Abstract
Acute Oak Decline (AOD) is a complex and rapidly progressing disease affecting several Quercus species across Europe. While previously reported in Quercus ilex in Italy, this study provides the first evidence of AOD symptoms and associated bacterial infection in Quercus coccifera (kermes oak). Symptomatic trees were identified in a Mediterranean forest in southern Italy, and bacterial isolation, qPCR detection, and 16S rRNA sequencing confirmed the presence of Brenneria goodwinii and Gibbsiella quercinecans. Phylogenetic analyses clustered the isolates closely with known AOD-related strains. Pathogenicity tests on excised Q. coccifera branches demonstrated that both bacteria induced wood necrosis and external exudates consistent with natural symptoms, confirming their virulence. These findings expand the known host range of AOD-related bacteria and highlight the potential threat to Mediterranean oak ecosystems. Early detection and monitoring of Q. coccifera decline are essential to inform conservation strategies and forest management practices aimed at mitigating AOD spread.
1. Introduction
Kermes oak (Quercus coccifera L.) is a shrub-like evergreen oak that generally does not exceed two meters in height [1]. Distinctive of this oak species are the leaves, which are 1.5–4 cm long and coriaceous with spiny margins, and also the acorns, which possess a characteristic cup that exhibits numerous elongated and reflexed scales [2,3]. Regarding its ecology, it mainly forms the garrigue [4], and it can also be associated with taller trees, like pines and other oak species, and with other shrubs [5]. It is a hardy and slow-growing oak, able to grow in harsh conditions, tolerating nutrient-poor and rocky soils, drought, and fire [6]. It is native to the Mediterranean basin and can be found in Portugal, Spain, Greece, Turkey, South France, and the Southernmost areas of Italy [7], where it is commonly found in Sardinia, Sicily, Basilicata, and Apulia [8]. Given its ecological importance and adaptive resilience in Mediterranean environments, investigating the susceptibility of Q. coccifera to emerging oak diseases has become increasingly relevant—particularly in light of recent reports of Acute Oak Decline (AOD) in neighboring oak species such as Q. ilex. In 2023, bacteria identified on holm oak in Apulia, specifically Salento, were reported as pathogenic and necrogenic for oaks in many European countries [9,10,11,12,13]. These include Brenneria goodwinii, Gibbsiella quercinecans, and Rahnella victoriana. These bacteria are known to be the biotic cause of a disease called “Acute Oak Decline” (AOD), a plant disease described for the first time in the UK in 2014, on Q. pertraea (Matt.) and Q. robur (L.) [14,15,16]. This disease is characterized by distinct symptoms such as brownish exudate bleeding from longitudinal bark cracks, rarefaction of the canopy due to the defoliation, and sprouting of epicormic shoots from the trunk and main branches. The disease is referred to as “acute” as it can quickly lead to the tree’s death, usually within a few years of symptoms, and is characterized by sudden regression in the health of oak trees [16,17,18]. After its first description, the disease has been observed in different European countries, such as Q. robur in Switzerland [9,19], Q. petraea in Spain [20], Q. ilex in Slovakia and Italy [11,13], Q. cerris in Poland [10], Q. suber Latvia [21], and Q. suber in Portugal [12].
This study presents novel insights into the interactions between Q. coccifera and bacterial species associated with Acute Oak Decline (AOD), specifically Brenneria goodwinii, Gibbsiella quercinecans, and Rahnella victoriana. The primary aim is to evaluate the pathogenic potential of these bacteria towards Q. coccifera and to elucidate their phylogenetic relationships with related taxa.
