The Complete Chloroplast Genome of Purdom’s Rhododendron (Rhododendron purdomii Rehder & E. H. Wilson): Genome Structure and Phylogenetic Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material, DNA Extraction, and Sequencing
2.2. Assembly and Annotation of Chloroplast Genome
2.3. Data Analysis
2.3.1. Repeat Sequence Analysis
2.3.2. Comparative Analysis of the Chloroplast Genome
2.3.3. Phylogenetic Analysis
3. Results
3.1. Chloroplast Genome Structure
3.2. Repeat Sequences
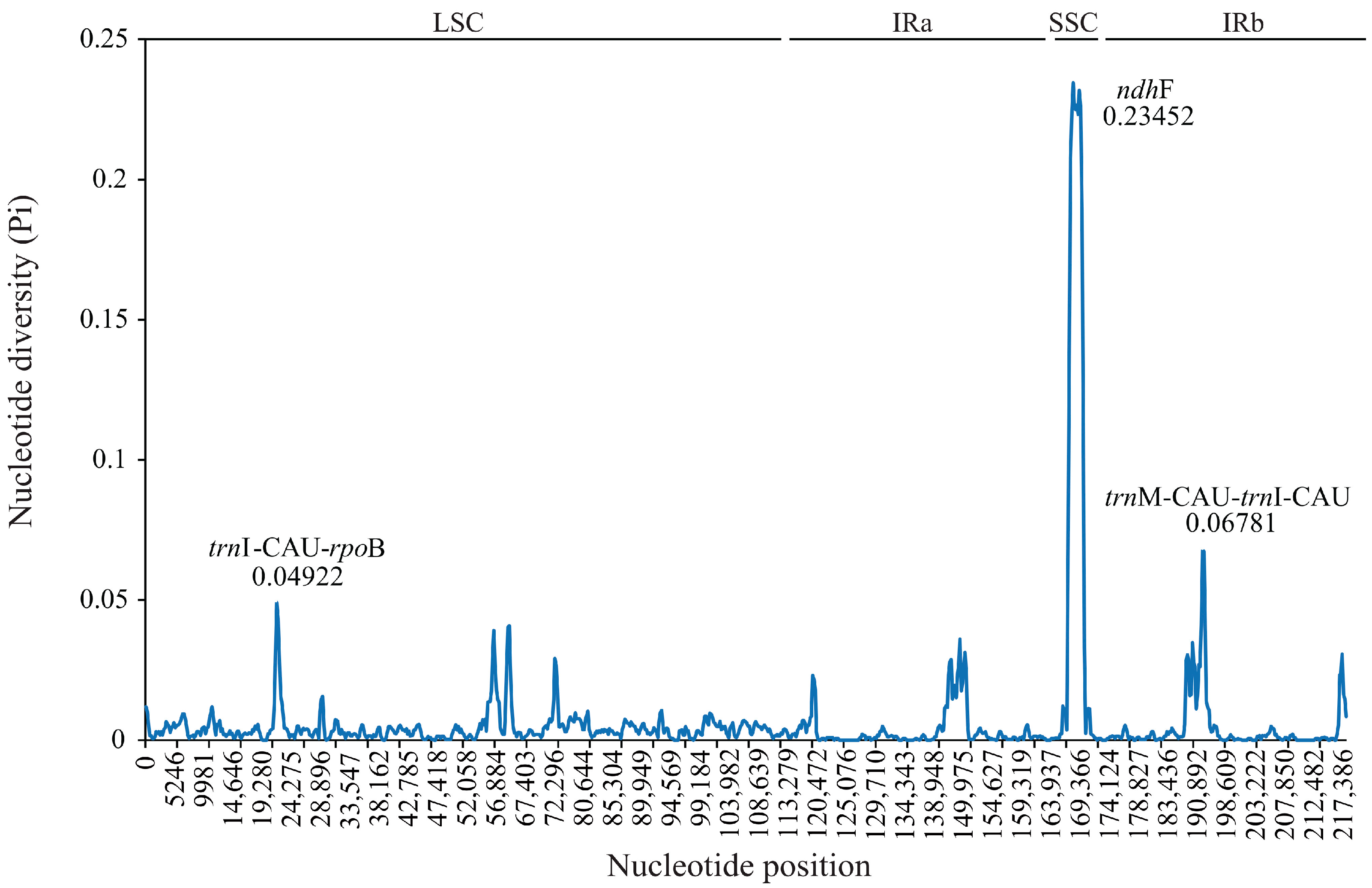
3.3. Genome Comparison and Nucleotide Diversity
3.4. Phylogenetic Relationship
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shaw, J.; Lickey, E.B.; Schilling, E.E.; Small, R.L. Comparison of Whole Chloroplast Genome Sequences to Choose Noncoding Regions for Phylogenetic Studies in Angiosperms: The Tortoise and the Hare III. Am. J. Bot. 2007, 94, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Wicke, S.; Schneeweiss, G.M.; dePamphilis, C.W.; Müller, K.F.; Quandt, D. The Evolution of the Plastid Chromosome in Land Plants: Gene Content, Gene Order, Gene Function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.K.; Ruhlman, T.A. Plastid Genomes of Seed Plants. In Genomics of Chloroplasts and Mitochondria; Bock, R., Knoop, V., Eds.; Springer: Dordrecht, The Netherlands, 2012; pp. 103–126. ISBN 978-94-007-2920-9. [Google Scholar]
- Jin, J.-J.; Yu, W.-B.; Yang, J.-B.; Song, Y.; dePamphilis, C.W.; Yi, T.-S.; Li, D.-Z. GetOrganelle: A Fast and Versatile Toolkit for Accurate de Novo Assembly of Organelle Genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.; Guo, W.; Gupta, S.; Fan, W.; Mower, J.P. Evolutionary Dynamics of the Plastid Inverted Repeat: The Effects of Expansion, Contraction, and Loss on Substitution Rates. New Phytol. 2016, 209, 1747–1756. [Google Scholar] [CrossRef]
- Rather, S.A.; Wang, K.; Wang, T.; Liu, H.; Schneider, H. Comparative Chloroplast Genome Analysis Reveals Powerful Barcodes for Combatting Illegal Logging of CITES-Listed Threatened Asian Rosewoods (Dalbergia, Leguminosae, Papilionoideae). Bot. J. Linn. Soc. 2024, boae086. [Google Scholar] [CrossRef]
- Mo, Z.-Q.; Fu, C.-N.; Zhu, M.-S.; Milne, R.I.; Yang, J.-B.; Cai, J.; Qin, H.-T.; Zheng, W.; Hollingsworth, P.M.; Li, D.-Z.; et al. Resolution, Conflict and Rate Shifts: Insights from a Densely Sampled Plastome Phylogeny for Rhododendron (Ericaceae). Ann. Bot. 2022, 130, 687–701. [Google Scholar] [CrossRef]
