Functional Potential and Network-Based Insights into the Rhizosphere Microbiomes of Quercus mongolica and Larix kaempferi Stands
Abstract
1. Introduction
2. Materials and Methods
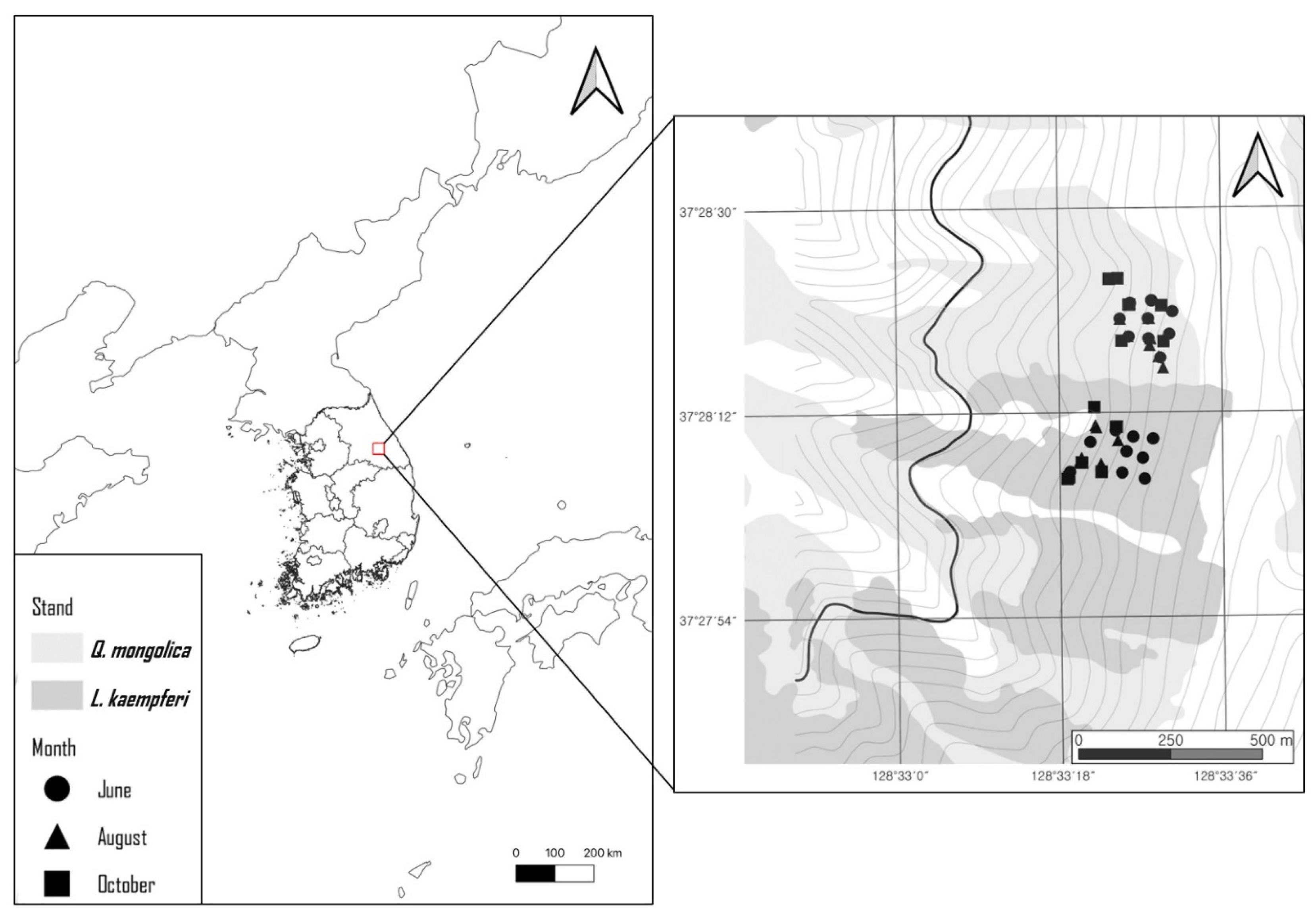
2.1. Sample Collection and Physicochemical Analysis
2.2. DNA Extraction and Sequencing
2.3. Diversity Analysis
2.4. Functional Potential Prediction
2.5. Microbial Network Analysis
3. Results
3.1. Soil Physicochemical Properties
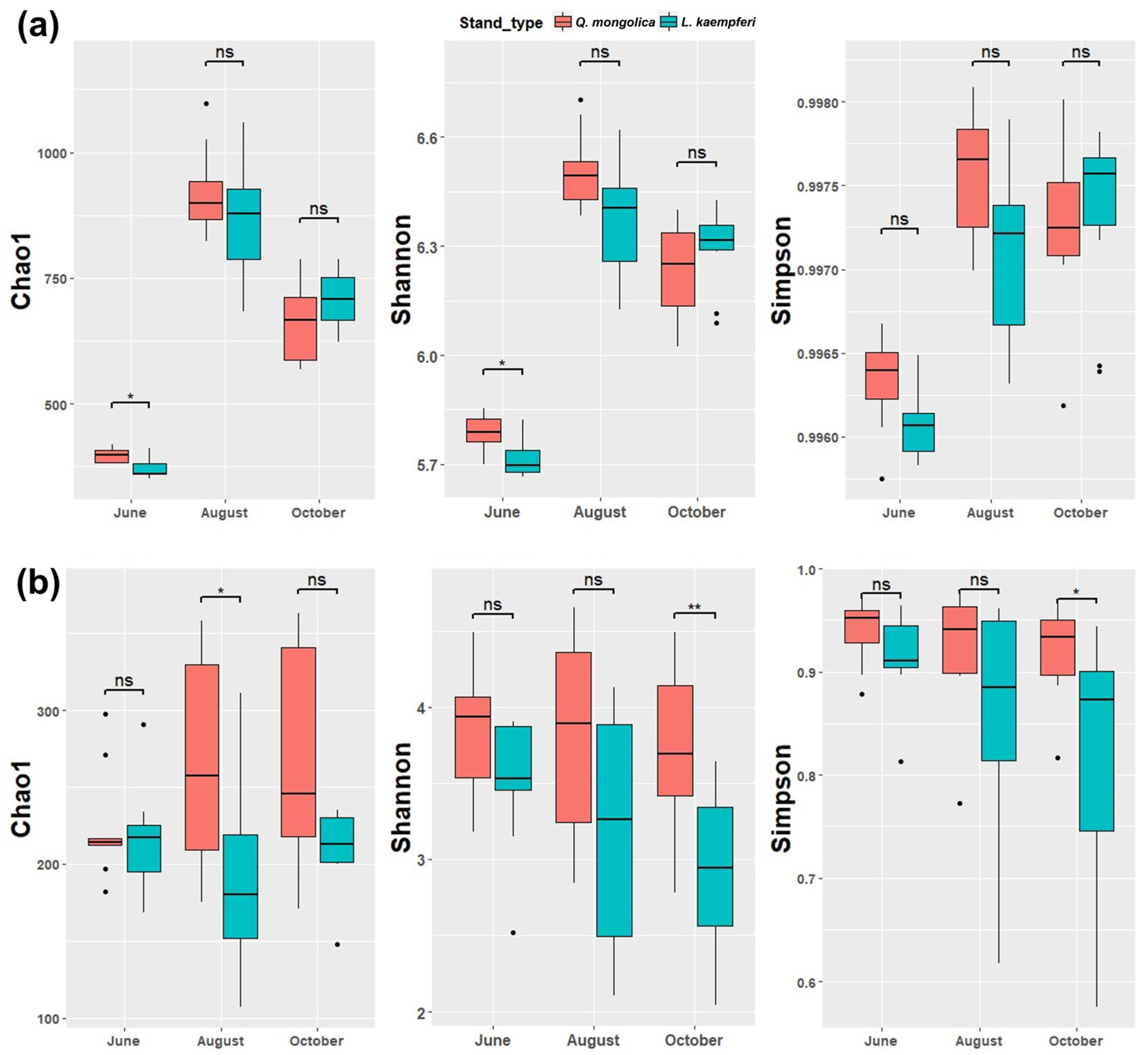
3.2. Alpha Diversity
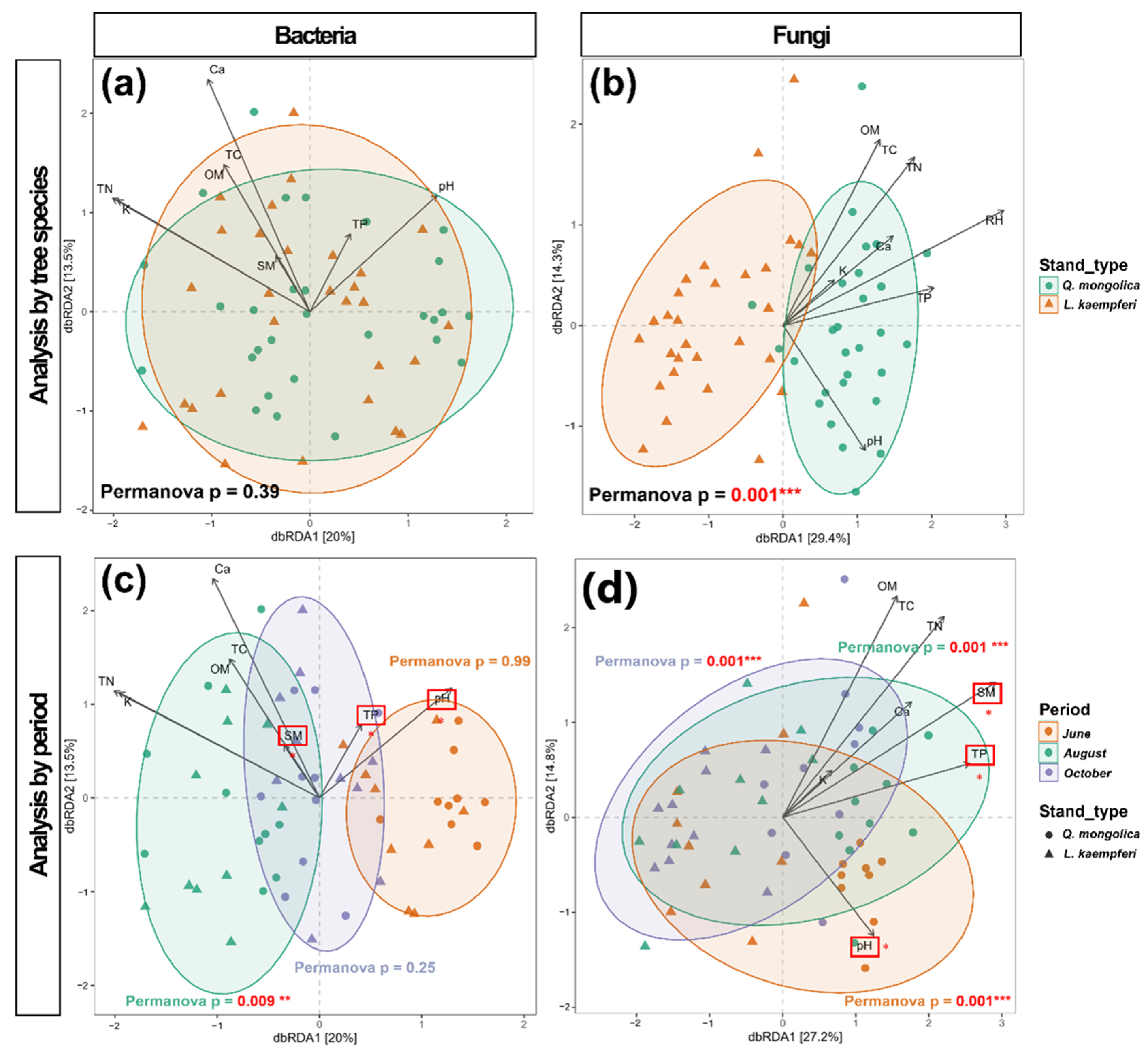
3.3. Beta Diversity
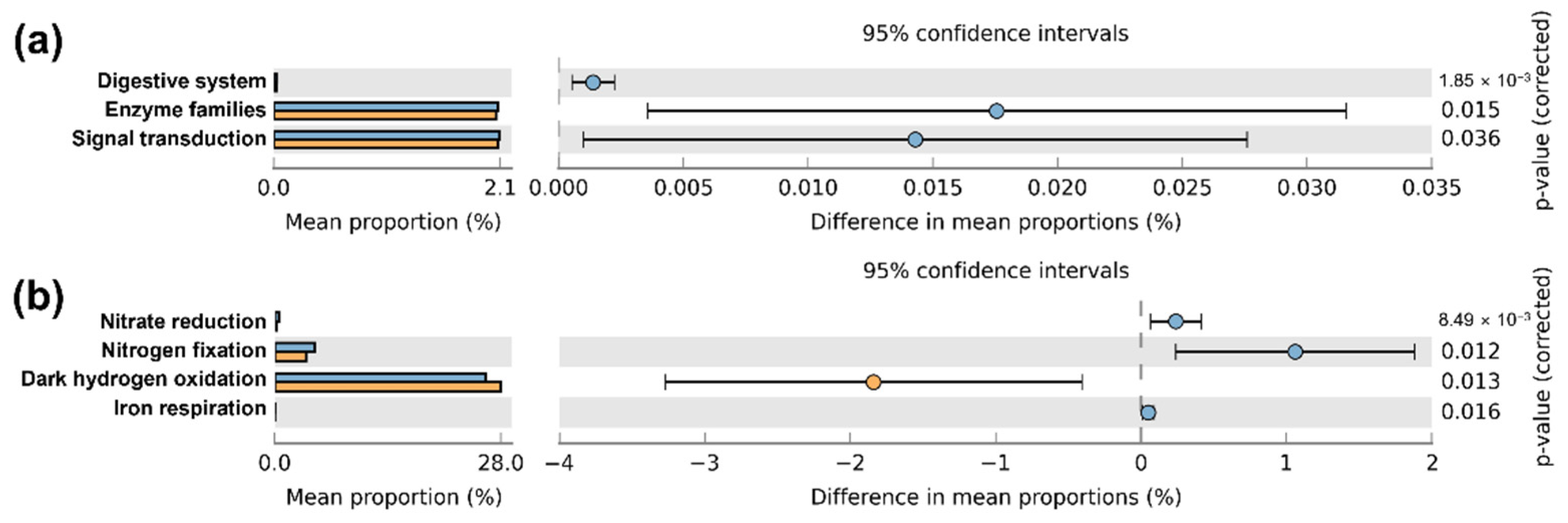
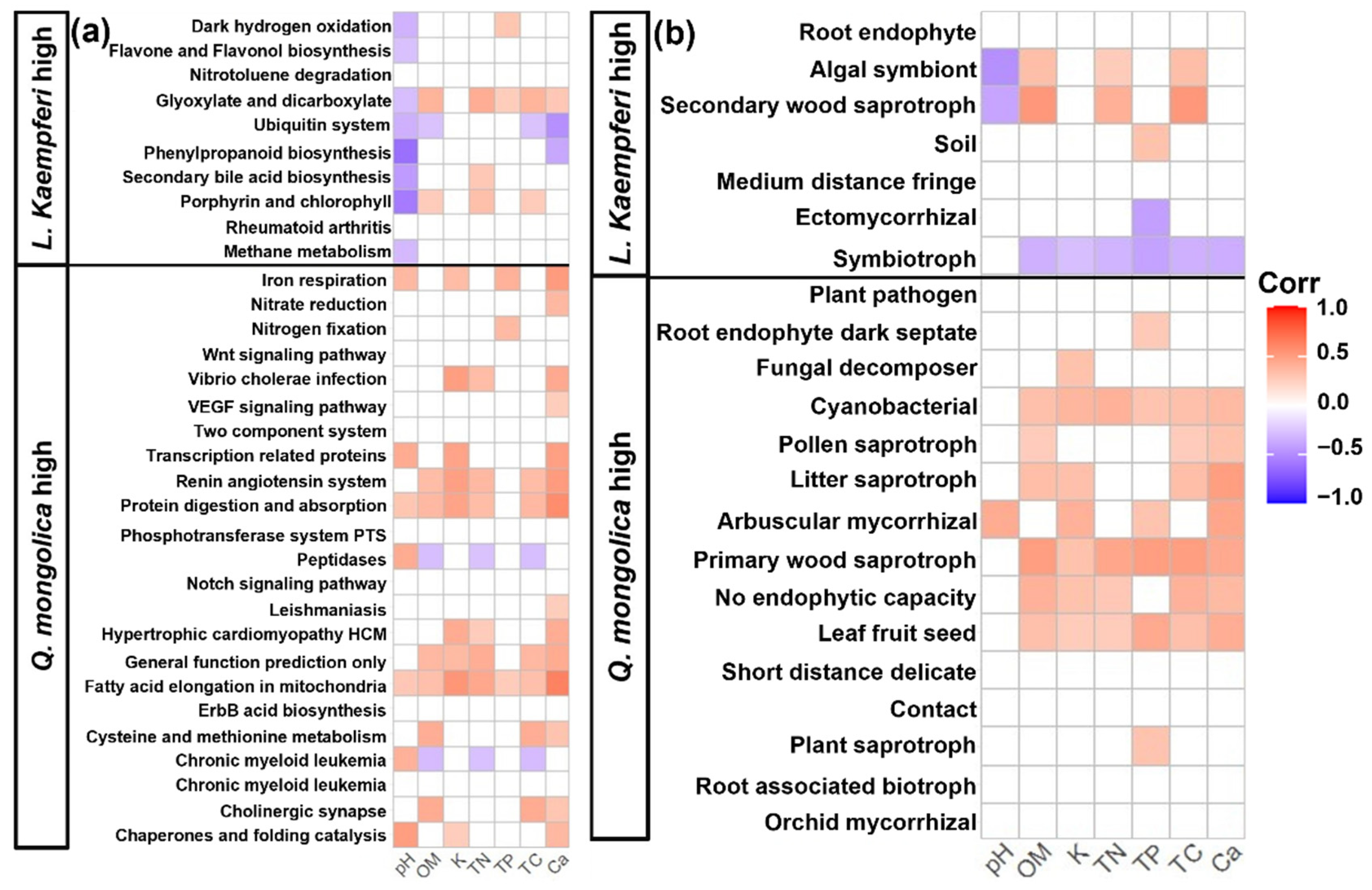
3.4. Functional Potential Prediction
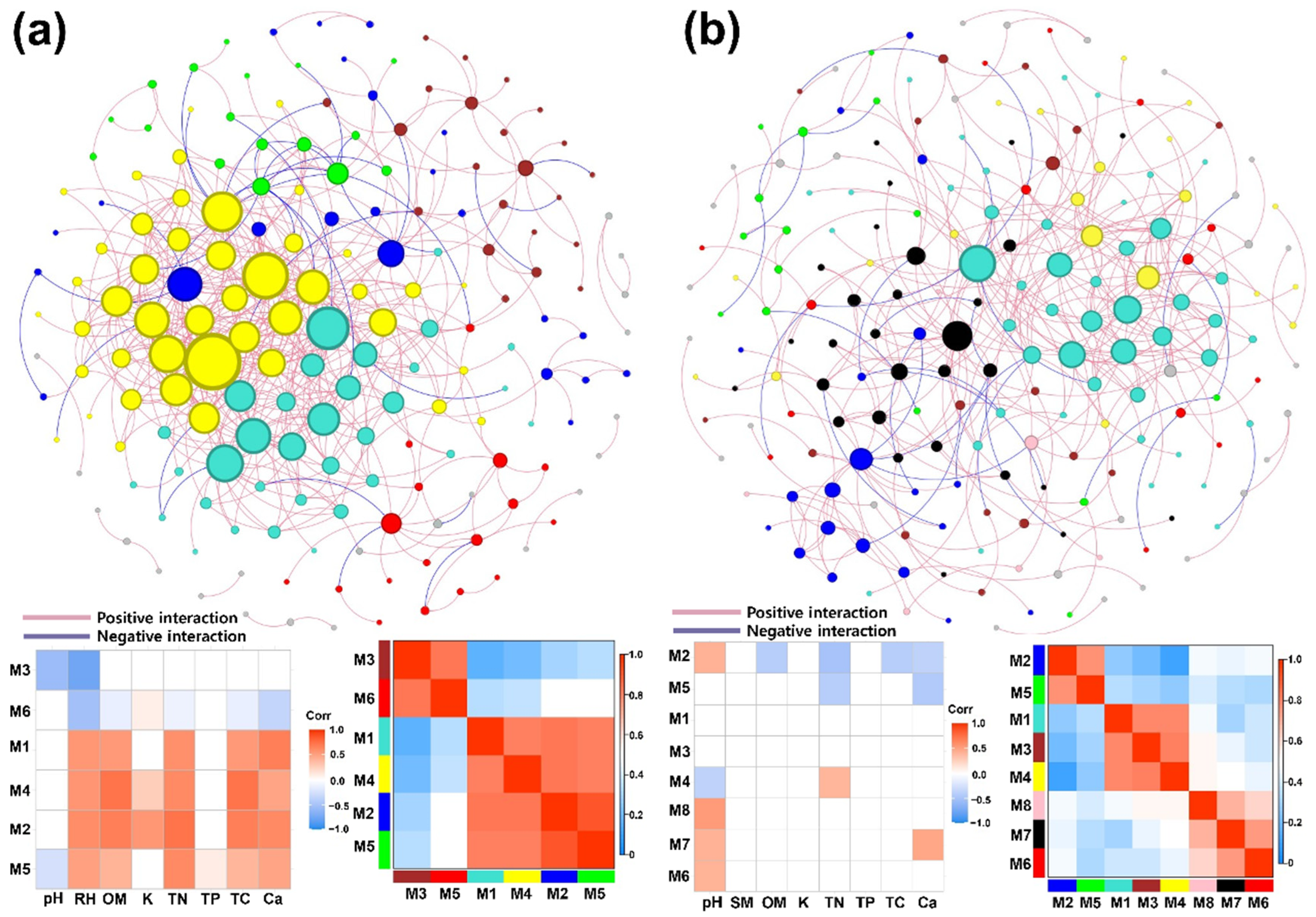
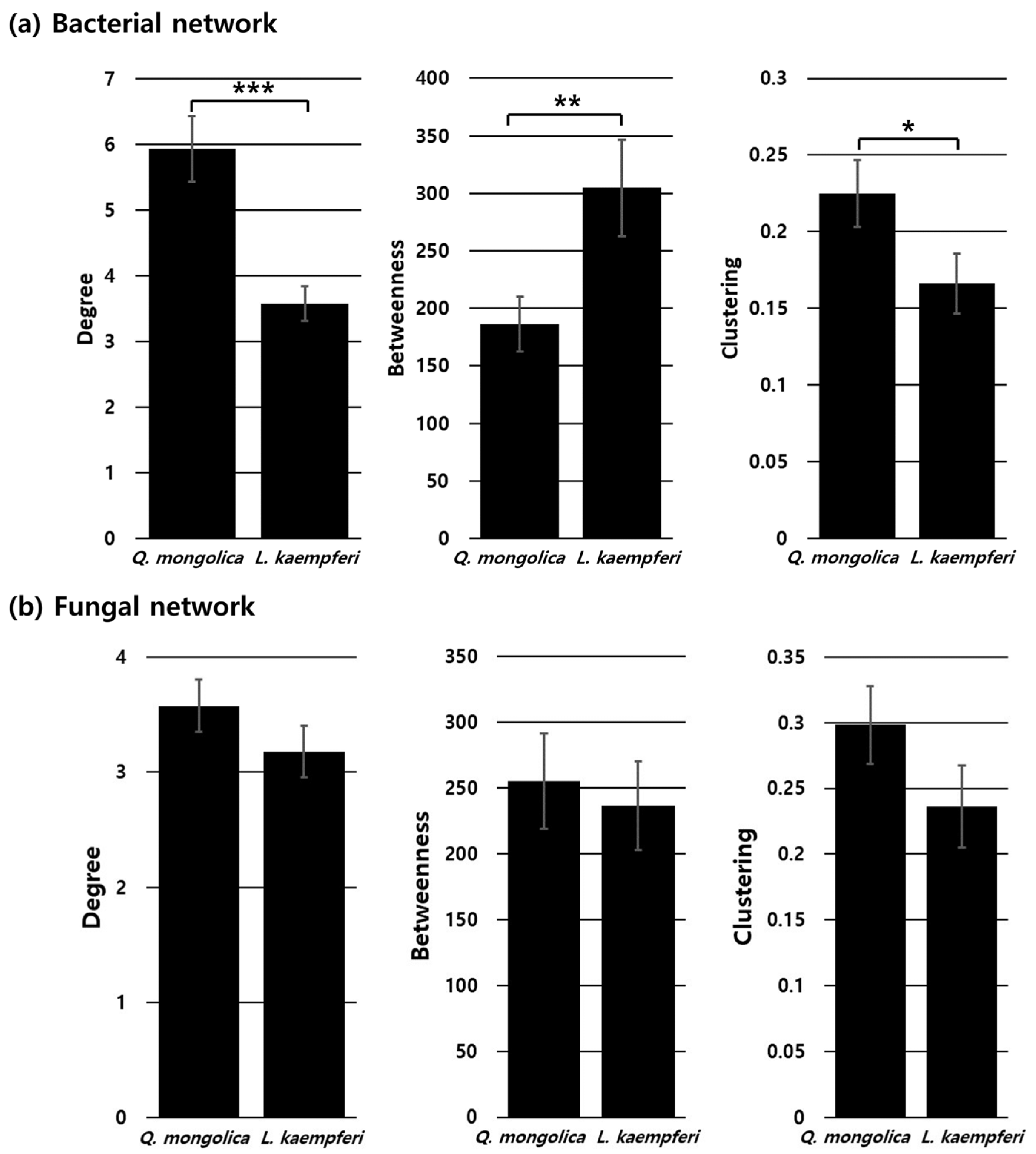
3.5. Network Analysis
4. Discussion
4.1. Difference in Community Composition
4.2. Difference in Functional Responses
4.3. Differences in Network Structure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Mather, A. The Forest Transition. Area 1992, 24, 367–379. Available online: https://www.jstor.org/stable/20003181 (accessed on 12 April 2025).
- Lee, S.-J.; Shin, D.-B.; Byeon, J.-G.; Oh, S.-H. Climate Change Vulnerability Assessment and ecological characteristics study of Abies nephrolepis in South Korea. Forests 2023, 14, 855. [Google Scholar] [CrossRef]