2. Materials and Methods
2.1. Sampling and Symptom Assessment
The sampled area, designated as “Bosco dei Romani” (39°53′41.514″ N, 18°10′4.393″ E), is located within the Regional Natural Park of the Ugento Coast in Salento, Italy (Figure 1). This area encompasses a dense mixed forest characterized by Mediterranean shrubland, such as Rosmarinus officinalis (L.), Cistus (L.), and Rhamnus alaternus (L.), and oak species, in particular Q. ilex and Q. coccifera. Within this forest, a group of several kermes oaks exhibiting symptoms of AOD were identified. The symptoms of each specimen were assessed with Finch’s pathogenicity index, which takes into account 31 descriptive parameters (Table S1) of the oak’s phenological and health status [22]. These parameters aim to evaluate each oak’s size, canopy, and trunk conditions. The data collected visually with a measuring tape or with the hypsometer Haglof Vertex 5 (Haglofs, Torsang, Sweden) were immitted in the online tool (https://jasenfinch.shinyapps.io/decliner/, accessed on 6 March 2025) to evaluate and indicate the type and severity of the decline through the Phenotypic Decline Index (PDI) and the Decline Acuteness Index (DAI). The PDI indicates the decline severity and ranges between 0 and 1, the maximum severity score. In contrast, the DAI indicates the differences between two types of decline, Chronic Oak Decline (COD) and Acute Oak Decline, and ranges between −1 and 1, where oaks with a more severe COD score are closer to −1 and oaks with a more severe AOD score are closer to 1.
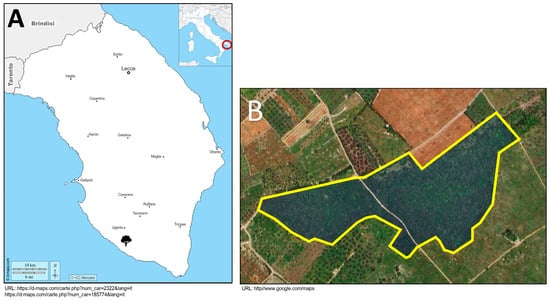
Figure 1.
(A) The figure presents the geographical map of Italy, with a focus on the Salento region. The red triangle indicates the geographic location of the sampled area. (B) The figure depicts the aerial view of “Bosco dei Romani”, with the area highlighted in blue.
After symptom evaluation, bark samples were collected from the bleeding points. To diagnose symptomatic plants, approximately 1 g of outer bark fragments was collected at the point where the exudates and brown patches were present, using a sterile blade. The wooden pieces were placed in a sterile plastic test tube and properly labelled; for each tree, at least five exudates were sampled.
For bacteria isolations, bark panels (approximately 3 cm × 4 cm), also in correspondence with the brownish patches with exudates, were excised using a sterile scalpel and placed into sterile plastic containers. The wood panels collected consisted of the outer bark, cambium, and xylem, which also exhibited necrotic areas. For isolations, three panels were collected for each tree.
For the pathogenicity test, segments of branches approximately 20–25 cm long and 3 cm in diameter were excised from healthy kermes oak trees that showed no symptoms of AOD, fungi body fruits, or other potential pathogens.
All samples were maintained at 4 °C and transported to the laboratory.
2.2. Plant Diagnosis
B. goodwinii, G. quercinecans, and R. victoriana were detected as described by Carluccio et al. [13], based on Crampton et al. [23] with modifications. Bark fragments for each sample were macerated with 1 mL of Sterile Distilled Water (SDW) and gently shaken for 5 min. Then, the suspension was transferred into a sterile 1.5 mL tube and left to settle for a few minutes in order to remove woody debris. After the supernatant was collected, it was centrifugated for 4 min at 9000× g. The resulting pellet was resuspended in 1 mL of SDW, vortexed, and centrifuged again for 4 min at 9000× g; this process was repeated four times in total. Finally, the pellet was resuspended in 50 µL of SDW and stored at −20 °C until use.
To improve specificity and sensitivity, real-time PCR was performed according to Crampton et al. (2020) [23], with species-specific primers validated against a panel of related bacterial species (Table 1). The reaction mixture for G. quercinecans consisted of 5 µL of TaqMan™ Environmental Master Mix 2.0 2× (Thermo Fisher, Waltham, MA, USA) and 0.1 μM of each primer and probe, while for the screening of B. goodwinii and R. victoriana, 0.25 μM of each primer was used. The final volume of the qPCR mixture was 10 µL.

Table 1.
Primers and probes used for screening colonies, according to Crampton et al. [23].