- López, M.G.; Fass, M.; Rivas, J.G.; Carbonell-Caballero, J.; Vera, P.; Puebla, A.; Defacio, R.; Dopazo, J.; Paniego, N.; Hopp, H.E.; et al. Plastome Genomics in South American Maize Landraces: Chloroplast Lineages Parallel the Geographical Structuring of Nuclear Gene Pools. Ann. Bot. 2021, 128, 115–125. [Google Scholar] [CrossRef]
- Fang, M.; Fang, R.; He, M.; Hu, L.; Yang, H.; Chamberlain, D. Flora of China—Apiaceae through Ericaceae; Zhengyi, W., Raven, P.H., Deyuan, H., Eds.; Science Press: Beijing, China, 2005; Volume 14, pp. 260–455. [Google Scholar]
- Ma, Y.; Nielsen, J.; Chamberlain, D.F.; Li, X.; Sun, W. The Conservation of Rhododendrons Is of Greater Urgency than Has Been Previously Acknowledged in China. Biodivers. Conserv. 2014, 23, 3149–3154. [Google Scholar] [CrossRef]
- Chamberlain, D.; Hyam, R.; Argent, G.; Fairweather, G.; Walter, K.S. The Genus Rhododendron: Its Classification and Synonymy; Royal Botanic Garden Edinburgh: Edinburgh, UK, 1996. [Google Scholar]
- Popescu, R.; Kopp, B. The Genus Rhododendron: An Ethnopharmacological and Toxicological Review. J. Ethnopharmacol. 2013, 147, 42–62. [Google Scholar] [CrossRef]
- Gibbs, D. The Red List of Rhododendrons; Botanic Gardens Conservation International: Richmond, UK, 2011. [Google Scholar]
- Yu, F.; Skidmore, A.K.; Wang, T.; Huang, J.; Ma, K.; Groen, T.A. Rhododendron Diversity Patterns and Priority Conservation Areas in China. Divers. Distrib. 2017, 23, 1143–1156. [Google Scholar] [CrossRef]
- Milne, R.I.; Abbott, R.J. Reproductive Isolation among Two Interfertile Rhododendron Species: Low Frequency of Post-F1 Hybrid Genotypes in Alpine Hybrid Zones. Mol. Ecol. 2008, 17, 1108–1121. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Mao, X.; Wang, J.; Zhang, L.; Jiang, Y.; Geng, Y.; Ma, T.; Cai, L.; Huang, S.; Hollingsworth, P.; et al. Pervasive Hybridization during Evolutionary Radiation of Rhododendron Subgenus Hymenanthes in Mountains of Southwest China. Natl. Sci. Rev. 2022, 9, nwac276. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.-M.; Yang, M.-Q.; Li, C.-L.; Huang, S.-X.; Jin, W.-T.; Shen, T.-T.; Wang, F.; Li, X.-H.; Yoichi, W.; Zhang, L.-H.; et al. Spatiotemporal Evolution of the Global Species Diversity of Rhododendron. Mol. Biol. Evol. 2022, 39, msab314. [Google Scholar] [CrossRef]
