Comprehensive Analysis of JAZ Family Genes Involved in Sex Differentiation in Areca catechu
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Characterization of JAZ Genes in A. catechu
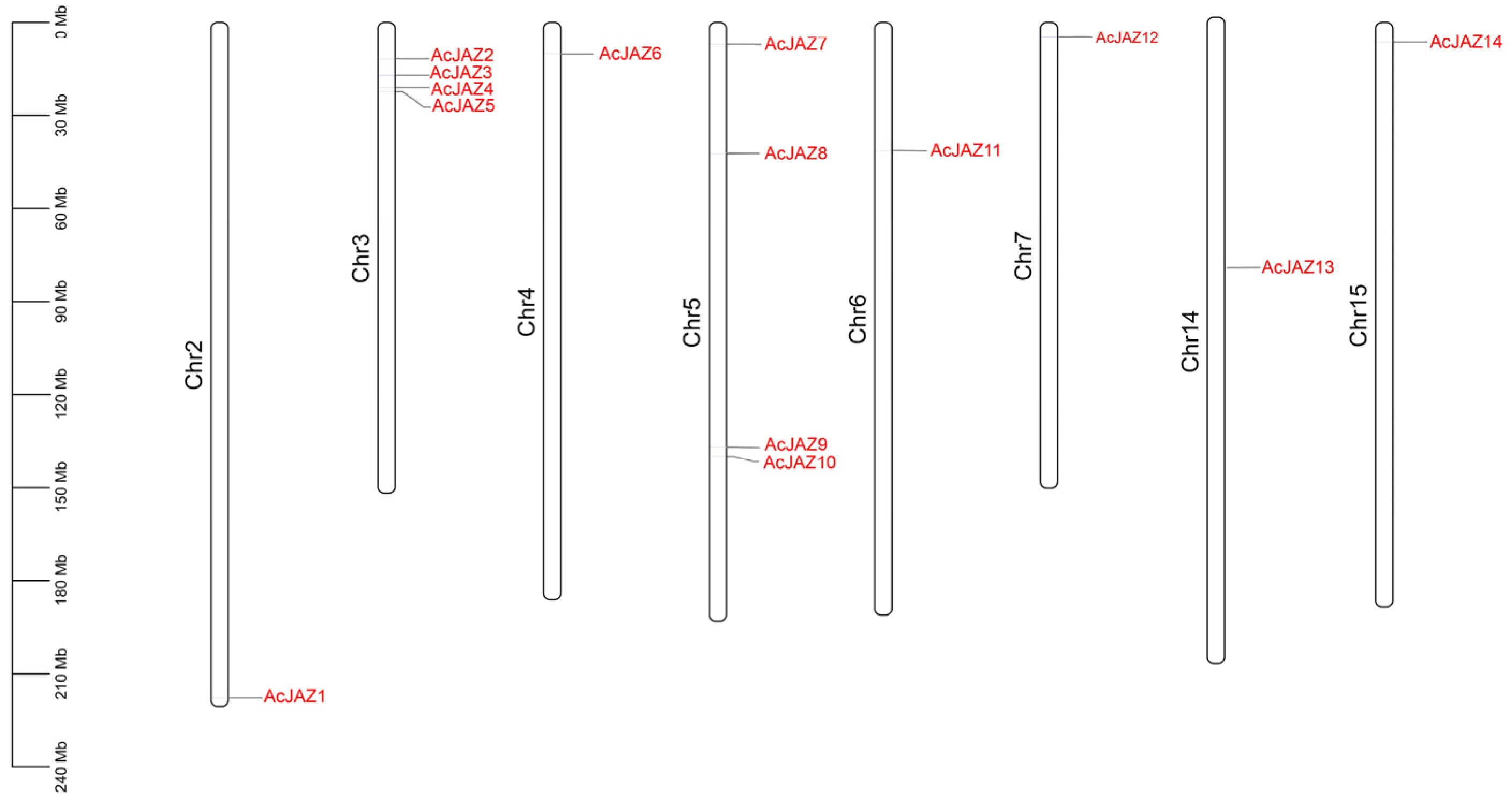
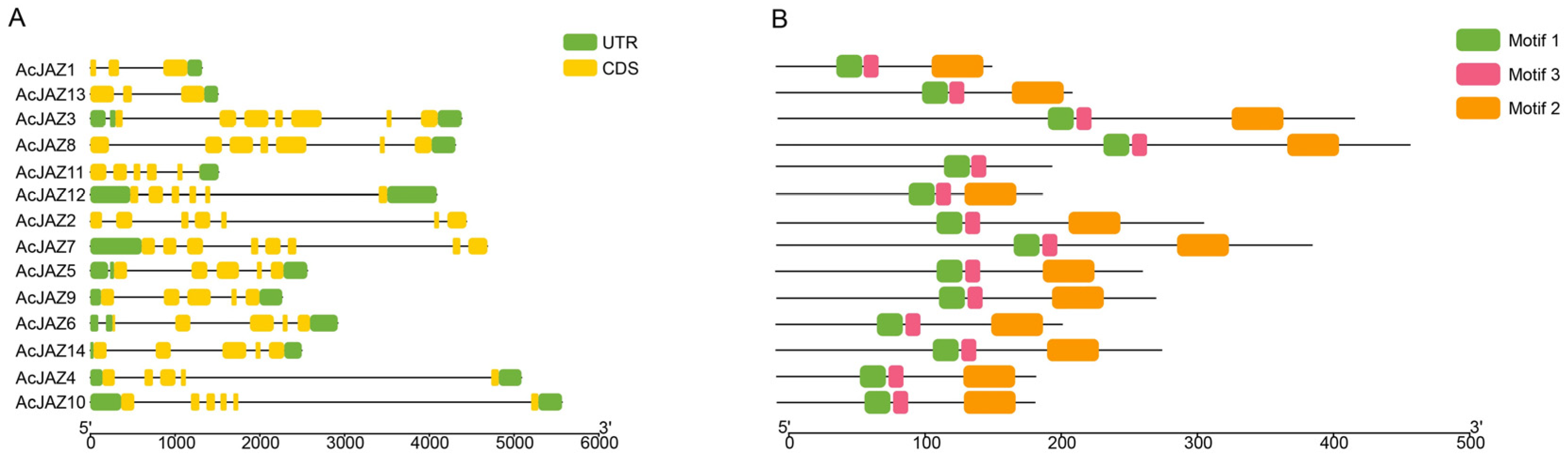
2.2. Chromosomal Location, Gene Structure, and Conserved Motif Analysis of AcJAZ Genes
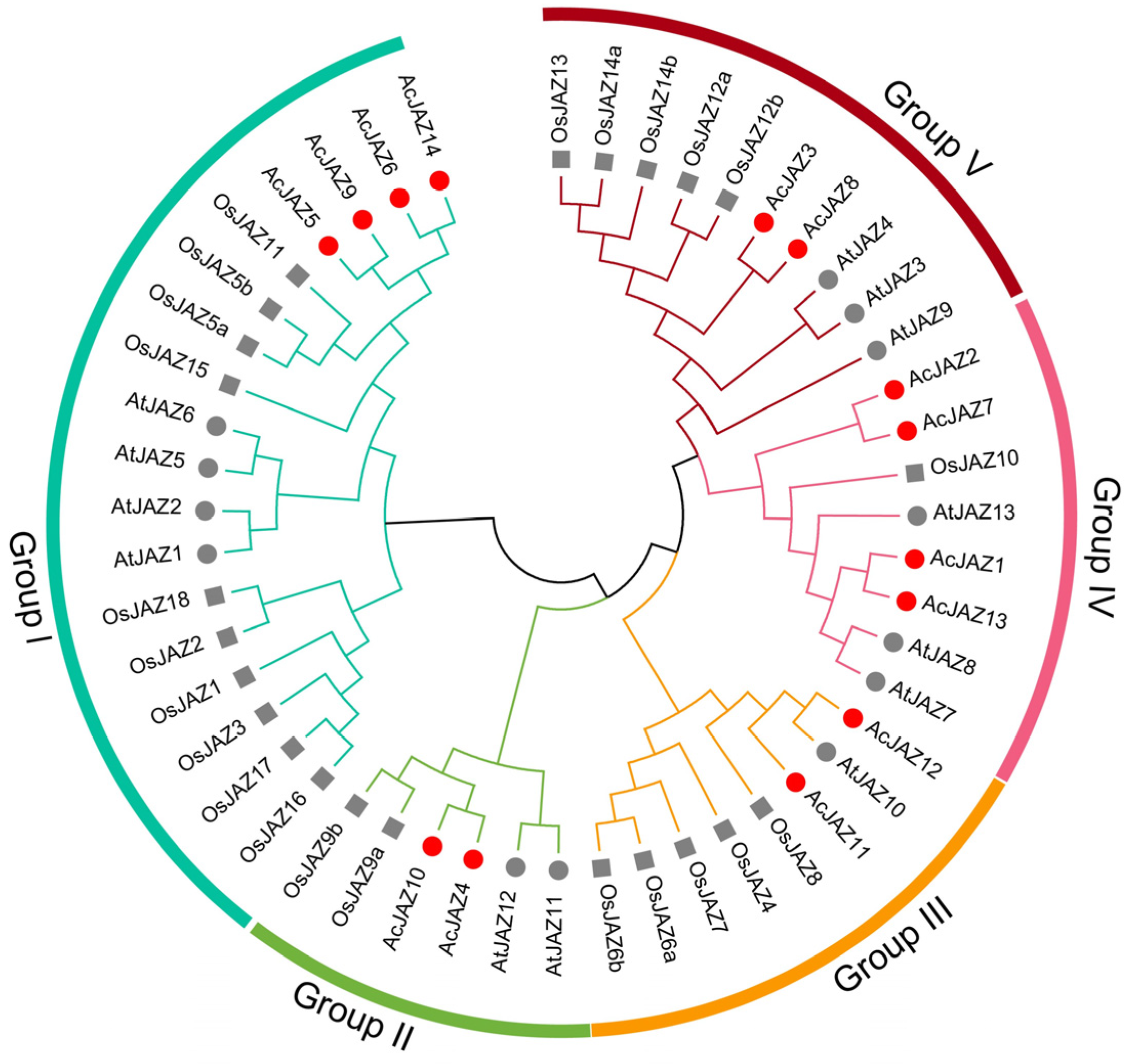
2.3. Phylogenetic Analysis of the JAZ Gene Family in A. catechu
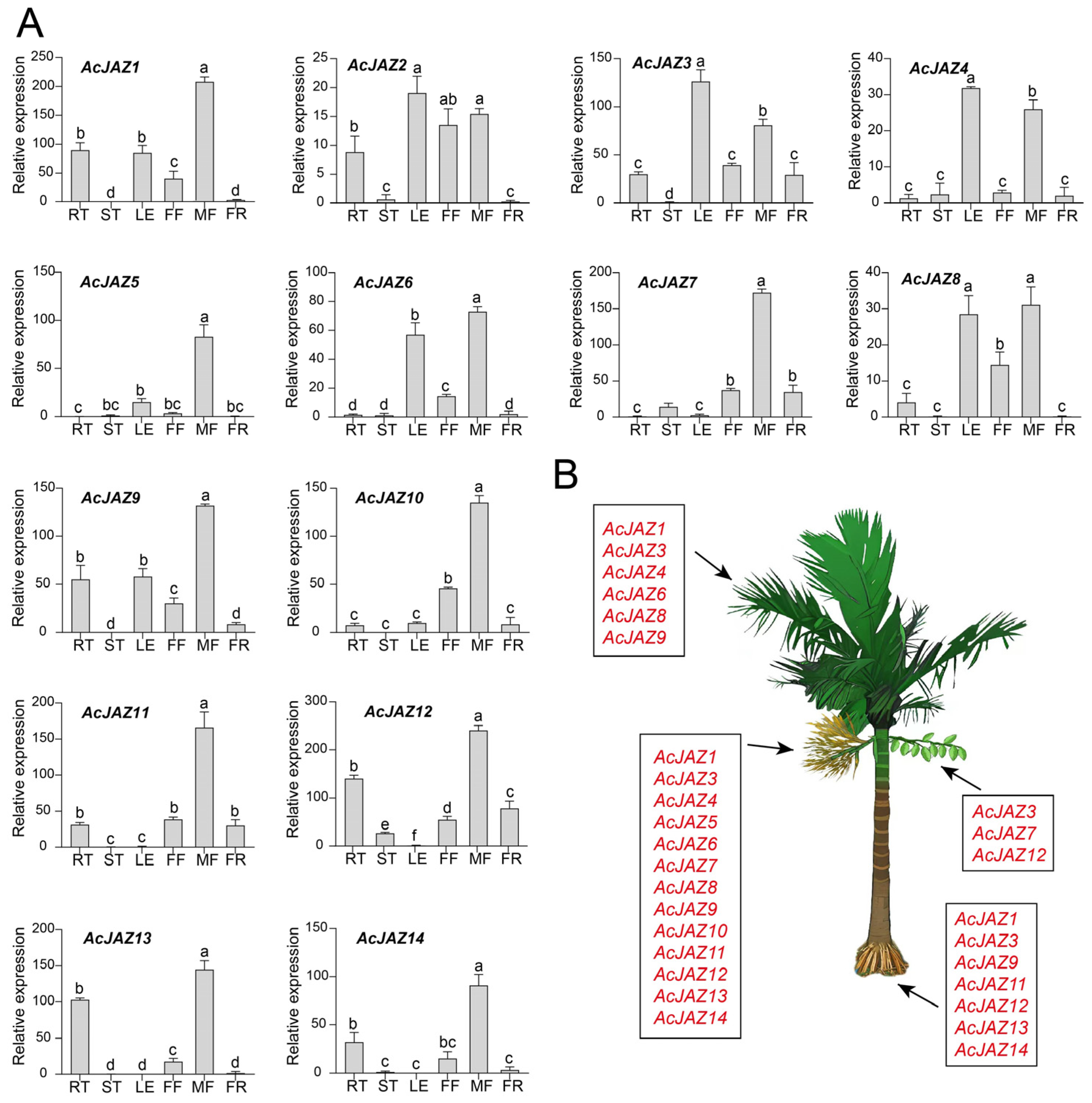
2.4. Expression Profiling of AcJAZ Genes by qRT-PCR
3. Results
3.1. Identification of the AcJAZ Genes
3.2. Gene Structure and Conserved Motif Analysis
3.3. Phylogenetic Analysis of AcJAZ Genes
3.4. Expression Profiling of AcJAZ Genes in Different Organs
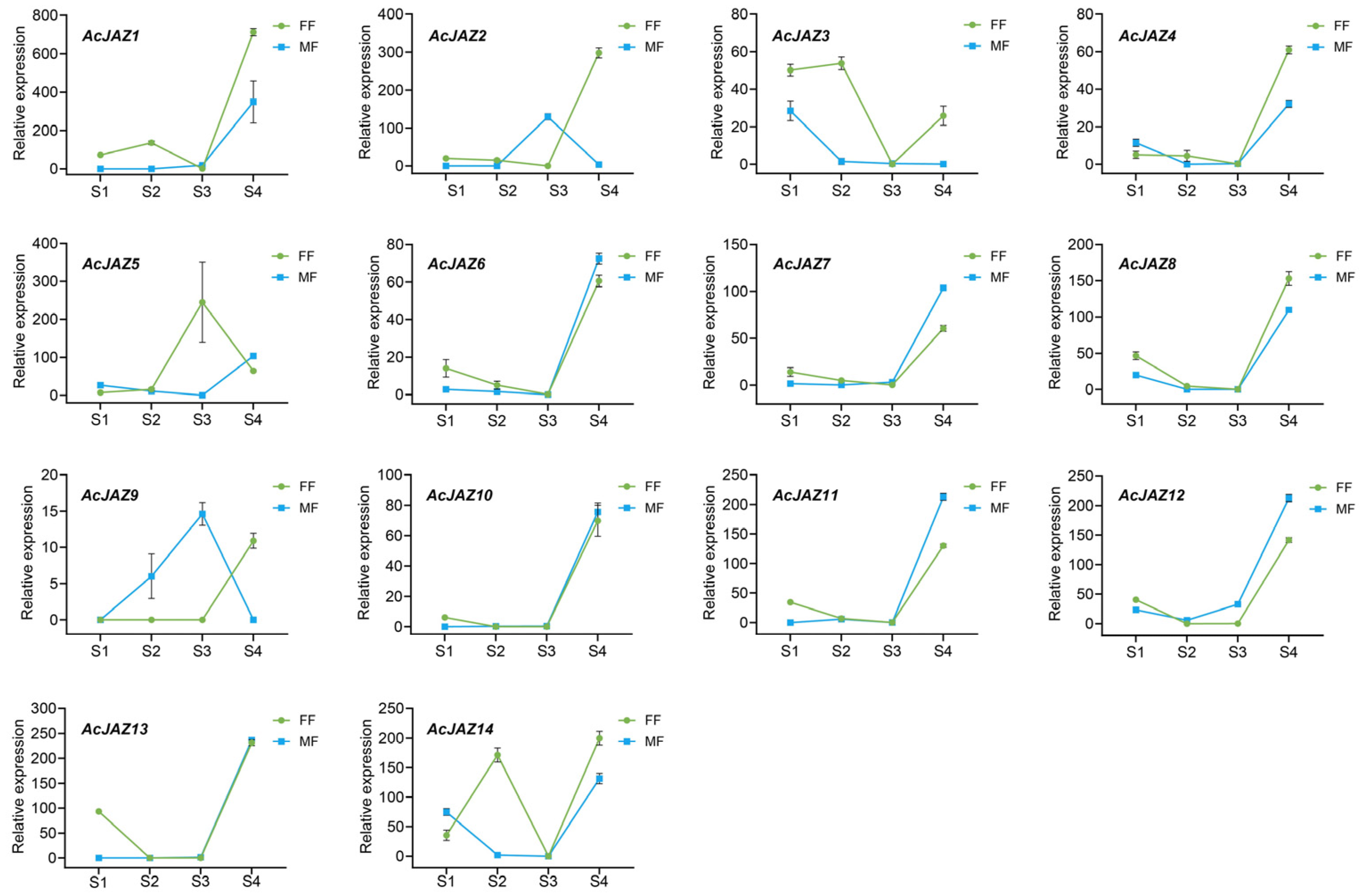
3.5. AcJAZ Genes in Male and Female Flower Development
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cai, K.; Zhu, S.; Jiang, Z.; Xu, K.; Sun, X.; Li, X. Biological macromolecules mediated by environmental signals affect flowering regulation in plants: A comprehensive review. Plant Physiol. Biochem. 2024, 214, 108931. [Google Scholar] [CrossRef] [PubMed]
- Renner, S.S. The relative and absolute frequencies of angiosperm sexual systems: Dioecy, monoecy, gynodioecy, and an updated online database. Am. J. Bot. 2014, 101, 1588–1596. [Google Scholar] [CrossRef] [PubMed]
