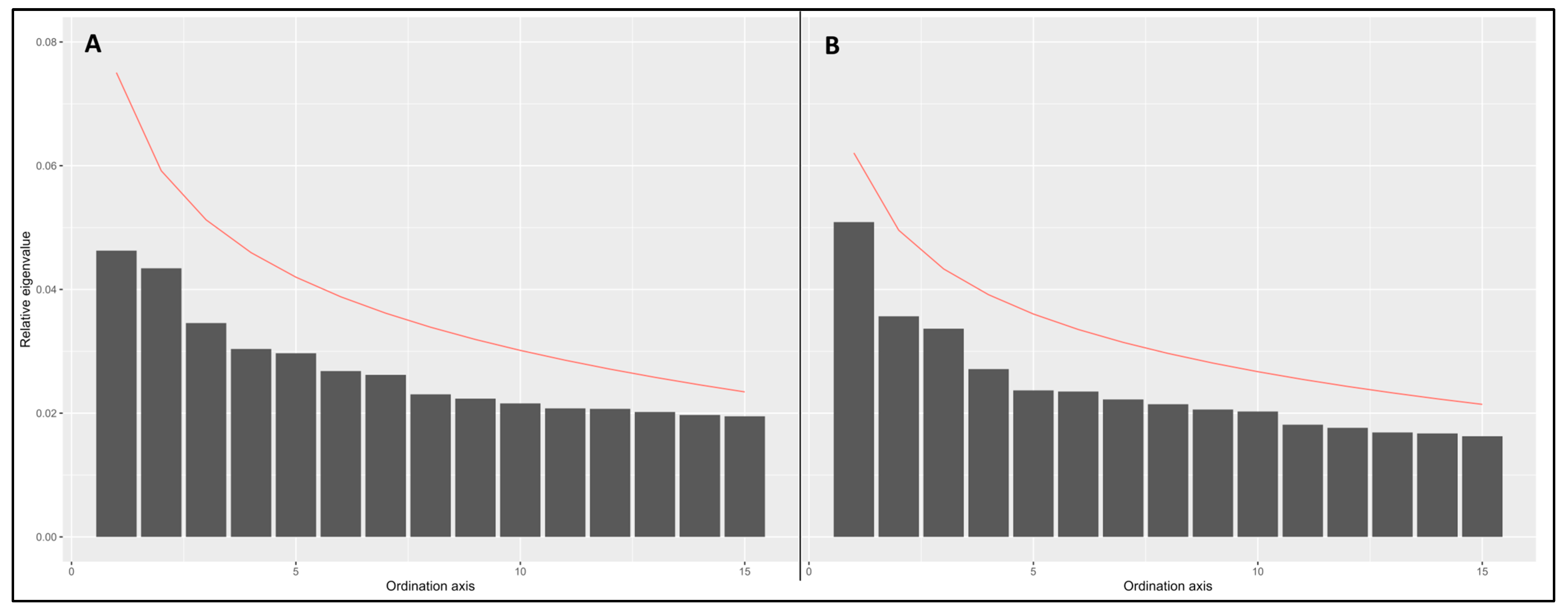
Appendix A.4. Scree Plots Used with the Broken Stick Criterion to Identify the Number of Population Clusters
Appendix A.5. Full List of Candidate Loci
| SNP ID | Chr. | Position | q-Value | Gene Annotation | Distance from Region (bp) |
|---|---|---|---|---|---|
| 5738_251 | 1 | 4940506 | 2.70 × 10−2 | ||
| 52577_276 | 1 | 44019104 | 4.89 × 10−3 | ||
| 55616_189 | 1 | 47495836 | 4.89 × 10−3 | Transportin-1 | 9254 |
| 72253_232 | 2 | 6326725 | 3.92 × 10−4 | ||
| 83272_136 | 2 | 15479089 | 2.10 × 10−2 | ||
| 99516_291 | 2 | 28807226 | 3.61 × 10−2 | ||
| 125851_39 | 2 | 50970570 | 8.53 × 10−4 | ||
| 125860_104 | 2 | 50970824 | 6.95 × 10−3 | ||
| 151636_115 | 2 | 72569754 | 4.05 × 10−2 | ||
| 164056_79 | 2 | 83613636 | 8.77 × 10−3 | ||
| 178467_290 | 2 | 95355512 | 2.70 × 10−2 | ||
| 178464_131 | 2 | 95355601 | 4.96 × 10−2 | ||
| 224662_285 | 3 | 37331722 | 8.43 × 10−4 | Small nucleolar RNA R71 | 4392 |
| 238225_54 | 3 | 49171843 | 4.43 × 10−2 | ||
| 262970_68 | 3 | 68389061 | 3.37 × 10−2 | ||
| 277265_315 | 4 | 12699248 | 3.32 × 10−2 | ||
| 305538_143 | 4 | 36255883 | 4.43 × 10−2 | ||
| 305941_99 | 4 | 36302491 | 2.10 × 10−2 | ||
| 316704_278 | 4 | 44095902 | 3.37 × 10−2 | ||
| 353121_310 | 4 | 73388711 | 2.70 × 10−2 | ||
| 428204_359 | 5 | 53793709 | 1.99 × 10−2 | ||
| 457620_63 | 5 | 77977948 | 4.43 × 10−2 | Uncharacterised LOC126727237 | 1582 |
| 540005_335 | 7 | 3787957 | 3.37 × 10−2 | Uncharacterised protein LOC126691808 | 13,272 |
| 614926_284 | 8 | 15323561 | 3.54 × 10−2 | (−)-Germacrene D synthase-like | 164 |
| 642304_134 | 8 | 38635600 | 7.06 × 10−3 | ||
| 645982_112 | 8 | 41689845 | 2.10 × 10−2 | ||
| 690066_287 | 9 | 11014634 | 8.77 × 10−3 | ||
| 693924_287 | 9 | 13741392 | 3.57 × 10−2 | rRNA 2′-O-methyltransferase fibrillarin 1-like | 6951 |
| 743550_323 | 10 | 1928227 | 3.55 × 10−2 | ||
| 751799_234 | 10 | 8893909 | 2.75 × 10−3 | ||
| 765481_239 | 10 | 20469263 | 2.70 × 10−2 | ||
| 773508_330 | 10 | 27709348 | 2.75 × 10−3 | ||
| 773518_376 | 10 | 27718161 | 2.70 × 10−2 | ||
| 775537_303 | 10 | 29436014 | 2.75 × 10−3 | ||
| 806456_18 | 10 | 53674173 | 2.18 × 10−2 | ||
| 809500_107 | 11 | 154845 | 1.87 × 10−2 | ||
| 809532_101 | 11 | 158944 | 2.12 × 10−2 | ||
| 837643_219 | 11 | 24896775 | 2.29 × 10−2 | MADS-box protein JOINTLESS-like | 19,473 |
| 861794_142 | 11 | 45831267 | 3.57 × 10−2 | ||
| 920002_285 | 12 | 39036922 | 3.37 × 10−2 | ||
| 39587_320 | 1 | 32902497 | 2.16 × 10−2 | ||
| 50170_23 | 1 | 41844157 | 1.29 × 10−2 | ||
| 52095_31 | 1 | 43584107 | 3.84 × 10−2 | Mediator of RNA polymerase II transcription subunit 25 | 1163 |
| 88039_54 | 2 | 19634956 | 7.76 × 10−3 | ||
| 99805_306 | 2 | 29029208 | 2.81 × 10−3 | ||
| 185314_48 | 3 | 3798649 | 1.10 × 10−2 | ||
| 269438_43 | 4 | 5348239 | 2.81 × 10−3 | ||
| 276966_142 | 4 | 12516864 | 2.77 × 10−2 | Uncharacterized LOC126721394 | 184 |
| 277111_32 | 4 | 12599017 | 3.21 × 10−3 | Uncharacterized LOC126720597 | 7172 |
| 277235_194 | 4 | 12685808 | 6.41 × 10−8 | G-type lectin S-receptor-like serine/threonine-protein kinase At1G67520 | 17,139 |
| 277259_148 | 4 | 12699073 | 3.83 × 10−7 | ||
| 277265_315 | 4 | 12699248 | 1.51 × 10−7 | ||
| 277258_119 | 4 | 12699337 | 2.81 × 10−3 | ||
| 277268_134 | 4 | 12700024 | 1.52 × 10−7 | ||
| 277302_93 | 4 | 12720241 | 2.74 × 10−10 | ||
| 318247_277 | 4 | 45176449 | 2.57 × 10−5 | ||
| 318272_16 | 4 | 45180545 | 1.15 × 10−4 | ||
| 318285_308 | 4 | 45182834 | 2.60 × 10−3 | ||
| 355472_275 | 4 | 75298409 | 6.57 × 10−3 | Putative disease resistance RPP13-like protein 1 | 0 |
| 396880_16 | 5 | 25781248 | 1.96 × 10−2 | ||
| 398425_213 | 5 | 27087039 | 2.81 × 10−3 | ||
| 448915_31 | 5 | 70661084 | 1.71 × 10−2 | ||
| 449028_6 | 5 | 70766439 | 5.62 × 10−6 | Putative calcium-transporting ATPase 13, plasma membrane-type | 4865 |
| 500170_149 | 6 | 24250569 | 4.04 × 10−2 | ||
| 525417_341 | 6 | 46452486 | 3.84 × 10−2 | ||
| 526007_195 | 6 | 46911021 | 1.81 × 10−2 | ||
| 526011_106 | 6 | 46911056 | 1.81 × 10−2 | ||
| 614447_285 | 8 | 15038558 | 1.92 × 10−2 | ||
| 614462_248 | 8 | 15039408 | 3.34 × 10−4 | ||
| 614940_70 | 8 | 15336382 | 2.18 × 10−3 | (−)-Germacrene D synthase-like | 12,985 |
| 621412_264 | 8 | 20775860 | 3.79 × 10−2 | ||
| 631293_23 | 8 | 29570266 | 4.22 × 10−2 | ||
| 663736_102 | 8 | 56030387 | 6.57 × 10−3 | ||
| 678922_284 | 9 | 148031 | 2.50 × 10−2 | ||
| 693321_211 | 9 | 13368039 | 3.84 × 10−2 | ||
| 732227_197 | 9 | 47910412 | 1.71 × 10−2 | ||
| 742782_127 | 10 | 1176690 | 1.10 × 10−2 | ||
| 745733_6 | 10 | 3566118 | 4.03 × 10−2 | ||
| 751799_234 | 10 | 8893909 | 4.16 × 10−2 | ||
| 766837_288 | 10 | 21665273 | 3.48 × 10−2 | ||
| 773466_213 | 10 | 27673363 | 3.73 × 10−2 | ||
| 784637_285 | 10 | 36590051 | 4.26 × 10−2 | ||