- Zheng, W.; Yan, L.-J.; Burgess, K.S.; Luo, Y.-H.; Zou, J.-Y.; Qin, H.-T.; Wang, J.-H.; Gao, L.-M. Natural Hybridization among Three Rhododendron Species (Ericaceae) Revealed by Morphological and Genomic Evidence. BMC Plant Biol. 2021, 21, 529. [Google Scholar] [CrossRef]
- Yan, L.-J.; Liu, J.; Möller, M.; Zhang, L.; Zhang, X.-M.; Li, D.-Z.; Gao, L.-M. DNA Barcoding of Rhododendron (Ericaceae), the Largest Chinese Plant Genus in Biodiversity Hotspots of the Himalaya-Hengduan Mountains. Mol. Ecol. Resour. 2015, 15, 932–944. [Google Scholar] [CrossRef]
- Fu, C.-N.; Mo, Z.-Q.; Yang, J.-B.; Cai, J.; Ye, L.-J.; Zou, J.-Y.; Qin, H.-T.; Zheng, W.; Hollingsworth, P.M.; Li, D.-Z.; et al. Testing Genome Skimming for Species Discrimination in the Large and Taxonomically Difficult Genus Rhododendron. Mol. Ecol. Resour. 2022, 22, 404–414. [Google Scholar] [CrossRef]
- Dong, H.; Zhang, N.; Shen, S.; Zhu, S.; Fan, S.; Lu, Y. Effects of Climate Change on the Spatial Distribution of the Threatened Species Rhododendron purdomii in Qinling-Daba Mountains of Central China: Implications for Conservation. Sustainability 2023, 15, 3181. [Google Scholar] [CrossRef]
- Zhang, N.; Qin, M.; Zhu, S.; Huang, Z.; Dong, H.; Yang, Y.; Yang, L.; Lu, Y. Development and Characterization of Microsatellite Markers for Rhododendron purdomii (Ericaceae) Using next-Generation Sequencing. Genes Genet. Syst. 2021, 96, 253–257. [Google Scholar] [CrossRef]
- Yang, J. Study on Chemical Constituents from Three Medical Plants and Their Bio-Activities. Ph.D. Thesis, College of Chemistry&Pharmacy, Northwest A&F University, Xianyang, China, 2025. (In Chinese). [Google Scholar] [CrossRef]
- Qin, H.; Yang, Y.; Dong, S.; He, Q.; Jia, Y.; Zhao, L.; Yu, S.; Liu, H.; Liu, B.; Yan, Y. Threatened Species List of China’s Higher Plants. Biodiv. Sci. 2017, 25, 696–744. [Google Scholar] [CrossRef]
- Si, G.; Zhang, Y.; Zhao, B.; Xu, H. Phenotypic Variation of Natural Populations in Rhododendron purdomii in Qinling Mountains. Acta Bot. Boreali-Occident. Sin. 2012, 32, 1560–1566. (In Chinese) [Google Scholar]
- Zhao, B.; Yin, Z.; Xu, M.; Wang, Q. AFLP Analysis of Genetic Variation in Wild Populations of Five Rhododendron Species in Qinling Mountain in China. Biochem. Syst. Ecol. 2012, 45, 198–205. [Google Scholar] [CrossRef]
- Chamberlain, D.F. A Revision of Rhododendron II. Subgenus Hymenanthes. Notes R. Bot. Gard. Edinb. 1982, 39, 209–486. [Google Scholar]