- Stehlik, I.; Friedman, J.; Barrett, S.C.H. Environmental influence on primary sex ratio in a dioecious plant. Proc. Natl. Acad. Sci. USA 2008, 105, 10847–10852. [Google Scholar] [CrossRef] [PubMed]
- Renner, S.S.; Müller, N.A. Plant sex chromosomes defy evolutionary models of expanding recombination suppression and genetic degeneration. Nat. Plants 2021, 7, 392–402. [Google Scholar] [CrossRef]
- Rashid, M.; Shamsi, S.; Zaman, R.; Ilahi, A. Areca catechu: Enfolding of historical and therapeutic traditional knowledge with modern update. Int. J. Phramacognosy 2015, 2, 221–228. [Google Scholar]
- Ansari, A.; Mahmood, T.; Bagga, P.; Ahsan, F.; Shamim, A.; Ahmad, S.; Shariq, M.; Parveen, S. Areca catechu: A phytopharmacological legwork. Food Front. 2021, 2, 163–183. [Google Scholar] [CrossRef]
- Zhou, G.; Yin, H.; Chen, F.; Wang, Y.; Gao, Q.; Yang, F.; He, C.; Zhang, L.; Wan, Y. The genome of Areca catechu provides insights into sex determination of monoecious plants. New Phytol. 2022, 236, 2327–2343. [Google Scholar] [CrossRef]
- Luo, H.; Lu, Z.; Guan, J.; Yan, M.; Liu, Z.; Wan, Y.; Zhou, G. Gene co-expression network analysis in areca floral organ and the potential role of the AcMADS17 and AcMADS23 in transgenic Arabidopsis. Plant Sci. 2024, 342, 112049. [Google Scholar] [CrossRef]