| 810514_104 | 11 | 978573 | 3.79 × 10−2 | ||
| 822414_51 | 11 | 11597339 | 6.57 × 10−3 | ||
| 840143_190 | 11 | 26874077 | 6.29 × 10−3 | ||
| 846236_271 | 11 | 31918101 | 2.81 × 10−3 | Vacuolar-sorting receptor 3-like | 15,453 |
| 884916_149 | 12 | 8517225 | 3.70 × 10−2 |
References
- Körner, C.; Möhl, P.; Hiltbrunner, E. Four Ways to Define the Growing Season. Ecol. Lett. 2023, 26, 1277–1292. [Google Scholar] [CrossRef]
- Chuine, I. Why Does Phenology Drive Species Distribution? Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 3149–3160. [Google Scholar] [CrossRef]
- Kollas, C.; Körner, C.; Randin, C.F. Spring Frost and Growing Season Length Co-Control the Cold Range Limits of Broad-Leaved Trees. J. Biogeogr. 2014, 41, 773–783. [Google Scholar] [CrossRef]
- Körner, C.; Basler, D.; Hoch, G.; Kollas, C.; Lenz, A.; Randin, C.F.; Vitasse, Y.; Zimmermann, N.E. Where, Why and How? Explaining the Low-Temperature Range Limits of Temperate Tree Species. J. Ecol. 2016, 104, 1076–1088. [Google Scholar] [CrossRef]
- Jump, A.S.; Peñuelas, J. Running to Stand Still: Adaptation and the Response of Plants to Rapid Climate Change. Ecol. Lett. 2005, 8, 1010–1020. [Google Scholar] [CrossRef]
- Jeong, S.-J.; Ho, C.-H.; Gim, H.-J.; Brown, M.E. Phenology Shifts at Start vs. End of Growing Season in Temperate Vegetation over the Northern Hemisphere for the Period 1982–2008. Glob. Change Biol. 2011, 17, 2385–2399. [Google Scholar] [CrossRef]
- Piao, S.; Liu, Q.; Chen, A.; Janssens, I.A.; Fu, Y.; Dai, J.; Liu, L.; Lian, X.; Shen, M.; Zhu, X. Plant Phenology and Global Climate Change: Current Progresses and Challenges. Glob. Change Biol. 2019, 25, 1922–1940. [Google Scholar] [CrossRef]
- Davis, M.B.; Shaw, R.G. Range Shifts and Adaptive Responses to Quaternary Climate Change. Science 2001, 292, 673–679. [Google Scholar] [CrossRef]
- Trew, B.T.; Maclean, I.M. Vulnerability of Global Biodiversity Hotspots to Climate Change. Glob. Ecol. Biogeogr. 2021, 30, 768–783. [Google Scholar] [CrossRef]
- Aitken, S.N.; Yeaman, S.; Holliday, J.A.; Wang, T.; Curtis-McLane, S. Adaptation, Migration or Extirpation: Climate Change Outcomes for Tree Populations. Evol. Appl. 2008, 1, 95–111. [Google Scholar] [CrossRef]
- Basler, D.; Körner, C. Photoperiod Sensitivity of Bud Burst in 14 Temperate Forest Tree Species. Agric. For. Meteorol. 2012, 165, 73–81. [Google Scholar] [CrossRef]
- Basler, D.; Körner, C. Photoperiod and Temperature Responses of Bud Swelling and Bud Burst in Four Temperate Forest Tree Species. Tree Physiol. 2014, 34, 377–388. [Google Scholar] [CrossRef]
- Kuster, T.M.; Dobbertin, M.; Günthardt-Goerg, M.S.; Schaub, M.; Arend, M. A Phenological Timetable of Oak Growth under Experimental Drought and Air Warming. PLoS ONE 2014, 9, e89724. [Google Scholar] [CrossRef]
- Morin, X.; Roy, J.; Sonié, L.; Chuine, I. Changes in Leaf Phenology of Three European Oak Species in Response to Experimental Climate Change. New Phytol. 2010, 186, 900–910. [Google Scholar] [CrossRef]
- Laube, J.; Sparks, T.H.; Estrella, N.; Menzel, A. Does Humidity Trigger Tree Phenology? Proposal for an Air Humidity Based Framework for Bud Development in Spring. New Phytol. 2014, 202, 350–355. [Google Scholar] [CrossRef]
- Murray, M.; Smith, R.; Leith, I.; Fowler, D.; Lee, H.; Friend, A.; Jarvis, P. Effects of Elevated CO2, Nutrition and Climatic Warming on Bud Phenology in Sitka Spruce (Picea Sitchensis) and Their Impact on the Risk of Frost Damage. Tree Physiol. 1994, 14, 691–706. [Google Scholar] [CrossRef]
- Norby, R.J.; Hartz-Rubin, J.S.; Verbrugge, M.J. Phenological Responses in Maple to Experimental Atmospheric Warming and CO₂ Enrichment. Glob. Change Biol. 2003, 9, 1792–1801. [Google Scholar] [CrossRef]
- Vitasse, Y.; Hoch, G.; Randin, C.F.; Lenz, A.; Kollas, C.; Scheepens, J.; Körner, C. Elevational Adaptation and Plasticity in Seedling Phenology of Temperate Deciduous Tree Species. Oecologia 2013, 171, 663–678. [Google Scholar] [CrossRef]
- Vitasse, Y.; Porté, A.J.; Kremer, A.; Michalet, R.; Delzon, S. Responses of Canopy Duration to Temperature Changes in Four Temperate Tree Species: Relative Contributions of Spring and Autumn Leaf Phenology. Oecologia 2009, 161, 187–198. [Google Scholar] [CrossRef]
- Estiarte, M.; Peñuelas, J. Alteration of the Phenology of Leaf Senescence and Fall in Winter Deciduous Species by Climate Change: Effects on Nutrient Proficiency. Glob. Change Biol. 2015, 21, 1005–1017. [Google Scholar] [CrossRef]
- Delpierre, N.; Vitasse, Y.; Chuine, I.; Guillemot, J.; Bazot, S.; Rutishauser, T.; Rathgeber, C.B. Temperate and Boreal Forest Tree Phenology: From Organ-Scale Processes to Terrestrial Ecosystem Models. Ann. For. Sci. 2016, 73, 5–25. [Google Scholar] [CrossRef]