- Geng, Y. The Genus Rhododendron of China; Shanghai Scientific and Technical Publishers: Shanghai, China, 2014. [Google Scholar]
- Doyle, J.J.; Dickson, E.E. Preservation of Plant Samples for Dna Restriction Endonuclease Analysis. TAXON 1987, 36, 715–722. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of Long, Error-Prone Reads Using Repeat Graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Zhao, Q.-Y.; Wang, Y.; Kong, Y.-M.; Luo, D.; Li, X.; Hao, P. Optimizing de Novo Transcriptome Assembly from Short-Read RNA-Seq Data: A Comparative Study. BMC Bioinform. 2011, 12, S2. [Google Scholar] [CrossRef]
- Liu, C.; Shi, L.; Zhu, Y.; Chen, H.; Zhang, J.; Lin, X.; Guan, X. CpGAVAS, an Integrated Web Server for the Annotation, Visualization, Analysis, and GenBank Submission of Completely Sequenced Chloroplast Genome Sequences. BMC Genom. 2012, 13, 715. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved Detection and Functional Classification of Transfer RNA Genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Zheng, S.; Poczai, P.; Hyvönen, J.; Tang, J.; Amiryousefi, A. Chloroplot: An Online Program for the Versatile Plotting of Organelle Genomes. Front. Genet. 2020, 11, 576124. [Google Scholar] [CrossRef]
- Sharp, P.M.; Li, W.-H. The Codon Adaptation Index-a Measure of Directional Synonymous Codon Usage Bias, and Its Potential Applications. Nucleic Acids Res. 1987, 15, 1281–1295. [Google Scholar] [CrossRef]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The Manifold Applications of Repeat Analysis on a Genomic Scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef] [PubMed]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-Web: A Web Server for Microsatellite Prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An Integrated and Scalable Desktop Platform for Streamlined Molecular Sequence Data Management and Evolutionary Phylogenetics Studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Bio Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational Tools for Comparative Genomics. Nucleic Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef]