- Ruan, J.; Zhou, Y.; Zhou, M.; Yan, J.; Khurshid, M.; Weng, W.; Cheng, J.; Zhang, K. Jasmonic Acid Signaling Pathway in Plants. Int. J. Mol. Sci. 2019, 20, 2479. [Google Scholar] [CrossRef]
- Guo, Q.; Yoshida, Y.; Major, I.T.; Wang, K.; Sugimoto, K.; Kapali, G.; Havko, N.E.; Benning, C.; Howe, G.A. JAZ repressors of metabolic defense promote growth and reproductive fitness in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E10768–E10777. [Google Scholar] [CrossRef]
- Bisht, N.; Anshu, A.; Singh, P.C.; Chauhan, P.S. Comprehensive analysis of OsJAZ gene family deciphers rhizobacteria-mediated nutrient stress modulation in rice. Int. J. Biol. Macromol. 2023, 253, 126832. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Ben-Romdhane, W.; Hassairi, A.; Aboul-Soud, M.A.M. Identification of TIFY/JAZ family genes in Solanum lycopersicum and their regulation in response to abiotic stresses. PLoS ONE 2017, 12, e0177381. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Luthe, D. Identification and evolution analysis of the JAZ gene family in maize. BMC Genom. 2021, 2, 256. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Wang, S.; Long, L.; Yu, X.; Cai, H.; Chen, P.; Gu, L.; Yang, M. Genome-wide identification and expression analysis of PtJAZ gene family in poplar (Populus trichocarpa). BMC Genom. Data 2023, 24, 55. [Google Scholar] [CrossRef]