- Estrella, N.; Menzel, A. Responses of Leaf Colouring in Four Deciduous Tree Species to Climate and Weather in Germany. Clim. Res. 2006, 32, 253–267. [Google Scholar] [CrossRef]
- Mutz, J.; McClory, R.; van Dijk, L.J.; Ehrlén, J.; Tack, A.J. Pathogen Infection Influences the Relationship between Spring and Autumn Phenology at the Seedling and Leaf Level. Oecologia 2021, 197, 447–457. [Google Scholar] [CrossRef]
- Delpierre, N.; Guillemot, J.; Dufrêne, E.; Cecchini, S.; Nicolas, M. Tree Phenological Ranks Repeat from Year to Year and Correlate with Growth in Temperate Deciduous Forests. Agric. For. Meteorol. 2017, 234, 1–10. [Google Scholar] [CrossRef]
- Crawley, M.; Akhteruzzaman, M. Individual Variation in the Phenology of Oak Trees and Its Consequences for Herbivorous Insects. Funct. Ecol. 1988, 2, 409–415. [Google Scholar] [CrossRef]
- Zohner, C.M.; Renner, S.S. Ongoing Seasonally Uneven Climate Warming Leads to Earlier Autumn Growth Cessation in Deciduous Trees. Oecologia 2019, 189, 549–561. [Google Scholar] [CrossRef]
- Zohner, C.M.; Mirzagholi, L.; Renner, S.S.; Mo, L.; Rebindaine, D.; Bucher, R.; Palouš, D.; Vitasse, Y.; Fu, Y.H.; Stocker, B.D.; et al. Effect of Climate Warming on the Timing of Autumn Leaf Senescence Reverses after the Summer Solstice. Science 2023, 381, eadf5098. [Google Scholar] [CrossRef]
- Bennie, J.; Kubin, E.; Wiltshire, A.; Huntley, B.; Baxter, R. Predicting Spatial and Temporal Patterns of Bud-Burst and Spring Frost Risk in North-West Europe: The Implications of Local Adaptation to Climate. Glob. Change Biol. 2010, 16, 1503–1514. [Google Scholar] [CrossRef]
- Baumgarten, F.; Gessler, A.; Vitasse, Y. No Risk—No Fun: Penalty and Recovery from Spring Frost Damage in Deciduous Temperate Trees. Funct. Ecol. 2023, 37, 648–663. [Google Scholar] [CrossRef]
- Zohner, C.M.; Mo, L.; Renner, S.S.; Svenning, J.-C.; Vitasse, Y.; Benito, B.M.; Ordonez, A.; Baumgarten, F.; Bastin, J.-F.; Sebald, V.; et al. Late-Spring Frost Risk between 1959 and 2017 Decreased in North America but Increased in Europe and Asia. Proc. Natl. Acad. Sci. USA 2020, 117, 12192–12200. [Google Scholar] [CrossRef]
- Augspurger, C.K. Reconstructing Patterns of Temperature, Phenology, and Frost Damage over 124 Years: Spring Damage Risk Is Increasing. Ecology 2013, 94, 41–50. [Google Scholar] [CrossRef]
- Bert, D.; Lasnier, J.-B.; Capdevielle, X.; Dugravot, A.; Desprez-Loustau, M.-L. Powdery Mildew Decreases the Radial Growth of Oak Trees with Cumulative and Delayed Effects over Years. PLoS ONE 2016, 11, e0155344. [Google Scholar] [CrossRef]
- Pearse, I.S.; Funk, K.A.; Kraft, T.S.; Koenig, W.D. Lagged Effects of Early-Season Herbivores on Valley Oak Fecundity. Oecologia 2015, 178, 361–368. [Google Scholar] [CrossRef]
- Renner, S.S.; Zohner, C.M. Climate Change and Phenological Mismatch in Trophic Interactions among Plants, Insects, and Vertebrates. Annu. Rev. Ecol. Evol. Syst. 2018, 49, 165–182. [Google Scholar] [CrossRef]
- Gallinat, A.S.; Primack, R.B.; Wagner, D.L. Autumn, the Neglected Season in Climate Change Research. Trends Ecol. Evol. 2015, 30, 169–176. [Google Scholar] [CrossRef]
- Alberto, F.; Bouffier, L.; Louvet, J.-M.; Lamy, J.-B.; Delzon, S.; Kremer, A. Adaptive Responses for Seed and Leaf Phenology in Natural Populations of Sessile Oak along an Altitudinal Gradient. J. Evol. Biol. 2011, 24, 1442–1454. [Google Scholar] [CrossRef]
- Baliuckas, V.; Pliura, A. Genetic Variation and Phenotypic Plasticity of Quercus Robur Populations and Open-Pollinated Families in Lithuania. Scand. J. For. Res. 2003, 18, 305–319. [Google Scholar] [CrossRef]
- Mousseau, T.A. Intra- and Interpopulation Genetic Variation. In Adaptive Genetic Variation in the Wild; Mousseau, T.A., Sinervo, B., Endler, J.A., Eds.; Oxford University Press: New York, NY, USA, 2000; pp. 219–250. [Google Scholar]
- Sork, V.L.; Stowe, K.A.; Hochwender, C. Evidence for Local Adaptation in Closely Adjacent Subpopulations of Northern Red Oak (Quercus rubra L.) Expressed as Resistance to Leaf Herbivores. Am. Nat. 1993, 142, 928–936. [Google Scholar] [CrossRef]
- Bartholomé, J.; Brachi, B.; Marçais, B.; Mougou-Hamdane, A.; Bodénès, C.; Plomion, C.; Robin, C.; Desprez-Loustau, M.-L. The Genetics of Exapted Resistance to Two Exotic Pathogens in Pedunculate Oak. New Phytol. 2020, 226, 1088–1103. [Google Scholar] [CrossRef]
- Carmona, D.; Lajeunesse, M.J.; Johnson, M.T. Plant Traits That Predict Resistance to Herbivores. Funct. Ecol. 2011, 25, 358–367. [Google Scholar] [CrossRef]
- Lesur, I.; Le Provost, G.; Bento, P.; Da Silva, C.; Leplé, J.-C.; Murat, F.; Ueno, S.; Bartholomé, J.; Lalanne, C.; Ehrenmann, F.; et al. The Oak Gene Expression Atlas: Insights into Fagaceae Genome Evolution and the Discovery of Genes Regulated during Bud Dormancy Release. BMC Genom. 2015, 16, 112. [Google Scholar] [CrossRef]