- Amiryousefi, A.; Hyvönen, J.; Poczai, P. IRscope: An Online Program to Visualize the Junction Sites of Chloroplast Genomes. Bioinformatics 2018, 34, 3030–3031. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v3: An Online Tool for the Display and Annotation of Phylogenetic and Other Trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Zhou, X.-J.; Wei, M.-J.; Zhang, K.; Han, J.-W.; Wang, H.-L.; Dong, S.-W. Characterization of the Complete Chloroplast Genome of Rhododendron henanense Subsp. Lingbaoense Fang. Mitochondrial DNA B 2021, 6, 3325–3326. [Google Scholar] [CrossRef]
- Liu, D.; Fu, C.; Yin, L.; Ma, Y. Complete Plastid Genome of Rhododendron Griersonianum, a Critically Endangered Plant with Extremely Small Populations (PSESP) from Southwest China. Mitochondrial DNA B 2020, 5, 3086–3087. [Google Scholar] [CrossRef]
- Li, H.; Guo, Q.; Li, Q.; Yang, L. Long-Reads Reveal That Rhododendron Delavayi Plastid Genome Contains Extensive Repeat Sequences, and Recombination Exists among Plastid Genomes of Photosynthetic Ericaceae. PeerJ 2020, 8, e9048. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Cao, Y.; Yao, Y.; Zhang, Y. The Complete Chloroplast Genome Sequence of Rhododendron oreodoxa Var. Fargesii (Ericaceae), an Ornamental Plant. Mitochondrial DNA B 2022, 7, 1234–1236. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Gagen, M.J. The Evolution of Controlled Multitasked Gene Networks: The Role of Introns and Other Noncoding RNAs in the Development of Complex Organisms. Mol. Biol. Evol. 2001, 18, 1611–1630. [Google Scholar] [CrossRef]
- Behura, S.K.; Severson, D.W. Codon Usage Bias: Causative Factors, Quantification Methods and Genome-Wide Patterns: With Emphasis on Insect Genomes. Biol. Rev. Camb. Philos. Soc. 2013, 88, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Li, X.; Zhu, X.; Huang, X.; Jin, S. The Complete Plastid Genome of Rhododendron Pulchrum and Comparative Genetic Analysis of Ericaceae Species. Forests 2020, 11, 158. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, M.; Song, L. Structural Characterization of Four Rhododendron Spp. Chloroplast Genomes and Comparative Analyses with Other Azaleas. Biocell 2023, 47, 657–668. [Google Scholar] [CrossRef]
- Morton, B.R. The Role of Context-Dependent Mutations in Generating Compositional and Codon Usage Bias in Grass Chloroplast DNA. J. Mol. Evol. 2003, 56, 616–629. [Google Scholar] [CrossRef]
- Cavalier-Smith, T. Chloroplast Evolution: Secondary Symbiogenesis and Multiple Losses. Curr. Biol. 2002, 12, R62–R64. [Google Scholar] [CrossRef]
- Jiang, D. The Complete Chloroplast Genome Sequence of Artemisia stechmanniana (Asteraceae): Genome Structure and Phylogenetic Analysis. Biologia 2024, 79, 715–728. [Google Scholar] [CrossRef]