- Zhao, X.; He, Y.; Liu, Y.; Wang, Z.; Zhao, J. JAZ proteins: Key regulators of plant growth and stress response. Crop J. 2024, 12, 1505–1516. [Google Scholar] [CrossRef]
- Pauwels, L.; Barbero, G.F.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Pérez, A.C.; Chico, J.M.; Bossche, R.V.; Sewell, J.; Gil, E.; et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 2010, 464, 788–791. [Google Scholar] [CrossRef]
- Zhang, F.; Yao, J.; Ke, J.; Zhang, L.; Lam, V.Q.; Xin, X.; Zhou, E.; Chen, J.; Brunzelle, J.; Griffin, P.R.; et al. Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling. Nature 2015, 525, 269–273. [Google Scholar] [CrossRef]
- Li, J.; Chen, Y.; Zhou, G.; Li, M. Phytohormones and candidate genes synergistically regulate fruitlet abscission in Areca catechu L. BMC Plant Biol. 2023, 23, 537. [Google Scholar] [CrossRef]
- Howe, G.A.; Yoshida, Y. Evolutionary Origin of JAZ Proteins and Jasmonate Signaling. Mol. Plant 2019, 12, 153–155. [Google Scholar] [CrossRef]
- Johnson, L.Y.D.; Major, I.T.; Chen, Y.; Yang, C.; Vanegas-Cano, L.J.; Howe, G.A. Diversification of JAZ-MYC signaling function in immune metabolism. New Phytol. 2023, 239, 2277–2291. [Google Scholar] [CrossRef]