- Lee, S.-J.; Shin, D.-B.; Lee, A.-R.; Oh, S.-H. Characteristics of Dieback of Pinus densiflora and Risk Assessment in the Wangpicheon Ecosystem and Landscape Conservation Area in Uljin, South Korea. Forests 2023, 14, 903. [Google Scholar] [CrossRef]
- Mori, A.S.; Lertzman, K.P.; Gustafsson, L. Biodiversity and ecosystem services in forest ecosystems: A research agenda for applied forest ecology. J. Appl. Ecol. 2017, 54, 12–27. [Google Scholar] [CrossRef]
- Xiong, C.; He, J.Z.; Singh, B.K.; Zhu, Y.G.; Wang, J.T.; Li, P.P.; Zhang, Q.B.; Han, L.L.; Shen, J.P.; Ge, A.H. Rare taxa maintain the stability of crop mycobiomes and ecosystem functions. Environ. Microbiol. 2021, 23, 1907–1924. [Google Scholar] [CrossRef]
- Bhattacharyya, P.N.; Jha, D.K. Plant growth-promoting rhizobacteria (PGPR): Emergence in agriculture. World J. Microbiol. Biotechnol. 2012, 28, 1327–1350. [Google Scholar] [CrossRef]
- Lee, S.H.; Jeon, S.H.; Park, J.Y.; Kim, D.S.; Kim, J.A.; Jeong, H.Y.; Kang, J.W. Isolation and evaluation of the antagonistic activity of Cnidium officinale rhizosphere bacteria against phytopathogenic fungi (Fusarium solani). Microorganisms 2023, 11, 1555. [Google Scholar] [CrossRef]
- Baldrian, P. Forest microbiome: Diversity, complexity and dynamics. FEMS Microbiol. Rev. 2017, 41, 109–130. [Google Scholar] [CrossRef]