2.3. Bacterial Isolation and 16S Barcoding
Isolation procedures followed the protocol described by Moradi-Amirabad et al. [24]. Bark sections were sterilized using 70% (v/v) ethanol and rinsed three times with Sterile Distilled Water (SDW). Fragments were then collected from the transition zone between necrotic and healthy tissues as well as from the xylem and outer bark. Each sample was placed in a Petri dish containing 1 mL of SDW and finely chopped using a sterile scalpel. After a 30 min incubation at room temperature, the suspension was streaked onto Nutrient Agar (NA) using a sterile loop. The plates were incubated at 28 °C for five days and monitored daily. DNA was extracted from the resulting colonies [25], and qPCR was performed using the protocol from Crampton et al. [23] to identify colonies of Brenneria goodwinii, Gibbsiella quercinecans, and Rahnella victoriana. Colonies testing positive were subsequently isolated and grown individually on NA.
To confirm the qPCR results, partial 16S rRNA gene sequencing was conducted [26]. PCR reactions were carried out in a final volume of 25 µL, consisting of 12.5 µL of EmeraldAmp® MAX PCR Master Mix 2× (Takara Bio, Kusatsu, Japan), 0.5 µL of each primer (EUB9F: 5′–GAGTTTGATCMTGGCTCAG–3′; EUB1492R: 5′–ACGGYTACCTTGTTACGACTT–3′), and 5 µL of DNA template, following the methods described by Pettifor [25] and Crampton et al. [23]. PCR conditions were as follows: an initial denaturation at 95 °C for 2 min; 35 cycles of 95 °C for 1 min, 53 °C for 1 min, and 68 °C for 90 s; and a final extension at 68 °C for 5 min.
The resulting amplicons were sequenced via Sanger sequencing (Eurofins, Ebersberg, Germany), aligned with Clustal W [27], and trimmed to equal lengths; positions with gaps or ambiguous bases were excluded before analysis; lastly, they were compared on the NCBI (National Center for Biotechnology Information) database using the Basic Local Alignment Search Tool for nucleotide (BLASTn) [28]. Phylogenetic relationships were reconstructed using partial 16S rRNA sequences in MEGA 11 software [29,30], employing the maximum likelihood method with the Tamura–Nei model and 1000 bootstrap replications to assess clade support [16,29].
2.4. Pathogenicity Tests
Pathogenicity tests were carried out based on Eichenlaub et al. [31]. The branch segments collected were exposed to UV rays for 2 h for superficial sterilization in order to avoid contamination. Subsequently, four inoculation points, representing four replicas for each isolate, were created on each segment by removing a bark disc with a sterile cork borer (1 cm in diameter) along the woody segment length. A one-day-old colony was applied using a sterile loop to each inoculation point. The loop was carefully rubbed on the xylem and on the internal part of the bark disc to ensure a uniform colony distribution. In total, two wood segments were infected for each bacterium plus one branch segment for mock inoculation. Additionally, as a reference, separate twigs were inoculated with isolates of B. goodwinii and G. quercinecans that were isolated from holm oak and have already been shown to be necrogenic on excised holm oak branches [32].
After repositioning the removed bark discs, the inoculation points and the upper end of each branch segment were labelled and sealed with a plastic film to maintain moisture. Additionally, the lower end of each segment was placed in contact with SDW to prevent the branches from drying out. The prepared segments were then incubated for 1 month at 28 °C with a 12 h photoperiod in a growth chamber.
Following the incubation period, bark was removed from the inoculation sites to examine the underlying tissue. Wood fragments were taken from the boundary between necrotic and healthy areas, specifically from the inner bark and xylem. These samples were surface-sterilized using 70% ethanol, rinsed twice with Sterile Distilled Water (SDW), and then suspended in 1 mL of SDW and further shredded with a sterile scalpel. After a 30 min incubation at room temperature, the suspension was streaked onto Nutrient Agar (NA) plates using a sterile loop. The plates were incubated at 27 °C for 48 h, after which, the emerging colonies were screened via qPCR as previously described.