- Chini, A.; Fonseca, S.; Fernández, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; García-Casado, G.; López-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Jiao, W.B.; Schneeberger, K. Chromosome-level assemblies of multiple Arabidopsis genomes reveal hotspots of rearrangements with altered evolutionary dynamics. Nat. Commun. 2020, 11, 989. [Google Scholar] [CrossRef] [PubMed]
- Stein, J.C.; Yu, Y.; Copetti, D.; Zwickl, D.J.; Zhang, L.; Zhang, C.; Chougule, K.; Gao, D.; Iwata, A.; Goicoechea, J.L.; et al. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat. Genet. 2018, 50, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zou, Z.; Xing, H.; Duan, Y.; Zhu, X.; Ma, Y.; Wang, Y.; Fang, W. Genome-Wide Analysis Reveals Stress and Hormone Responsive Patterns of JAZ Family Genes in Camellia sinensis. Int. J. Mol. Sci. 2020, 21, 2433. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Ma, Y.; Qin, T.; Tang, W.; Qi, X.; Wang, X.; Liu, R.; Fang, H.; Chen, Z.; Liang, C.; et al. Transcriptome-Wide Identification and Characterization of the JAZ Gene Family in Mentha canadensis L. Int. J. Mol. Sci. 2021, 220, 8859. [Google Scholar] [CrossRef]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef]