- Behjati, S.; Tarpey, P.S. What is next generation sequencing? Arch. Dis. Child.-Educ. Pract. 2013, 98, 236–238. [Google Scholar] [CrossRef]
- Simon, C.; Daniel, R. Metagenomic analyses: Past and future trends. Appl. Environ. Microbiol. 2011, 77, 1153–1161. [Google Scholar] [CrossRef]
- Kanokratana, P.; Uengwetwanit, T.; Rattanachomsri, U.; Bunterngsook, B.; Nimchua, T.; Tangphatsornruang, S.; Plengvidhya, V.; Champreda, V.; Eurwilaichitr, L. Insights into the phylogeny and metabolic potential of a primary tropical peat swamp forest microbial community by metagenomic analysis. Microb. Ecol. 2011, 61, 518–528. [Google Scholar] [CrossRef] [PubMed]
- Castañeda, L.E.; Barbosa, O. Metagenomic analysis exploring taxonomic and functional diversity of soil microbial communities in Chilean vineyards and surrounding native forests. PeerJ 2017, 5, e3098. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Jiang, Y.-H.; Yang, Y.; He, Z.; Luo, F.; Zhou, J. Molecular ecological network analyses. BMC Bioinform. 2012, 13, 113. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-H.; Chu, H.-L.; Dou, Q.; Feng, H.; Tang, M.; Zhang, S.-X.; Wang, C.-Y. Seasonal changes in Pinus tabuliformis root-associated fungal microbiota drive N and P cycling in terrestrial ecosystem. Front. Microbiol. 2021, 11, 526898. [Google Scholar] [CrossRef]