- Derory, J.; Léger, P.; Garcia, V.; Schaeffer, J.; Hauser, M.-T.; Salin, F.; Luschnig, C.; Plomion, C.; Glössl, J.; Kremer, A. Transcriptome Analysis of Bud Burst in Sessile Oak (Quercus petraea). New Phytol. 2006, 170, 723–738. [Google Scholar] [CrossRef]
- Derory, J.; Scotti-Saintagne, C.; Bertocchi, E.; Le Dantec, L.; Graignic, N.; Jauffres, A.; Casasoli, M.; Chancerel, E.; Bodénès, C.; Alberto, F.; et al. Contrasting Relationships between the Diversity of Candidate Genes and Variation of Bud Burst in Natural and Segregating Populations of European Oaks. Heredity 2010, 104, 438–448. [Google Scholar] [CrossRef]
- Alberto, F.J.; Derory, J.; Boury, C.; Frigerio, J.-M.; Zimmermann, N.E.; Kremer, A. Imprints of Natural Selection along Environmental Gradients in Phenology-Related Genes of Quercus petraea. Genetics 2013, 195, 495–512. [Google Scholar] [CrossRef]
- Diekmann, M. Deciduous Forest Vegetation in Boreo-Nemoral Scandinavia. Ph.D. Thesis, Uppsala University, Uppsala, Sweden, 1994. [Google Scholar]
- Vranckx, G.; Jacquemyn, H.; Mergeay, J.; Cox, K.; Kint, V.; Muys, B.; Honnay, O. Transmission of Genetic Variation from the Adult Generation to Naturally Established Seedling Cohorts in Small Forest Stands of Pedunculate Oak (Quercus robur L.). For. Ecol. Manag. 2014, 312, 19–27. [Google Scholar] [CrossRef]
- Moracho, E.; Moreno, G.; Jordano, P.; Hampe, A. Unusually Limited Pollen Dispersal and Connectivity of Pedunculate Oak (Quercus robur) Refugial Populations at the Species’ Southern Range Margin. Mol. Ecol. 2016, 25, 3319–3331. [Google Scholar] [CrossRef]
- Buschbom, J.; Yanbaev, Y.; Degen, B. Efficient Long-Distance Gene Flow into an Isolated Relict Oak Stand. J. Hered. 2011, 102, 464–472. [Google Scholar] [CrossRef]
- Eaton, E.; Caudullo, G.; Oliveira, S.; de Rigo, D. Quercus Robur and Quercus Petraea in Europe: Distribution, Habitat, Usage and Threats. In European Atlas of Forest Tree Species; San-Miguel-Ayanz, J., de Rigo, D., Caudullo, G., Houston Durrant, T., Mauri, A., Eds.; Publications Office of the EU: Luxembourg, 2016; p. e01c6df+. [Google Scholar]
- Sunde, J.; Franzén, M.; Betzholtz, P.-E.; Francioli, Y.; Pettersson, L.B.; Pöyry, J.; Ryrholm, N.; Forsman, A. Century-Long Butterfly Range Expansions in Northern Europe Depend on Climate, Land Use and Species Traits. Commun. Biol. 2023, 6, 601. [Google Scholar] [CrossRef]
- Forsman, A.; Sunde, J.; Salis, R.; Franzén, M. Latitudinal Gradients of Biodiversity and Ecosystem Services in Protected and Non-Protected Oak Forest Areas Can Inform Climate Smart Conservation. Geogr. Sustain. 2024, 5, 647–659. [Google Scholar] [CrossRef]
- Hall, M.; Franzén, M.; Forsman, A.; Sunde, J. Spatially Varying Selection and Contrasting Patterns in Neutral and Adaptive Genetic Variation towards the Cold-Limited Northern Range Margin in Quercus Robur. Submitt. Evol. Appl. 2025. [Google Scholar]
- Black-Samuelsson, S. Den Skogliga Genbanken—Från Storhetstid Till Framtid; Skogsstyrelsen: Jönköping, Sweden, 2019. [Google Scholar]
- Johansson, V.; Forsman, A.; Gustafsson, L.; Hall, M.; Edvardsson, J.; Salis, R.; Sunde, J.; Franzén, M. Low Cross-Taxon Congruence and Weak Stand-Age Effects on Biodiversity in Swedish Oak Forests. Biodivers. Conserv. 2025, 34, 2739–2750. [Google Scholar] [CrossRef]
- Faticov, M.; Ekholm, A.; Roslin, T.; Tack, A.J. Climate and Host Genotype Jointly Shape Tree Phenology, Disease Levels and Insect Attacks. Oikos 2020, 129, 391–401. [Google Scholar] [CrossRef]
- Mariën, B.; Balzarolo, M.; Dox, I.; Leys, S.; Lorène, M.J.; Geron, C.; Portillo-Estrada, M.; AbdElgawad, H.; Asard, H.; Campioli, M. Detecting the Onset of Autumn Leaf Senescence in Deciduous Forest Trees of the Temperate Zone. New Phytol. 2019, 224, 166–176. [Google Scholar] [CrossRef]
- Johansson, B. Areal Precipitation and Temperature in the Swedish Mountains: An Evaluation from a Hydrological Perspective. Hydrol. Res. 2000, 31, 207–228. [Google Scholar] [CrossRef]
- Brooks, M.E.; Kristensen, K.; van Benthem, K.J.; Magnusson, A.; Berg, C.W.; Nielsen, A.; Skaug, H.J.; Mächler, M.; Bolker, B.M. glmmTMB Balances Speed and Flexibility among Packages for Zero-Inflated Generalized Linear Mixed Modeling. R J. 2017, 9, 378–400. [Google Scholar] [CrossRef]
- Weisberg, F.J. An R Companion to Applied Regression, 3rd ed.; Sage: Thousand Oaks, CA, USA, 2019. [Google Scholar]
- Bartoń, K. “MuMIn”: Multi-Model Inference 2025. Available online: https://CRAN.R-project.org/package=MuMIn (accessed on 10 July 2025).
- Rochette, N.C.; Rivera-Colón, A.G.; Catchen, J.M. Stacks 2: Analytical Methods for Paired-End Sequencing Improve RADseq-Based Population Genomics. Mol. Ecol. 2019, 28, 4737–4754. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 10 July 2025).