- Shen, J.; Huang, L.; Rong, X. Characteristics and Analysis of the Chloroplast Genome in Rhododendron farrerae. Mol. Plant Breed. 2025, 23, 600–607. [Google Scholar]
- Zong, D.; Gan, P.; Zhou, A.; Li, J.; Xie, Z.; Duan, A.; He, C. Comparative Analysis of the Complete Chloroplast Genomes of Seven Populus Species: Insights into Alternative Female Parents of Populus Tomentosa. PLoS ONE 2019, 14, e0218455. [Google Scholar] [CrossRef] [PubMed]
- Lin, N.; Zhang, X.; Deng, T.; Zhang, J.; Meng, A.; Wang, H.; Sun, H.; Sun, Y. Plastome Sequencing of Myripnois Dioica and Comparison within Asteraceae. Plant Divers. 2019, 41, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Nguyen, V.B.; Dong, J.; Wang, Y.; Park, J.Y.; Lee, S.-C.; Yang, T.-J. Evolution of the Araliaceae Family Inferred from Complete Chloroplast Genomes and 45S nrDNAs of 10 Panax-Related Species. Sci. Rep. 2017, 7, 4917. [Google Scholar] [CrossRef] [PubMed]
- Turmel, M.; Otis, C.; Lemieux, C. The Complete Chloroplast DNA Sequence of the Green Alga Nephroselmis Olivacea: Insights into the Architecture of Ancestral Chloroplast Genomes. Proc. Natl. Acad. Sci. USA 1999, 96, 10248–10253. [Google Scholar] [CrossRef]
- Wang, T.-R.; Wang, Z.-W.; Song, Y.-G.; Kozlowski, G. The Complete Chloroplast Genome Sequence of Quercus Ningangensis and Its Phylogenetic Implication. Plant Fungal Syst. 2021, 66, 155–165. [Google Scholar] [CrossRef]
- Kadam, S.K.; Youn, J.-S.; Tamboli, A.S.; Yang, J.; Pak, J.H.; Choo, Y.-S. Complete Chloroplast Genome Sequence of Artemisia Littoricola (Asteraceae) from Dokdo Island Korea: Genome Structure, Phylogenetic Analysis, and Biogeography Study. Funct. Integr. Genom. 2024, 24, 181. [Google Scholar] [CrossRef]
- Tamboli, A.S.; Yang, J.; Youn, J.-S.; Pak, J.H.; Choo, Y.-S. Intraspecific Variation within the Portulaca Oleracea L. from Republic of Korea Based on Chloroplast Genomes. Plant Biotechnol. Rep. 2024, 18, 939–951. [Google Scholar] [CrossRef]
- Park, J.; Min, J.; Kim, Y.; Chung, Y. The Comparative Analyses of Six Complete Chloroplast Genomes of Morphologically Diverse Chenopodium Album L. (Amaranthaceae) Collected in Korea. Int. J. Genom. 2021, 2021, e6643444. [Google Scholar] [CrossRef]