- Terhonen, E.; Blumenstein, K.; Kovalchuk, A.; Asiegbu, F.O. Forest tree microbiomes and associated fungal endophytes: Functional roles and impact on forest health. Forests 2019, 10, 42. [Google Scholar] [CrossRef]
- Singh, A.; Kumar, M.; Verma, S.; Choudhary, P.; Chakdar, H. Plant microbiome: Trends and prospects for sustainable agriculture. In Plant Microbe Symbiosis; Springer: Cham, Switzerland, 2020; pp. 129–151. [Google Scholar] [CrossRef]
- Lee, W.; Choi, J.; Lee, W.; Lee, Y.; Kim, S.; Jung, D. Comparison of growth characteristics for Korea red pine, Korea white pine, Japanese larch in Gang Won province. In Proceedings of the 2009 Meeting of the Korean Forest Society, Jeju Island, Republic of Korea, 30 November 2009; pp. 410–413. [Google Scholar]
- Kim, S.S.; Kim, R.Y.; Kim, M.S.; Kim, Y.H.; Song, G.C.; Yoon, H.B.; Lee, Y.J.; Jeon, Y.T.; Jo, H.S. Soil Chemical Analysis Method; NAAS (National Institute of Agricultural Sciences): Wanju, Republic of Korea, 2010. [Google Scholar] [CrossRef]
- Liu, D.; Chater, C.C.C.; Yu, F.; Perez-Moreno, J. Tuber pseudohimalayense ascomata-compartments strongly select their associated bacterial microbiome from nearby pine forest soils independently of their maturation stage. Pedobiologia 2021, 87–88, 150743. [Google Scholar] [CrossRef]