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize Analysis Results for Multiple Tools and Samples in a Single Report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows–Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Paris, J.R.; Stevens, J.R.; Catchen, J.M. Lost in Parameter Space: A Road Map for Stacks. Methods Ecol. Evol. 2017, 8, 1360–1373. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Browning, B.L.; Zhou, Y.; Browning, S.R. A One-Penny Imputed Genome from next-Generation Reference Panels. Am. J. Hum. Genet. 2018, 103, 338–348. [Google Scholar] [CrossRef]
- Vitasse, Y.; Bresson, C.C.; Kremer, A.; Michalet, R.; Delzon, S. Quantifying Phenological Plasticity to Temperature in Two Temperate Tree Species. Funct. Ecol. 2010, 24, 1211–1218. [Google Scholar] [CrossRef]
- Kang, H.M.; Zaitlen, N.A.; Wade, C.M.; Kirby, A.; Heckerman, D.; Daly, M.J.; Eskin, E. Efficient Control of Population Structure in Model Organism Association Mapping. Genetics 2008, 178, 1709–1723. [Google Scholar] [CrossRef]
- Lin, J.; Berveiller, D.; François, C.; Hänninen, H.; Morfin, A.; Vincent, G.; Zhang, R.; Rathgeber, C.; Delpierre, N. A Model of the Within-Population Variability of Budburst in Forest Trees. Geosci. Model Dev. 2024, 17, 865–879. [Google Scholar] [CrossRef]
- Frontiér, S. Étude de La Décroissance Des Valeurs Propres Dans Une Analyse En Composantes Principales: Comparaison Avec Le Modéle Du Batôn Brisé. J. Exp. Mar. Biol. Ecol. 1976, 25, 67–75. [Google Scholar] [CrossRef]
- Jackson, D.A. Stopping Rules in Principal Components Analysis: A Comparison of Heuristical and Statistical Approaches. Ecology 1993, 74, 2204–2214. [Google Scholar] [CrossRef]
- Cattell, R.B. The Scree Test for the Number of Factors. Multivar. Behav. Res. 1966, 1, 245–276. [Google Scholar] [CrossRef]
- Raj, A.; Stephens, M.; Pritchard, J.K. fastSTRUCTURE: Variational Inference of Population Structure in Large SNP Data Sets. Genetics 2014, 197, 573–589. [Google Scholar] [CrossRef]
- Hamilton, M.B. Population Genetics; Wiley-Blackwell: Hoboken, NJ, USA, 2009. [Google Scholar]
- Zhang, C.; Dong, S.-S.; Xu, J.-Y.; He, W.-M.; Yang, T.-L. PopLDdecay: A Fast and Effective Tool for Linkage Disequilibrium Decay Analysis Based on Variant Call Format Files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef]
- Otyama, P.I.; Wilkey, A.; Kulkarni, R.; Assefa, T.; Chu, Y.; Clevenger, J.; O’Connor, D.J.; Wright, G.C.; Dezern, S.W.; MacDonald, G.E.; et al. Evaluation of Linkage Disequilibrium, Population Structure, and Genetic Diversity in the US Peanut Mini Core Collection. BMC Genom. 2019, 20, 481. [Google Scholar] [CrossRef]
- Pang, Y.; Liu, C.; Wang, D.; Amand, P.S.; Bernardo, A.; Li, W.; He, F.; Li, L.; Wang, L.; Yuan, X.; et al. High-Resolution Genome-Wide Association Study Identifies Genomic Regions and Candidate Genes for Important Agronomic Traits in Wheat. Mol. Plant 2020, 13, 1311–1327. [Google Scholar] [CrossRef]
- Legendre, P.; Legendre, L. Numerical Ecology, 3rd ed.; Elsevier B.V.: Oxford, UK, 2012. [Google Scholar]
- Forester, B.R.; Lasky, J.R.; Wagner, H.H.; Urban, D.L. Comparing Methods for Detecting Multilocus Adaptation with Multivariate Genotype–Environment Associations. Mol. Ecol. 2018, 27, 2215–2233. [Google Scholar] [CrossRef]
- Le Corre, V.; Kremer, A. The Genetic Differentiation at Quantitative Trait Loci under Local Adaptation. Mol. Ecol. 2012, 21, 1548–1566. [Google Scholar] [CrossRef]
- Kremer, A.; Le Corre, V.; Petit, R.J.; Ducousso, A. Historical and Contemporary Dynamics of Adaptive Differentiation in European Oaks. In Molecular Approaches in Natural Resource Conservation and Management; DeWoody, J., Bickham, J., Michler, C., Nichols, K., Rhodes, G., Woeste, K., Eds.; Cambridge University Press: Cambridge, UK, 2010; pp. 101–122. [Google Scholar]
- Oksanen, J.; Simpson, G.; Blanchet, F.; Kindt, R.; Legendre, P.; Minchin, P.; O’Hara, R.; Solymos, P.; Stevens, M.; Szoecs, E.; et al. Vegan: Community Ecology Package. 2022. Available online: https://CRAN.R-project.org/package=vegan (accessed on 10 July 2025).
- Storey, J.D.; Bass, A.J.; Dabney, A.; Robinson, D. Qvalue: Q-Value Estimation for False Discovery Rate Control. 2023. Available online: https://www.bioconductor.org/packages/devel/bioc/manuals/qvalue/man/qvalue.pdf (accessed on 21 July 2025).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer-Verlag: New York, NY, USA, 2016. [Google Scholar]
- Bodénès, C.; Chancerel, E.; Ehrenmann, F.; Kremer, A.; Plomion, C. High-Density Linkage Mapping and Distribution of Segregation Distortion Regions in the Oak Genome. DNA Res. 2016, 23, 115–124. [Google Scholar] [CrossRef]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of Molecular Variance Inferred from Metric Distances among DNA Haplotypes: Application to Human Mitochondrial DNA Restriction Data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef]
- Paradis, E. Pegas: An R Package for Population Genetics with an Integrated–Modular Approach. Bioinformatics 2010, 26, 419–420. [Google Scholar] [CrossRef]
- Holsinger, K.E.; Weir, B.S. Genetics in Geographically Structured Populations: Defining, Estimating and Interpreting FST. Nat. Rev. Genet. 2009, 10, 639–650. [Google Scholar] [CrossRef]
- Ziemienowicz, A.; Haasen, D.; Staiger, D.; Merkle, T. Arabidopsis Transportin1 Is the Nuclear Import Receptor for the Circadian Clock-Regulated RNA-Binding Protein AtGRP7. Plant Mol. Biol. 2003, 53, 201–212. [Google Scholar] [CrossRef]
- Yan, B.; Wang, X.; Wang, Z.; Chen, N.; Mu, C.; Mao, K.; Han, L.; Zhang, W.; Liu, H. Identification of Potential Cargo Proteins of Transportin Protein AtTRN1 in Arabidopsis thaliana. Plant Cell Rep. 2016, 35, 629–640. [Google Scholar] [CrossRef]
- Bhattacharya, D.P. Study of Small Non-Coding RNAs in Plants by Developing Novel Pipelines. Ph.D. Thesis, Universitäts-und Landesbibliothek Sachsen-Anhalt, Halle, Germany, 2017. [Google Scholar]
- Osuna-Caballero, S.; Rubiales, D.; Rispail, N. Genome-Wide Association Study Uncovers Pea Candidate Genes and Metabolic Pathways Involved in Rust Resistance. Plant Genome 2024, 17, e20510. [Google Scholar] [CrossRef]
- Arimura, G.; Huber, D.P.; Bohlmann, J. Forest Tent Caterpillars (Malacosoma disstria) Induce Local and Systemic Diurnal Emissions of Terpenoid Volatiles in Hybrid Poplar (Populus trichocarpa × Deltoides): cDNA Cloning, Functional Characterization, and Patterns of Gene Expression of (-)-Germacrene D Synthase, PtdTPS1. Plant J. 2004, 37, 603–616. [Google Scholar] [CrossRef]