- Shyu, C.; Figueroa, P.; DePew, C.L.; Cooke, T.F.; Sheard, L.B.; Moreno, J.E.; Katsir, L.; Zheng, N.; Browse, J.; Howe, G.A. JAZ8 Lacks a Canonical Degron and Has an EAR Motif That Mediates Transcriptional Repression of Jasmonate Responses in Arabidopsis. Plant Cell 2012, 24, 536–550. [Google Scholar] [CrossRef]
- Thatcher, L.F.; Cevik, V.; Grant, M.; Zhai, B.; Jones, J.D.G.; Manners, J.M.; Kazan, K. Characterization of aJAZ7 activation-tagged Arabidopsis mutant with increased susceptibility to the fungal pathogen Fusarium oxysporum. J. Exp. Bot. 2016, 67, 2367–2386. [Google Scholar] [CrossRef]
- Yuan, Z.; Zhang, D. Roles of jasmonate signalling in plant inflorescence and flower development. Curr. Opin. Plant Biol. 2015, 27, 44–51. [Google Scholar] [CrossRef]
- Pauwels, L.; Goossens, A. The JAZ Proteins: A Crucial Interface in the Jasmonate Signaling Cascade. Plant Cell 2011, 23, 3089–3100. [Google Scholar] [CrossRef]
- Acosta, I.F.; Laparra, H.; Romero, S.P.; Schmelz, E.; Hamberg, M.; Mottinger, J.P.; Moreno, M.A.; Dellaporta, S.L. tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science 2009, 323, 262–265. [Google Scholar] [CrossRef] [PubMed]
- DeLong, A.; Calderon-Urrea, A.; Dellaporta, S.L. Sex determination gene TASSELSEED2 of maize encodes a short-chain alcohol dehydrogenase required for stage-specific floral organ abortion. Cell 1993, 74, 757–768. [Google Scholar] [CrossRef] [PubMed]