- Naumova, N.B.; Belanov, I.P.; Alikina, T.Y.; Kabilov, M.R. Undisturbed soil pedon under birch forest: Characterization of microbiome in genetic horizons. Soil Syst. 2021, 5, 14. [Google Scholar] [CrossRef]
- Odendaal, M.-L.; Groot, J.A.; Hasrat, R.; Chu, M.L.J.; Franz, E.; Bogaert, D.; Bosch, T.; de Steenhuijsen Piters, W.A. Higher off-target amplicon detection rate in MiSeq v3 compared to v2 reagent kits in the context of 16S-rRNA-sequencing. Sci. Rep. 2022, 12, 16489. [Google Scholar] [CrossRef]
- Glenn, T.C.; Nilsen, R.A.; Kieran, T.J.; Sanders, J.G.; Bayona-Vásquez, N.J.; Finger, J.W.; Pierson, T.W.; Bentley, K.E.; Hoffberg, S.L.; Louha, S. Adapterama I: Universal stubs and primers for 384 unique dual-indexed or 147,456 combinatorially-indexed Illumina libraries (iTru & iNext). PeerJ 2019, 7, e7755. [Google Scholar] [CrossRef]
- Amplicon, P.C.R.; Clean-Up, P.C.R.; Index, P.C.R. 16s Metagenomic Sequencing Library Preparation; Illumina: San Diego, CA, USA, 2013; p. 21. [Google Scholar]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Robeson, M.S.; O’Rourke, D.R.; Kaehler, B.D.; Ziemski, M.; Dillon, M.R.; Foster, J.T.; Bokulich, N.A. RESCRIPt: Reproducible sequence taxonomy reference database management. PLoS Comput. Biol. 2021, 17, e1009581. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Abarenkov, K.; Zirk, A.; Piirmann, T.; Pöhönen, R.; Ivanov, F.; Nilsson, R.H.; Kõljalg, U. UNITE QIIME Release for Fungi; UNITE Community: Saline, MI, USA, 2020. [Google Scholar] [CrossRef]
- Mirarab, S.; Nguyen, N.; Warnow, T. SEPP: SATé-enabled phylogenetic placement. In Biocomputing 2012; World Scientific: Singapore, 2012; pp. 247–258. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Morris, E.K.; Caruso, T.; Buscot, F.; Fischer, M.; Hancock, C.; Maier, T.S.; Meiners, T.; Müller, C.; Obermaier, E.; Prati, D. Choosing and using diversity indices: Insights for ecological applications from the German Biodiversity Exploratories. Ecol. Evol. 2014, 4, 3514–3524. [Google Scholar] [CrossRef]
- Sorenson, T. A method of establishing groups of equal amplitude in plant sociology based on similarity of species content and its application to analyses of the vegetation on Danish commons. Biol. Skrifter. 1948, 5, 1. [Google Scholar]
- French, A.; Macedo, M.; Poulsen, J.; Waterson, T.; Yu, A.; Multivariate Analysis of Variance (MANOVA). San Francisco State University 2008. Available online: https://www.academia.edu/7751963/Multivariate_Analysis_of_Variance_MANOVA (accessed on 12 April 2025).