- Muñoz-Diaz, E.; Fuenzalida-Valdivia, I.; Darriere, T.; de Bures, A.; Blanco-Herrera, F.; Rompais, M.; Carapito, C.; Saez-Vasquez, J. Proteomic Profiling of Arabidopsis Nuclei Reveals Distinct Protein Accumulation Kinetics upon Heat Stress. Sci. Rep. 2024, 14, 18914. [Google Scholar] [CrossRef]
- Cohen, O.; Borovsky, Y.; David-Schwartz, R.; Paran, I. CaJOINTLESS Is a MADS-Box Gene Involved in Suppression of Vegetative Growth in All Shoot Meristems in Pepper. J. Exp. Bot. 2012, 63, 4947–4957. [Google Scholar] [CrossRef]
- Mao, L.; Begum, D.; Chuang, H.; Budiman, M.A.; Szymkowiak, E.J.; Irish, E.E.; Wing, R.A. JOINTLESS Is a MADS-Box Gene Controlling Tomato Flower Abscission Zone Development. Nature 2000, 406, 910–913. [Google Scholar] [CrossRef]
- Fernandes, P.M. Molecular and Histological Approaches to Unravel Castanea spp. Responses to Phytophthora cinnamomi Infection. Ph.D. Thesis, Universidade NOVA de Lisboa, Lisbon, Portugal, 2022. [Google Scholar]
- Iizasa, S.; Iizasa, E.; Watanabe, K.; Nagano, Y. Transcriptome Analysis Reveals Key Roles of AtLBR-2 in LPS-Induced Defense Responses in Plants. BMC Genom. 2017, 18, 995. [Google Scholar] [CrossRef]
- Chandran, N.K.; Sriram, S.; Prakash, T.; Budhwar, R. Transcriptome Changes in Resistant and Susceptible Rose in Response to Powdery Mildew. J. Phytopathol. 2021, 169, 556–569. [Google Scholar] [CrossRef]
- Szajko, K.; Sołtys-Kalina, D.; Heidorn-Czarna, M.; Smyda-Dajmund, P.; Wasilewicz-Flis, I.; Jańska, H.; Marczewski, W. Transcriptomic and Proteomic Data Provide New Insights into Cold-Treated Potato Tubers with T-and D-Type Cytoplasm. Planta 2022, 255, 97. [Google Scholar] [CrossRef]
- Shen, C.; Shi, X.; Xie, C.; Li, Y.; Yang, H.; Mei, X.; Xu, Y.; Dong, C. The Change in Microstructure of Petioles and Peduncles and Transporter Gene Expression by Potassium Influences the Distribution of Nutrients and Sugars in Pear Leaves and Fruit. J. Plant Physiol. 2019, 232, 320–333. [Google Scholar] [CrossRef]
- Fernandes, I.; Paulo, O.S.; Marques, I.; Sarjkar, I.; Sen, A.; Graça, I.; Pawlowski, K.; Ramalho, J.C.; Ribeiro-Barros, A.I. Salt Stress Tolerance in Casuarina Glauca: Insights from the Branchlets Transcriptome. Plants 2022, 11, 2942. [Google Scholar] [CrossRef]
- Figueiredo, J.; Santos, R.B.; Guerra-Guimarães, L.; Leclercq, C.C.; Renaut, J.; Sousa, L.; Figueiredo, A. Revealing the Secrets beneath Grapevine and Plasmopara Viticola Early Communication: A Picture of Host and Pathogen Proteomes. bioRxiv 2021, preprint. [Google Scholar] [CrossRef]
- Hu, X.; Wu, L.; Zhao, F.; Zhang, D.; Li, N.; Zhu, G.; Li, C.; Wang, W. Phosphoproteomic Analysis of the Response of Maize Leaves to Drought, Heat and Their Combination Stress. Front. Plant Sci. 2015, 6, 298. [Google Scholar] [CrossRef]
- Lenz, A.; Hoch, G.; Vitasse, Y.; Körner, C. European Deciduous Trees Exhibit Similar Safety Margins against Damage by Spring Freeze Events along Elevational Gradients. New Phytol. 2013, 200, 1166–1175. [Google Scholar] [CrossRef]
- Schaber, J.; Badeck, F.-W. Physiology-Based Phenology Models for Forest Tree Species in Germany. Int. J. Biometeorol. 2003, 47, 193–201. [Google Scholar] [CrossRef]
- Zohner, C.M.; Benito, B.M.; Svenning, J.-C.; Renner, S.S. Day Length Unlikely to Constrain Climate-Driven Shifts in Leaf-out Times of Northern Woody Plants. Nat. Clim. Change 2016, 6, 1120–1123. [Google Scholar] [CrossRef]
- Dantec, C.F.; Ducasse, H.; Capdevielle, X.; Fabreguettes, O.; Delzon, S.; Desprez-Loustau, M.-L. Escape of Spring Frost and Disease through Phenological Variations in Oak Populations along Elevation Gradients. J. Ecol. 2015, 103, 1044–1056. [Google Scholar] [CrossRef]
- Zohner, C.M.; Mo, L.; Sebald, V.; Renner, S.S. Leaf-out in Northern Ecotypes of Wide-Ranging Trees Requires Less Spring Warming, Enhancing the Risk of Spring Frost Damage at Cold Range Limits. Glob. Ecol. Biogeogr. 2020, 29, 1065–1072. [Google Scholar] [CrossRef]
- Olsson, C.; Bolmgren, K.; Lindström, J.; Jönsson, A.M. Performance of Tree Phenology Models along a Bioclimatic Gradient in Sweden. Ecol. Model. 2013, 266, 103–117. [Google Scholar] [CrossRef]
- Ducousso, A.; Guyon, J.; Krémer, A. Latitudinal and Altitudinal Variation of Bud Burst in Western Populations of Sessile Oak (Quercus petraea (Matt) Liebl). In Proceedings of the Annales des Sciences Forestières; EDP Sciences: Les Ulis, France, 1996; Volume 53, pp. 775–782. [Google Scholar]
- Delpierre, N.; Dufrêne, E.; Soudani, K.; Ulrich, E.; Cecchini, S.; Boé, J.; François, C. Modelling Interannual and Spatial Variability of Leaf Senescence for Three Deciduous Tree Species in France. Agric. For. Meteorol. 2009, 149, 938–948. [Google Scholar] [CrossRef]
- Mariën, B.; Dox, I.; De Boeck, H.J.; Willems, P.; Leys, S.; Papadimitriou, D.; Campioli, M. Does Drought Advance the Onset of Autumn Leaf Senescence in Temperate Deciduous Forest Trees? Biogeosciences 2021, 18, 3309–3330. [Google Scholar] [CrossRef]
- Wang, H.; Gao, C.; Ge, Q. Low Temperature and Short Daylength Interact to Affect the Leaf Senescence of Two Temperate Tree Species. Tree Physiol. 2022, 42, 2252–2265. [Google Scholar] [CrossRef]
- Zani, D.; Crowther, T.W.; Mo, L.; Renner, S.S.; Zohner, C.M. Increased Growing-Season Productivity Drives Earlier Autumn Leaf Senescence in Temperate Trees. Science 2020, 370, 1066–1071. [Google Scholar] [CrossRef]
- Firmat, C.; Delzon, S.; Louvet, J.-M.; Parmentier, J.; Kremer, A. Evolutionary Dynamics of the Leaf Phenological Cycle in an Oak Metapopulation along an Elevation Gradient. J. Evol. Biol. 2017, 30, 2116–2131. [Google Scholar] [CrossRef]
- Desprez-Loustau, M.-L.; Vitasse, Y.; Delzon, S.; Capdevielle, X.; Marais, B.; Kremer, A. Are Plant Pathogen Populations Adapted for Encounter with Their Host? A Case Study of Phenological Synchrony between Oak and an Obligate Fungal Parasite along an Altitudinal Gradient. J. Evol. Biol. 2010, 23, 87–97. [Google Scholar] [CrossRef]
- Desprez-Loustau, M.-L.; Saint-Jean, G.; Barrès, B.; Dantec, C.F.; Dutech, C. Oak Powdery Mildew Changes Growth Patterns in Its Host Tree: Host Tolerance Response and Potential Manipulation of Host Physiology by the Parasite. Ann. For. Sci. 2014, 71, 563–573. [Google Scholar] [CrossRef]