- Lunde, C.; Kimberlin, A.; Leiboff, S.; Koo, A.J.; Hake, S. Tasselseed5 overexpresses a wound-inducible enzyme, ZmCYP94B1, that affects jasmonate catabolism, sex determination, and plant architecture in maize. Commun. Biol. 2019, 2, 114. [Google Scholar] [CrossRef] [PubMed]
- Hayward, A.P.; Moreno, M.A.; Howard, T.P., 3rd; Hague, J.; Nelson, K.; Heffelfinger, C.; Romero, S.; Kausch, A.P.; Glauser, G.; Acosta, I.F.; et al. Control of sexuality by the sk1-encoded UDP-glycosyltransferase of maize. Sci. Adv. 2016, 2, e1600991. [Google Scholar] [CrossRef]
- Riemann, M.; Haga, K.; Shimizu, T.; Okada, K.; Ando, S.; Mochizuki, S.; Nishizawa, Y.; Yamanouchi, U.; Nick, P.; Yano, M.; et al. Identification of rice Allene Oxide Cyclase mutants and the function of jasmonate for defence against Magnaporthe oryzae. Plant J. 2013, 74, 226–238. [Google Scholar] [CrossRef]
| Rename | Gene ID | Chr | AA Length | MW (KDa) | pI |
|---|---|---|---|---|---|
| AcJAZ1 | AC02G143000.1 | 2 | 158 | 17.92 | 9.33 |
| AcJAZ2 | AC03G008620.1 | 3 | 313 | 33.66 | 9.98 |
| AcJAZ3 | AC03G012030.1 | 3 | 423 | 44.45 | 8.99 |
| AcJAZ4 | AC03G015370.1 | 3 | 190 | 19.95 | 5.54 |
| AcJAZ5 | AC03G016350.1 | 3 | 269 | 29.25 | 9.05 |
| AcJAZ6 | AC04G007780.1 | 4 | 210 | 22.86 | 7.93 |