- Legendre, P.; Anderson, M.J. Distance-based redundancy analysis: Testing multispecies responses in multifactorial ecological experiments. Ecol. Monogr. 1999, 69, 1–24. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Cui, Y.; Li, X.; Yao, M. microeco: An R package for data mining in microbial community ecology. FEMS Microbiol. Ecol. 2021, 97, fiaa255. [Google Scholar] [CrossRef]
- Põlme, S.; Abarenkov, K.; Henrik Nilsson, R.; Lindahl, B.D.; Clemmensen, K.E.; Kauserud, H.; Nguyen, N.; Kjøller, R.; Bates, S.T.; Baldrian, P. FungalTraits: A user-friendly traits database of fungi and fungus-like stramenopiles. Fungal Divers. 2020, 105, 1–16. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef]
- Edelman, A.; Rao, N.R. Random matrix theory. Acta Numer. 2005, 14, 233–297. [Google Scholar] [CrossRef]
- Newman, M.E. Fast algorithm for detecting community structure in networks. Phys. Rev. E—Stat. Nonlinear Soft Matter Phys. 2004, 69, 066133. [Google Scholar] [CrossRef]
- Yuan, M.M.; Guo, X.; Wu, L.; Zhang, Y.; Xiao, N.; Ning, D.; Shi, Z.; Zhou, X.; Wu, L.; Yang, Y. Climate warming enhances microbial network complexity and stability. Nat. Clim. Change 2021, 11, 343–348. [Google Scholar] [CrossRef]
- Diniz-Filho, J.A.F.; Soares, T.N.; Lima, J.S.; Dobrovolski, R.; Landeiro, V.L.; Telles, M.P.d.C.; Rangel, T.F.; Bini, L.M. Mantel test in population genetics. Genet. Mol. Biol. 2013, 36, 475–485. [Google Scholar] [CrossRef]
- Guimera, R.; Amaral, L.A.N. Cartography of complex networks: Modules and universal roles. J. Stat. Mech. Theory Exp. 2005, 2005, P02001. [Google Scholar] [CrossRef]
- Jiao, S.; Yang, Y.; Xu, Y.; Zhang, J.; Lu, Y. Balance between community assembly processes mediates species coexistence in agricultural soil microbiomes across eastern China. ISME J. 2020, 14, 202–216. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Duan, L.; Kim, B.; Mitchell, M.J.; Shibata, H. Potential effects of climate change and variability on watershed biogeochemical processes and water quality in Northeast Asia. Environ. Int. 2010, 36, 212–225. [Google Scholar] [CrossRef] [PubMed]
- Sohng, J.; Han, A.R.; Jeong, M.A.; Park, Y.; Park, B.B.; Park, P.S. Seasonal pattern of decomposition and N, P, and C dynamics in leaf litter in a Mongolian oak forest and a Korean pine plantation. Forests 2014, 5, 2561–2580. [Google Scholar] [CrossRef]
- Nakayama, M.; Tateno, R. Solar radiation strongly influences the quantity of forest tree root exudates. Trees 2018, 32, 871–879. [Google Scholar] [CrossRef]
- Jakoby, G.; Rog, I.; Megidish, S.; Klein, T. Enhanced root exudation of mature broadleaf and conifer trees in a Mediterranean forest during the dry season. Tree Physiol. 2020, 40, 1595–1605. [Google Scholar] [CrossRef]
- Sun, S.; Li, S.; Avera, B.N.; Strahm, B.D.; Badgley, B.D. Soil bacterial and fungal communities show distinct recovery patterns during forest ecosystem restoration. Appl. Environ. Microbiol. 2017, 83, e00966-17. [Google Scholar] [CrossRef]
- Urbanová, M.; Šnajdr, J.; Baldrian, P. Composition of fungal and bacterial communities in forest litter and soil is largely determined by dominant trees. Soil Biol. Biochem. 2015, 84, 53–64. [Google Scholar] [CrossRef]
- Li, W.-Q.; Huang, Y.-X.; Chen, F.-S.; Liu, Y.-Q.; Lin, X.-F.; Zong, Y.-Y.; Wu, G.-Y.; Yu, Z.-R.; Fang, X.-M. Mixing with broad-leaved trees shapes the rhizosphere soil fungal communities of coniferous tree species in subtropical forests. For. Ecol. Manag. 2021, 480, 118664. [Google Scholar] [CrossRef]
- Hobbie, E.A.; Agerer, R. Nitrogen isotopes in ectomycorrhizal sporocarps correspond to belowground exploration types. Plant Soil 2010, 327, 71–83. [Google Scholar] [CrossRef]
- López-Mondéjar, R.; Zühlke, D.; Becher, D.; Riedel, K.; Baldrian, P. Cellulose and hemicellulose decomposition by forest soil bacteria proceeds by the action of structurally variable enzymatic systems. Sci. Rep. 2016, 6, 25279. [Google Scholar] [CrossRef]
- Waters, C.M.; Bassler, B.L. Quorum sensing: Cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Wang, Z.; Sun, H.; Yang, W.; Xu, H. The response patterns of arbuscular mycorrhizal and ectomycorrhizal symbionts under elevated CO2: A meta-analysis. Front. Microbiol. 2018, 9, 1248. [Google Scholar] [CrossRef] [PubMed]
- Nuccio, E.E.; Hodge, A.; Pett-Ridge, J.; Herman, D.J.; Weber, P.K.; Firestone, M.K. An arbuscular mycorrhizal fungus significantly modifies the soil bacterial community and nitrogen cycling during litter decomposition. Environ. Microbiol. 2013, 15, 1870–1881. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Zhang, X.; Zhao, G.; Zhang, X.; Dong, L.; Chen, Y. Aerobic hydrogen-oxidizing bacteria in soil: From cells to ecosystems. Rev. Environ. Sci. Bio/Technol. 2022, 21, 877–904. [Google Scholar] [CrossRef]
- Ling, N.; Zhu, C.; Xue, C.; Chen, H.; Duan, Y.; Peng, C.; Guo, S.; Shen, Q. Insight into how organic amendments can shape the soil microbiome in long-term field experiments as revealed by network analysis. Soil Biol. Biochem. 2016, 99, 137–149. [Google Scholar] [CrossRef]
- Chen, Z.-J.; Liu, Y.-Q.; Li, Y.-Y.; Lin, L.-A.; Zheng, B.-H.; Ji, M.-F.; Li, B.L.; Han, X.-M. The seasonal patterns, ecological function and assembly processes of bacterioplankton communities in the Danjiangkou Reservoir, China. Front. Microbiol. 2022, 13, 884765. [Google Scholar] [CrossRef]
- Siles, J.A.; Margesin, R. Seasonal soil microbial responses are limited to changes in functionality at two Alpine forest sites differing in altitude and vegetation. Sci. Rep. 2017, 7, 2204. [Google Scholar] [CrossRef]