- Ovaskainen, O.; Laine, A.-L. Inferring Evolutionary Signals from Ecological Data in a Plant–Pathogen Metapopulation. Ecology 2006, 87, 880–891. [Google Scholar] [CrossRef]
- Bertić, M.; Schroeder, H.; Kersten, B.; Fladung, M.; Orgel, F.; Buegger, F.; Schnitzler, J.-P.; Ghirardo, A. European Oak Chemical Diversity–from Ecotypes to Herbivore Resistance. New Phytol. 2021, 232, 818–834. [Google Scholar] [CrossRef]
- Hunter, M.D. A Variable Insect–Plant Interaction: The Relationship between Tree Budburst Phenology and Population Levels of Insect Herbivores among Trees. Ecol. Entomol. 1992, 17, 91–95. [Google Scholar] [CrossRef]
- Wesołowski, T.; Rowiński, P. Late Leaf Development in Pedunculate Oak (Quercus Robur): An Antiherbivore Defence? Scand. J. For. Res. 2008, 23, 386–394. [Google Scholar] [CrossRef]
- Ekholm, A.; Tack, A.J.; Bolmgren, K.; Roslin, T. The Forgotten Season: The Impact of Autumn Phenology on a Specialist Insect Herbivore Community on Oak. Ecol. Entomol. 2019, 44, 425–435. [Google Scholar] [CrossRef]
- Valdés-Correcher, E.; Kadiri, Y.; Bourdin, A.; Mrazova, A.; Bălăcenoiu, F.; Branco, M.; Bogdziewicz, M.; Bjørn, M.C.; Damestoy, T.; Dobrosavljević, J.; et al. Effects of Climate on Leaf Phenolics, Insect Herbivory, and Their Relationship in Pedunculate Oak (Quercus Robur) across Its Geographic Range in Europe. Oecologia 2025, 207, 61. [Google Scholar] [CrossRef]
- Čehulić, I.; Sever, K.; Katičić Bogdan, I.; Jazbec, A.; Škvorc, Ž.; Bogdan, S. Drought Impact on Leaf Phenology and Spring Frost Susceptibility in a Quercus Robur L. Provenance Trial. Forests 2019, 10, 50. [Google Scholar] [CrossRef]
- Vander Mijnsbrugge, K.; Turcsán, A.; Maes, J.; Duchêne, N.; Meeus, S.; Steppe, K.; Steenackers, M. Repeated Summer Drought and Re-Watering during the First Growing Year of Oak (Quercus petraea) Delay Autumn Senescence and Bud Burst in the Following Spring. Front. Plant Sci. 2016, 7, 419. [Google Scholar] [CrossRef]
- Misson, L.; Limousin, J.-M.; Rodriguez, R.; Letts, M.G. Leaf Physiological Responses to Extreme Droughts in Mediterranean Quercus Ilex Forest. Plant Cell Environ. 2010, 33, 1898–1910. [Google Scholar] [CrossRef]
- Misson, L.; Degueldre, D.; Collin, C.; Rodriguez, R.; Rocheteau, A.; Ourcival, J.-M.; Rambal, S. Phenological Responses to Extreme Droughts in a Mediterranean Forest. Glob. Change Biol. 2011, 17, 1036–1048. [Google Scholar] [CrossRef]
- Bréda, N.; Badeau, V. Forest Tree Responses to Extreme Drought and Some Biotic Events: Towards a Selection According to Hazard Tolerance? Comptes Rendus Géoscience 2008, 340, 651–662. [Google Scholar] [CrossRef]
- Bose, A.K.; Scherrer, D.; Camarero, J.J.; Ziche, D.; Babst, F.; Bigler, C.; Bolte, A.; Dorado-Liñán, I.; Etzold, S.; Fonti, P.; et al. Climate Sensitivity and Drought Seasonality Determine Post-Drought Growth Recovery of Quercus petraea and Quercus robur in Europe. Sci. Total Environ. 2021, 784, 147222. [Google Scholar] [CrossRef]
- Grossman, J.J. Phenological Physiology: Seasonal Patterns of Plant Stress Tolerance in a Changing Climate. New Phytol. 2023, 237, 1508–1524. [Google Scholar] [CrossRef]
- Koenig, W.D.; Funk, K.A.; Kraft, T.S.; Carmen, W.J.; Barringer, B.C.; Knops, J.M. Stabilizing Selection for Within-Season Flowering Phenology Confirms Pollen Limitation in a Wind-Pollinated Tree. J. Ecol. 2012, 100, 758–763. [Google Scholar] [CrossRef]
- Ismail, S.A.; Kokko, H. An Analysis of Mating Biases in Trees. Mol. Ecol. 2020, 29, 184–198. [Google Scholar] [CrossRef]
- Alberto, F.J.; Aitken, S.N.; Alía, R.; González-Martínez, S.C.; Hänninen, H.; Kremer, A.; Lefèvre, F.; Lenormand, T.; Yeaman, S.; Whetten, R.; et al. Potential for Evolutionary Responses to Climate Change–Evidence from Tree Populations. Glob. Change Biol. 2013, 19, 1645–1661. [Google Scholar] [CrossRef]
- Rellstab, C.; Gugerli, F.; Eckert, A.J.; Hancock, A.M.; Holderegger, R. A Practical Guide to Environmental Association Analysis in Landscape Genomics. Mol. Ecol. 2015, 24, 4348–4370. [Google Scholar] [CrossRef]
- Hewitt, G. The Genetic Legacy of the Quaternary Ice Ages. Nature 2000, 405, 907–913. [Google Scholar] [CrossRef]
- Kremer, A.; Hipp, A.L. Oaks: An Evolutionary Success Story. New Phytol. 2020, 226, 987–1011. [Google Scholar] [CrossRef]
- Gerber, S.; Chadøeuf, J.; Gugerli, F.; Lascoux, M.; Buiteveld, J.; Cottrell, J.; Dounavi, A.; Fineschi, S.; Forrest, L.L.; Fogelqvist, J.; et al. High Rates of Gene Flow by Pollen and Seed in Oak Populations across Europe. PLoS ONE 2014, 9, e85130. [Google Scholar] [CrossRef]
- Slatkin, M. Gene Flow and the Geographic Structure of Natural Populations. Science 1987, 236, 787–792. [Google Scholar] [CrossRef]
- Pringle, J.M.; Wares, J.P. Going against the Flow: Maintenance of Alongshore Variation in Allele Frequency in a Coastal Ocean. Mar. Ecol. Prog. Ser. 2007, 335, 69–84. [Google Scholar] [CrossRef]
- Koski, V. A Study of Pollen Dispersal as a Mechanism of Gene Flow in Conifers. Commun. Inst. For. Fenn. 1970, 70, 78. [Google Scholar]
- Persson, B.; Ståhl, E.G. Survival and Yield of Pinus Sylvestris L. as Related to Provenance Transfer and Spacing at High Altitudes in Northern Sweden. Scand. J. For. Res. 1990, 5, 381–395. [Google Scholar] [CrossRef]
- Dalongeville, A.; Andrello, M.; Mouillot, D.; Lobreaux, S.; Fortin, M.-J.; Lasram, F.; Belmaker, J.; Rocklin, D.; Manel, S. Geographic Isolation and Larval Dispersal Shape Seascape Genetic Patterns Differently According to Spatial Scale. Evol. Appl. 2018, 11, 1437–1447. [Google Scholar] [CrossRef]
- Kleinschmit, J. Intraspecific Variation of Growth and Adaptive Traits in European Oak Species. Ann. Des Sci. For. 1993, 50, 166–185. [Google Scholar] [CrossRef]
- Eckert, C.G.; Samis, K.E.; Lougheed, S.C. Genetic Variation across Species’ Geographical Ranges: The Central–Marginal Hypothesis and Beyond. Mol. Ecol. 2008, 17, 1170–1188. [Google Scholar] [CrossRef]
- Templ, B.; Koch, E.; Bolmgren, K.; Ungersböck, M.; Paul, A.; Scheifinger, H.; Rutishauser, T.; Busto, M.; Chmielewski, F.-M.; Hájková, L.; et al. Pan European Phenological Database (PEP725): A Single Point of Access for European Data. Int. J. Biometeorol. 2018, 62, 1109–1113. [Google Scholar] [CrossRef]
- Chau, J. Gslnls: GSL Nonlinear Least-Squares Fitting 2023. Available online: https://CRAN.R-project.org/package=gslnls (accessed on 10 July 2025).