- Silva, S.R.; Pinheiro, D.G.; Penha, H.A.; Płachno, B.J.; Michael, T.P.; Meer, E.J.; Miranda, V.F.O.; Varani, A.M. Intraspecific Variation within the Utricularia Amethystina Species Morphotypes Based on Chloroplast Genomes. Int. J. Mol. Sci. 2019, 20, 6130. [Google Scholar] [CrossRef]
- Wang, J.; Luo, J.; Ma, Y.-Z.; Mao, X.-X.; Liu, J.-Q. Nuclear Simple Sequence Repeat Markers Are Superior to DNA Barcodes for Identification of Closely Related Rhododendron Species on the Same Mountain. J. Syst. Evol. 2019, 57, 278–286. [Google Scholar] [CrossRef]
- Kurashige, Y.; Etoh, J.-I.; Handa, T.; Takayanagi, K.; Yukawa, T. Sectional Relationships in the Genus Rhododendron (Ericaceae): Evidence from matK and trnK Intron Sequences. Plant Syst. Evol. 2001, 228, 1–14. [Google Scholar] [CrossRef]
- Gao, L.; Li, D.; Zhang, C. Phylogenetic relationships of Rhododendron section Azaleastrum (Ericaceae) based on ITS sequence. Acta Phytotax. Sin. 2003, 41, 173–179. [Google Scholar]
- Shen, J.; Li, X.; Li, M.; Cheng, H.; Huang, X.; Jin, S. Characterization, Comparative Phylogenetic, and Gene Transfer Analyses of Organelle Genomes of Rhododendron × Pulchrum. Front. Plant Sci. 2022, 13, 969765. [Google Scholar] [CrossRef] [PubMed]
- Meanchaipiboon, S.; Kobayashi, N.; Nakatsuka, A. Genetic Relationships Among Hirado Azalea Cultivars and Their Putative Parents Inferred from Flavonoid 3′, 5′ Hydroxylase Gene Sequences. Hortic. J. 2021, 90, 114–121. [Google Scholar] [CrossRef]
- Khan, G.; Nolzen, J.; Schepker, H.; Albach, D.C. Incongruent Phylogenies and Their Implications for the Study of Diversification, Taxonomy, and Genome Size Evolution of Rhododendron. Am. J. Bot. 2021, 108, 1957–1981. [Google Scholar] [CrossRef]

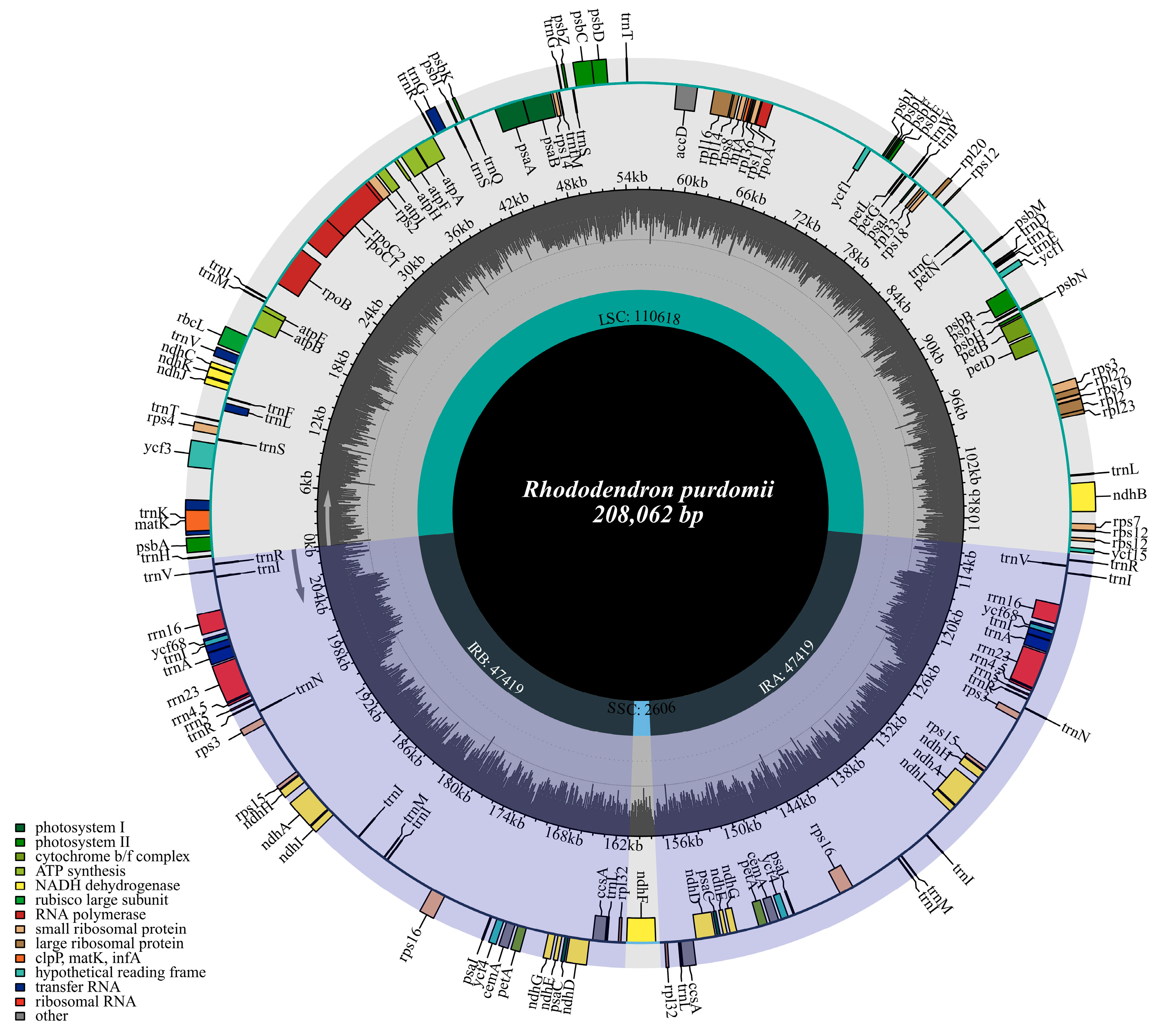
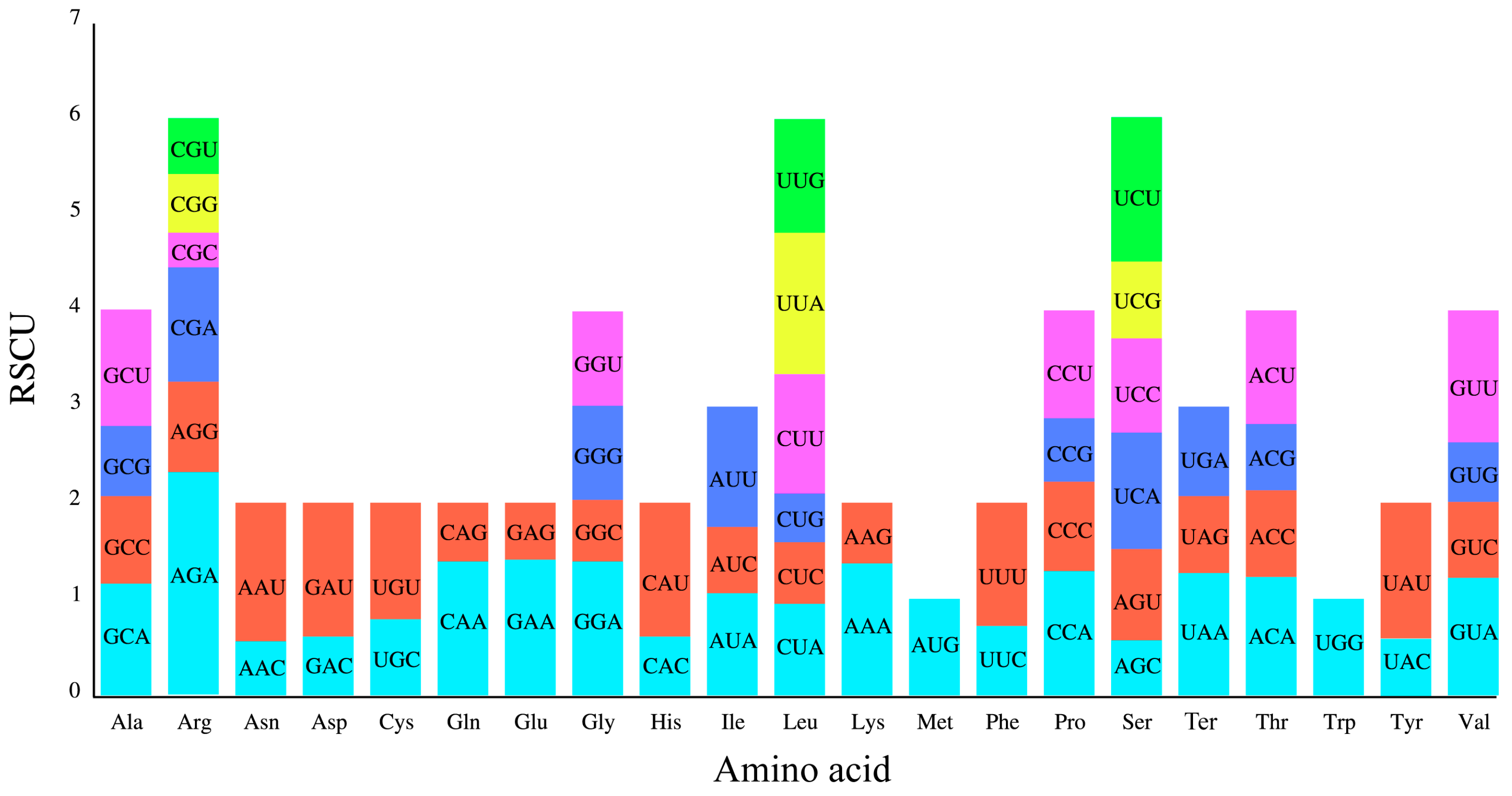
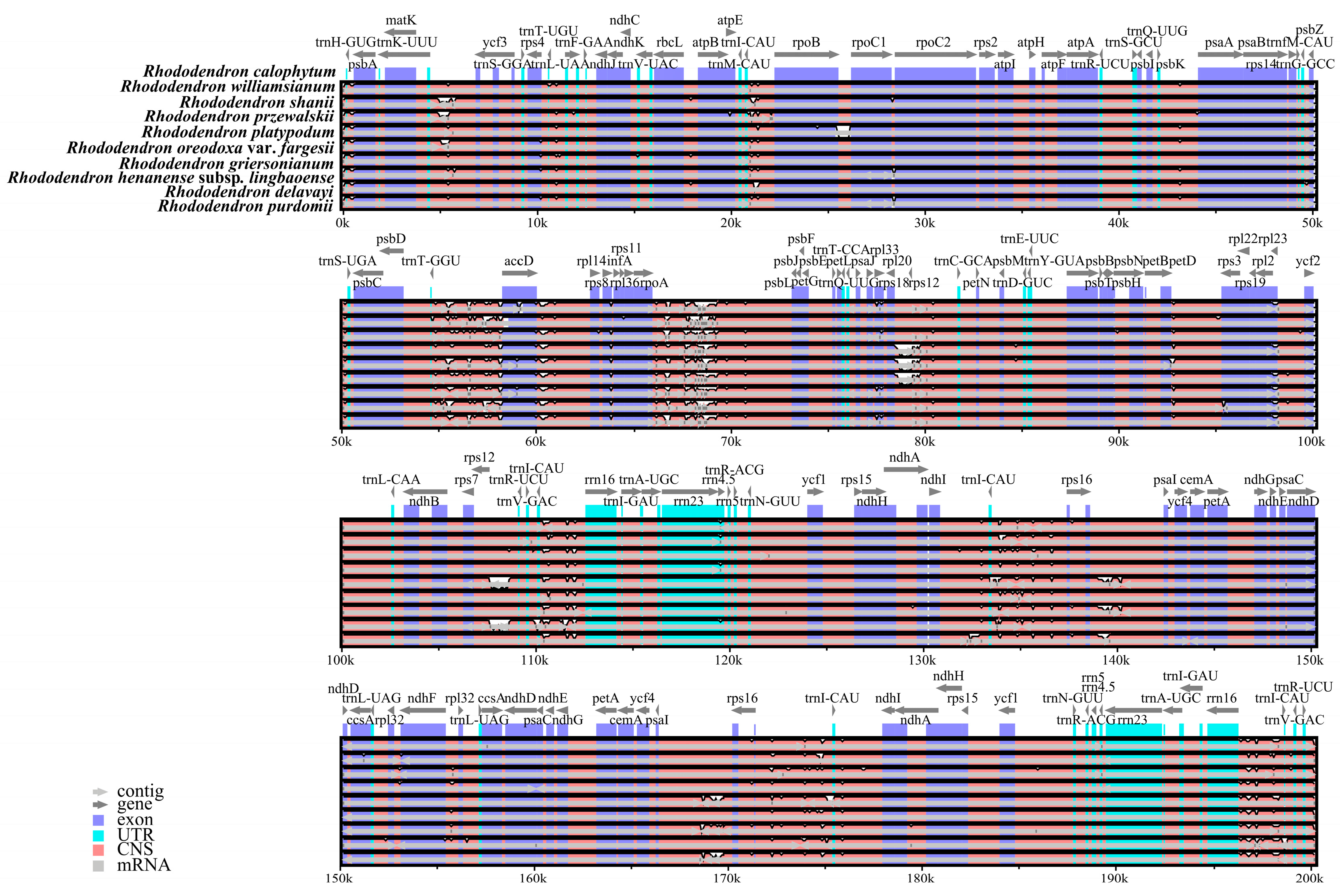
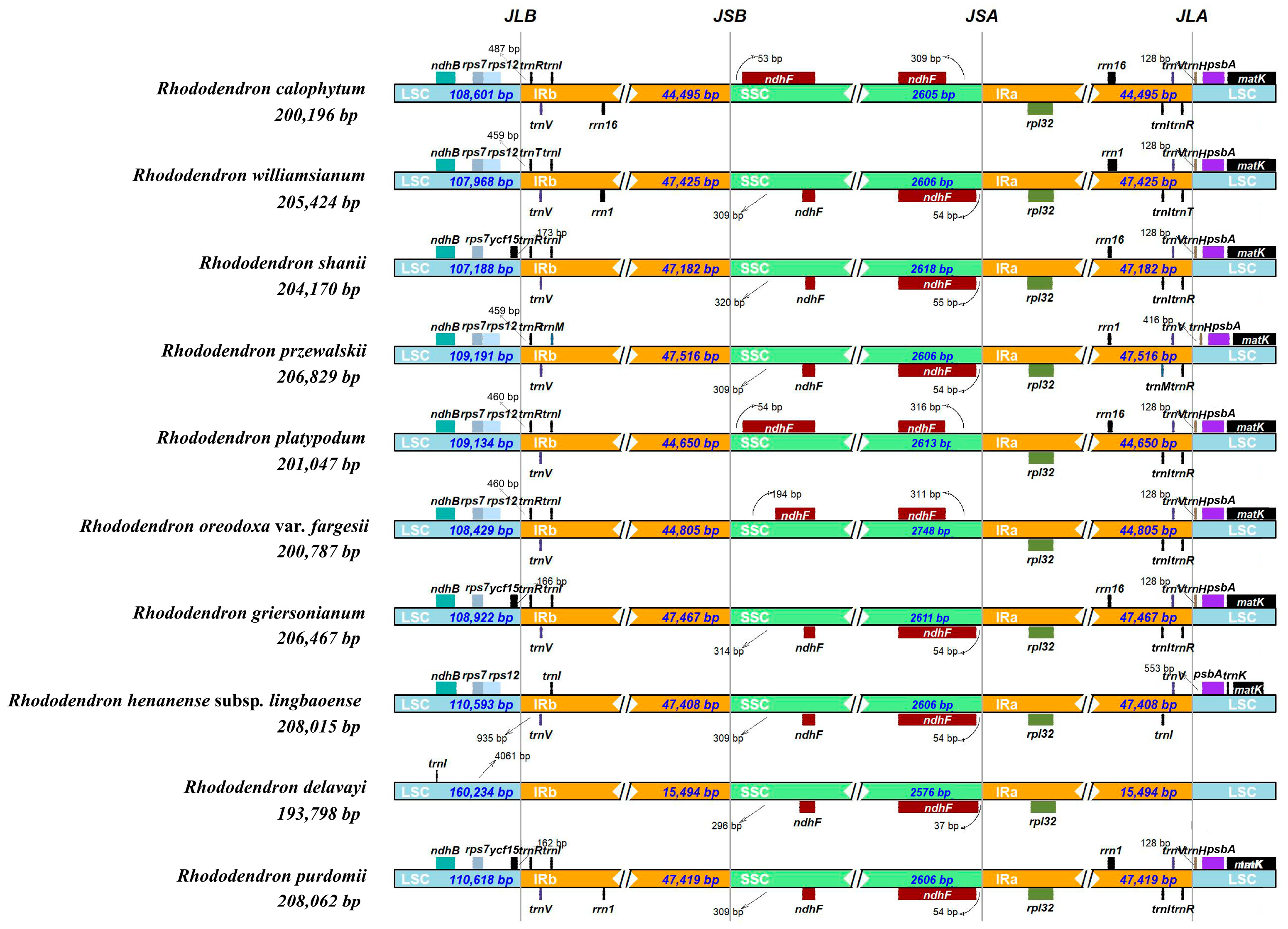

| Genome Features | Rh. purdomii |
|---|---|
| Genome size (bp)/GC content (%) | 208,062/35.81 |
| LSC size (bp)/GC content (%) | 110,618/35.27 |
| SSC size (bp)/GC content (%) | 2606/30.05 |
| IR size (bp)/GC content (%) | 47,419/36.59 |
| Total gene number | 146 |
| Protein-coding genes | 96 |
| tRNAs | 42 |
| rRNAs | 8 |
| Category of Genes | Group of Genes | Name of Genes | ||||
|---|---|---|---|---|---|---|
| Photosynthesis-related genes | photosystem I | psaA | psaB | psaC | psaI | psaJ |
| photosystem II | psbA | psbB | psbC | psbD | psbE | |
| psbF | psbI | psbJ | psbK | psbL | ||