| AcJAZ7 | AC05G004750.1 | 5 | 393 | 42.99 | 9.30 |
| AcJAZ8 | AC05G022110.1 | 5 | 465 | 48.66 | 9.55 |
| AcJAZ9 | AC05G066040.1 | 5 | 278 | 29.78 | 9.19 |
| AcJAZ10 | AC05G067940.1 | 5 | 189 | 19.63 | 5.93 |
| AcJAZ11 | AC06G024110.1 | 6 | 202 | 21.72 | 10.52 |
| AcJAZ12 | AC07G003060.1 | 7 | 195 | 21.33 | 9.77 |
| AcJAZ13 | AC14G051720.1 | 14 | 217 | 24.78 | 9.05 |
| AcJAZ14 | AC15G004960.1 | 15 | 283 | 30.90 | 9.13 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, J.; Ji, C.; Wen, X.; Li, H.; Yang, F. Comprehensive Analysis of JAZ Family Genes Involved in Sex Differentiation in Areca catechu. Forests 2025, 16, 1133. https://doi.org/10.3390/f16071133
Du J, Ji C, Wen X, Li H, Yang F. Comprehensive Analysis of JAZ Family Genes Involved in Sex Differentiation in Areca catechu. Forests. 2025; 16(7):1133. https://doi.org/10.3390/f16071133
Chicago/Turabian StyleDu, Jin, Changlei Ji, Xinyu Wen, Han Li, and Fusun Yang. 2025. "Comprehensive Analysis of JAZ Family Genes Involved in Sex Differentiation in Areca catechu" Forests 16, no. 7: 1133. https://doi.org/10.3390/f16071133
APA StyleDu, J., Ji, C., Wen, X., Li, H., & Yang, F. (2025). Comprehensive Analysis of JAZ Family Genes Involved in Sex Differentiation in Areca catechu. Forests, 16(7), 1133. https://doi.org/10.3390/f16071133