- R Core Team. R: A Language and Environment for Statistical Computing 2023; R Foundation for Statistical Computing: Vienna, Austria, 2023; Available online: https://www.R-project.org/ (accessed on 10 July 2025).
- Kikuchi, S.; Bheemanahalli, R.; Jagadish, K.S.; Kumagai, E.; Masuya, Y.; Kuroda, E.; Raghavan, C.; Dingkuhn, M.; Abe, A.; Shimono, H. Genome-Wide Association Mapping for Phenotypic Plasticity in Rice. Plant Cell Environ. 2017, 40, 1565–1575. [Google Scholar] [CrossRef]


| χ2 | Est. | p-Value | R2M | R2C | ||
|---|---|---|---|---|---|---|
| Budburst | April temp. | 7.55 | −2.89 | 0.001 | 0.36 | 0.79 |
| Latitude | 3.90 | 1.30 | 0.048 | 0.24 | 0.79 | |
| Leaf senescence | Budburst | 0.18 | 0.21 | 0.67 | 0.01 | 0.34 |
| August temp. | 3.60 | 10.38 | 0.06 | 0.11 | 0.32 | |
| Latitude | 10.73 | −3.42 | 0.001 | 0.21 | 0.32 |
| SNP ID | Chr. | Position | q-Value | Gene Annotation | Possibly Associated with | Distance from Region (bp) |
|---|---|---|---|---|---|---|
| 55616_189 | 1 | 47495836 | 4.89 × 10−3 | Transportin-1 | Several functions | 9254 |
| 224662_285 | 3 | 37331722 | 8.43 × 10−4 | Small nucleolar RNA R71 | Pathogen defence | 4392 |
| 614926_284 | 8 | 15323561 | 3.54 × 10−2 | (−)-Germacrene D synthase-like | Herbivory defence | 164 |
| 693924_287 | 9 | 13741392 | 3.57 × 10−2 | rRNA 2′-O-methyltransferase serine/threonine-fibrillarin 1-like | Heat stress recovery | 6951 |
| 837643_219 | 11 | 24896775 | 2.29 × 10−2 | MADS-box protein JOINTLESS-like | Rate of vegetal development | 19,473 |
| 52095_31 | 1 | 43584107 | 3.84 × 10−2 | Mediator of RNA polymerase II transcription subunit 25 | Flowering time | 1163 |
| 277235_194 | 4 | 12685808 | 6.41 × 10−8 | G-type lectin S-receptor-like serine/threonine-protein kinase At1G67520 | Pathogen defence | 17,139 |
| 355472_275 | 4 | 75298409 | 6.57 × 10−3 | Putative disease resistance RPP13-like protein 1 | Pathogen defence | 0 |
| 449028_6 | 5 | 70766439 | 5.62 × 10−6 | Putative calcium-transporting ATPase 13, plasma membrane-type | Several functions | 4865 |
| 614940_70 | 8 | 15336382 | 2.18 × 10−3 | (−)-Germacrene D synthase-like | Herbivory defence | 12,985 |
| 846236_271 | 11 | 31918101 | 2.81 × 10−3 | Vacuolar-sorting receptor 3-like | Drought and heat stress | 15,453 |
| Full | Budburst | Leaf Senescence | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Stand | Lat. (°N) | Long. (°E) | Min. April Temp. (°C) | Min. August Temp. (°C) | n | PA | P | Dist. | P | Dist. | P | Dist. |
| Bellinga | 55.52 | 13.68 | 1.15 | 12.87 | 9 | 66 | 0.6 | 3244.04 | 0.95 | 19.02 | 0.96 | 23.24 |
| Björnstorp | 55.62 | 13.43 | 1.05 | 12.64 | 10 | 91 | 0.69 | 3839.84 | 0.98 | 21.35 | 0.96 | 26.08 |
| Tranemåla | 56.36 | 14.78 | 0.57 | 11.99 | 6 | 67 | 0.53 | 3302.74 | 0.95 | 19.61 | 1.00 | 28.18 |
| Strömsrum | 56.93 | 16.39 | 0.58 | 12.65 | 10 | 75 | 0.65 | 3555.13 | 0.95 | 19.95 | 1.00 | 27.04 |
| Tånnö | 57.06 | 14.01 | −0.06 | 11.32 | 10 | 84 | 0.64 | 3434.44 | 0.98 | 20.73 | 1.00 | 26.36 |
| Vårgårda | 57.96 | 12.83 | −0.08 | 11.20 | 9 | 54 | 0.58 | 3160.35 | 0.93 | 16.89 | 0.94 | 21.28 |
| Vagnhärad | 58.96 | 17.60 | −0.26 | 12.56 | 10 | 116 | 0.66 | 3693.39 | 0.95 | 19.22 | 0.98 | 22.11 |
| Vinala | 59.13 | 15.38 | −0.48 | 11.65 | 9 | 101 | 0.54 | 2890.13 | 0.88 | 18.61 | 0.91 | 21.22 |
| Testeboån | 60.77 | 16.98 | −1.89 | 10.98 | 8 | 229 | 0.58 | 3591.91 | 0.90 | 17.45 | 0.98 | 25.70 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Isaksson, J.; Hall, M.; Rula, I.; Franzén, M.; Forsman, A.; Sunde, J. Genetic Variation Associated with Leaf Phenology in Pedunculate Oak (Quercus robur L.) Implicates Pathogens, Herbivores, and Heat Stress as Selective Drivers. Forests 2025, 16, 1233. https://doi.org/10.3390/f16081233
Isaksson J, Hall M, Rula I, Franzén M, Forsman A, Sunde J. Genetic Variation Associated with Leaf Phenology in Pedunculate Oak (Quercus robur L.) Implicates Pathogens, Herbivores, and Heat Stress as Selective Drivers. Forests. 2025; 16(8):1233. https://doi.org/10.3390/f16081233
Chicago/Turabian StyleIsaksson, Jonatan, Marcus Hall, Iryna Rula, Markus Franzén, Anders Forsman, and Johanna Sunde. 2025. "Genetic Variation Associated with Leaf Phenology in Pedunculate Oak (Quercus robur L.) Implicates Pathogens, Herbivores, and Heat Stress as Selective Drivers" Forests 16, no. 8: 1233. https://doi.org/10.3390/f16081233
APA StyleIsaksson, J., Hall, M., Rula, I., Franzén, M., Forsman, A., & Sunde, J. (2025). Genetic Variation Associated with Leaf Phenology in Pedunculate Oak (Quercus robur L.) Implicates Pathogens, Herbivores, and Heat Stress as Selective Drivers. Forests, 16(8), 1233. https://doi.org/10.3390/f16081233