- Landi, P.; Minoarivelo, H.O.; Brännström, Å.; Hui, C.; Dieckmann, U. Complexity and stability of ecological networks: A review of the theory. Popul. Ecol. 2018, 60, 319–345. [Google Scholar] [CrossRef]
- Zhang, C.; Lei, S.; Wu, H.; Liao, L.; Wang, X.; Zhang, L.; Liu, G.; Wang, G.; Fang, L.; Song, Z. Simplified microbial network reduced microbial structure stability and soil functionality in alpine grassland along a natural aridity gradient. Soil Biol. Biochem. 2024, 191, 109366. [Google Scholar] [CrossRef]
- Hu, A.; Nie, Y.; Yu, G.; Han, C.; He, J.; He, N.; Liu, S.; Deng, J.; Shen, W.; Zhang, G. Diurnal temperature variation and plants drive latitudinal patterns in seasonal dynamics of soil microbial community. Front. Microbiol. 2019, 10, 674. [Google Scholar] [CrossRef]
- Yang, H.; Li, J.; Xiao, Y.; Gu, Y.; Liu, H.; Liang, Y.; Liu, X.; Hu, J.; Meng, D.; Yin, H. An integrated insight into the relationship between soil microbial community and tobacco bacterial wilt disease. Front. Microbiol. 2017, 8, 2179. [Google Scholar] [CrossRef] [PubMed]
- Tiessen, A.; Pérez-Rodríguez, P.; Delaye-Arredondo, L.J. Mathematical modeling and comparison of protein size distribution in different plant, animal, fungal and microbial species reveals a negative correlation between protein size and protein number, thus providing insight into the evolution of proteomes. BMC Res. Notes 2012, 5, 85. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Zhou, L.; Liang, T.; Man, J.; Wang, Y.; Li, Y.; Chen, H.; Zhang, T. Deciphering rhizosphere microbiome assembly of Castanea henryi in plantation and natural forest. Microorganisms 2022, 10, 42. [Google Scholar] [CrossRef] [PubMed]
- Van Der Heijden, M.G.; Hartmann, M. Networking in the plant microbiome. PLoS Biol. 2016, 14, e1002378. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, R.; Lu, B.; Guerin-Laguette, A.; He, X.; Yu, F. Mycorrhization of Quercus mongolica seedlings by Tuber melanosporum alters root carbon exudation and rhizosphere bacterial communities. Plant Soil 2021, 467, 391–403. [Google Scholar] [CrossRef]
- Deng, J.; Yin, Y.; Zhu, W.; Zhou, Y. Variations in soil bacterial community diversity and structures among different revegetation types in the Baishilazi Nature Reserve. Front. Microbiol. 2018, 9, 2874. [Google Scholar] [CrossRef]
- Lin, W.; Liu, L.; Liang, J.; Tang, X.; Shi, J.; Zhang, L.; Wu, P.; Lan, S.; Wang, S.; Zhou, Y. Changes of endophytic microbial community in Rhododendron simsii roots under heat stress and its correlation with leaf physiological indicators. Front. Microbiol. 2022, 13, 1006686. [Google Scholar] [CrossRef]
- Muratova, A.; Golubev, S.; Romanova, V.; Sungurtseva, I.; Nurzhanova, A. Effect of heavy-metal-resistant PGPR inoculants on growth, rhizosphere microbiome and remediation potential of miscanthus× giganteus in Zinc-contaminated Soil. Microorganisms 2023, 11, 1516. [Google Scholar] [CrossRef]
- Chen, Y.; Xi, J.; Xiao, M.; Wang, S.; Chen, W.; Liu, F.; Shao, Y.; Yuan, Z. Soil fungal communities show more specificity than bacteria for plant species composition in a temperate forest in China. BMC Microbiol. 2022, 22, 208. [Google Scholar] [CrossRef]

| Month | Stand | pH | SM (%) | OM (g/kg) | TN (%) | TP (mg/kg) | TC (%) | K+ (cmolc/kg) | Ca2+ (cmolc/kg) |
|---|---|---|---|---|---|---|---|---|---|
| June | Q. mongolica | 5.26 a (0.03) | - | 124.18 a (8.56) | 0.65 b (0.04) | 679.89 a (27.30) | 7.20 a (0.50) | 0.33 c (0.08) | 3.57 ab (0.47) |
| L. kaempferi | 4.81 c (0.15) | - | 132.76 a (23.12) | 0.63 b (0.10) | 568.36 bc (50.73) | 7.70 a (1.34) | 0.53 b (0.29) | 3.08 b (0.50) | |
| August | Q. mongolica | 5.11 ab (0.08) | 84.51 a (4.13) | 141.49 a (13.44) | 0.86 a (0.06) | 669.53 c (36.00) | 8.20 a (0.78) | 0.77 b (0.10) | 6.48 ab (1.44) |
| L. kaempferi | 4.80 c (0.09) | 73.36 a(4.20) | 140.89 a (16.71) | 0.76 ab (0.07) | 525.82 ab (38.24) | 8.17 a (0.97) | 0.53 a (0.05) | 3.73 a (1.01) | |
| October | Q. mongolica | 4.87 bc (0.08) | 74.70 a (4.21) | 149.78 a (18.31) | 0.59 ab (0.06) | 536.87 c (23.27) | 8.69 a (1.06) | 0.65 ab (0.07) | 4.08 ab (0.77) |
| L. kaempferi | 5.01 abc (0.05) | 63.92 a (2.58) | 113.13 a (10.32) | 0.70 b (0.05) | 530.22 c (37.98) | 6.56 a (0.60) | 0.46 bc (0.03) | 3.61 ab (0.89) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.H.; Park, J.Y.; Jeon, S.H.; Kim, D.S.; Lee, S.H.; Park, Y.D.; Kang, J.W. Functional Potential and Network-Based Insights into the Rhizosphere Microbiomes of Quercus mongolica and Larix kaempferi Stands. Forests 2025, 16, 883. https://doi.org/10.3390/f16060883
Lee SH, Park JY, Jeon SH, Kim DS, Lee SH, Park YD, Kang JW. Functional Potential and Network-Based Insights into the Rhizosphere Microbiomes of Quercus mongolica and Larix kaempferi Stands. Forests. 2025; 16(6):883. https://doi.org/10.3390/f16060883
Chicago/Turabian StyleLee, Seok Hui, Jun Young Park, Su Hong Jeon, Dae Sol Kim, Su Ho Lee, Yeong Dae Park, and Jun Won Kang. 2025. "Functional Potential and Network-Based Insights into the Rhizosphere Microbiomes of Quercus mongolica and Larix kaempferi Stands" Forests 16, no. 6: 883. https://doi.org/10.3390/f16060883
APA StyleLee, S. H., Park, J. Y., Jeon, S. H., Kim, D. S., Lee, S. H., Park, Y. D., & Kang, J. W. (2025). Functional Potential and Network-Based Insights into the Rhizosphere Microbiomes of Quercus mongolica and Larix kaempferi Stands. Forests, 16(6), 883. https://doi.org/10.3390/f16060883