| psbM | psbN | psbT | psbZ | psbH | ||
| NADH dehydrogenase | ndhA * | ndhB * | ndhC | ndhD | ndhE | |
| ndhF | ndhG | ndhI | ndhJ | ndhK | ||
| ndhH | ||||||
| ATP synthase | atpA atpI | atpB | atpE | atpF * | atpH | |
| rubisco | rbcL | |||||
| cytochrome b/f complex | petB * | petD * | petG | petL | petN | |
| petA | ||||||
| cytochrome c synthesis | ccsA | |||||
| Transcription- and translation-related genes | ribosome proteins | rpl2 rpl23 rps3 rps12 ** rps19 | rpl14 rpl32 rps4 rps14 | rpl16 * rpl33 rps7 rps15 | rpl20 rpl36 rps8 rps16 * | rpl22 rps2 rps11 rps18 |
| transcription | rpoA | rpoB | rpoC1 | rpoC2 | ||
| translational initiation factor | infA | |||||
| RNA genes | ribosomal RNA | rrn4.5 | rrn5 | rrn16 | rrn23 | |
| transfer RNA | trnA-UGC * | trnC-GCA | trnD-GUC | trnE-UUC | trnF-GAA | |
| trnfM-CAU | trnG-GCC | trnG-UCC * | trnH-GUG | trnI-CAU | ||
| trnI-GAU * | trnK-UUU * | trnL-CAA | trnL-UAA * | trnL-UAG | ||
| trnM-CAU | trnN-GUU | trnP-UGG | trnQ-UUG | trnR-ACG | ||
| trnR-UCU | trnS-GCU | trnS-GGA | trnS-UGA | trnT-GGU | ||
| trnT-UGU | trnV-GAC | trnV-UAC * | trnW-CCA | trnY-GUA | ||
| Other genes | fatty acid synthesis | accD | ||||
| carbon metabolism | cemA | |||||
| RNA processing | matK | |||||
| Unknown | conserved reading frames | ycf1 | ycf3 ** | ycf4 | ycf15 | ycf68 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, L.; Zhang, N.; Zhu, S.; Lu, Y. The Complete Chloroplast Genome of Purdom’s Rhododendron (Rhododendron purdomii Rehder & E. H. Wilson): Genome Structure and Phylogenetic Analysis. Forests 2025, 16, 1120. https://doi.org/10.3390/f16071120
Yuan L, Zhang N, Zhu S, Lu Y. The Complete Chloroplast Genome of Purdom’s Rhododendron (Rhododendron purdomii Rehder & E. H. Wilson): Genome Structure and Phylogenetic Analysis. Forests. 2025; 16(7):1120. https://doi.org/10.3390/f16071120
Chicago/Turabian StyleYuan, Lu, Ningning Zhang, Shixin Zhu, and Yang Lu. 2025. "The Complete Chloroplast Genome of Purdom’s Rhododendron (Rhododendron purdomii Rehder & E. H. Wilson): Genome Structure and Phylogenetic Analysis" Forests 16, no. 7: 1120. https://doi.org/10.3390/f16071120
APA StyleYuan, L., Zhang, N., Zhu, S., & Lu, Y. (2025). The Complete Chloroplast Genome of Purdom’s Rhododendron (Rhododendron purdomii Rehder & E. H. Wilson): Genome Structure and Phylogenetic Analysis. Forests, 16(7), 1120. https://doi.org/10.3390/f16071120