3. Results
3.1. Symptomatology and Diagnosis
During the sampling of Bosco dei Romani, an area where symptomatic and AOD-positive holm oaks had previously been identified [13], Q. coccifera oaks exhibiting characteristic symptoms of AOD were also observed (Figure 2). These plants exhibited brown exudates on the bark of the trunk and branches, dark patches on the bark due to the wood’s impregnation with exudate, and larval galleries corresponding to the exudate. At the canopy level, the foliage appeared sparse. All the kermes oaks sampled were infected with B. goodwinii and G. quercinecans, but no sample tested positive for R. victoriana.
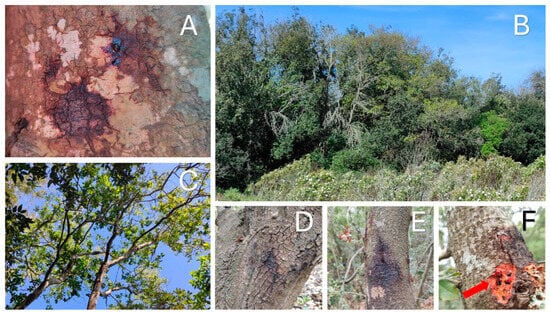
Figure 2.
The figure shows the symptoms and conditions observed: (A) brown exudate and patches observed on the bark; (B,C) crown dieback and defoliation; (D,E) dark patches on the bark, given by the necrotic activity of bacteria; (F) red arrow indicates insect galleries under the bark.
Regarding the analysis of symptomatology, the use of the Finch’s index revealed that all the plants were affected by AOD, with an average PDI of 0.81 and a DAI of 0.53. These values thus indicate that the oaks examined experience an acute type of decline (DAI close to 1) and similar severity, except for one specimen with a PDI of 0.88, thus showing more severe symptoms (Figure 3).
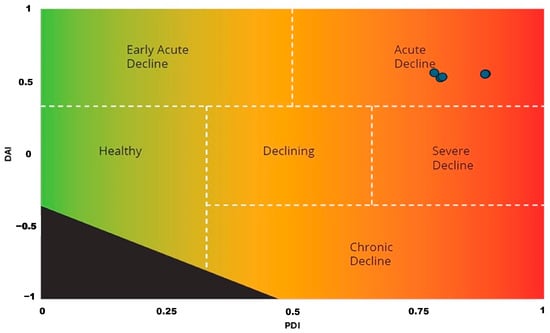
Figure 3.
Quantification and classification of decline symptoms on kermes oak: the trees have a positive Decline Acuteness Index (DAI) value, between 0.5 and 1, indicating acute decline, while the Phenotypic Decline Index (PDI) closer to 1 means high severity of symptoms. The blue circles represent samples where AOD symptoms were observed and confirmed. The black area at the bottom left indicates the undefined decline indexes.
3.2. Isolation, Sequencing, and Phylogenetic Analysis
After plating, white-colored colonies developed on the NA agar between the third and fifth days of incubation. The identity of the colonies was initially verified by qPCR, and then B. goodwinii and G. quercinecans colonies were plated individually on NA. No colony tested positive for R. victoriana.
Sequencing of the target bacteria by partial 16S rRNA gene revealed that the two isolates were similar to the reference strains, with 99.78% with isolate BY5 (Accession number KY231164) and 99.64% with isolate FRB 185 (Accession number GU562340) for B. goodwinii and G. quercinecans, respectively. The nucleotide sequences of the isolates were then deposited on GeneBank with the following accession numbers: PQ373172 for B. goodwinii (isolate BRUG24) and PQ373180 for G. quercinecans (isolate BRQS24); based on those sequences, deposited on NCBI, phylogenetic trees were then constructed for each bacterium (Figure 4 and Figure 5). For B. goodwinii, the loci OM287486 and MZ612851, respectively, of the strains KKP 3652 and S1T1 of Serratia fonticola were used as outliers, while for G. quercinecans, the loci NR112555 and EF122435 of the strains DSM 12163 and ICMP 14143 of Erwinia pyrifoliae were used as outliers.
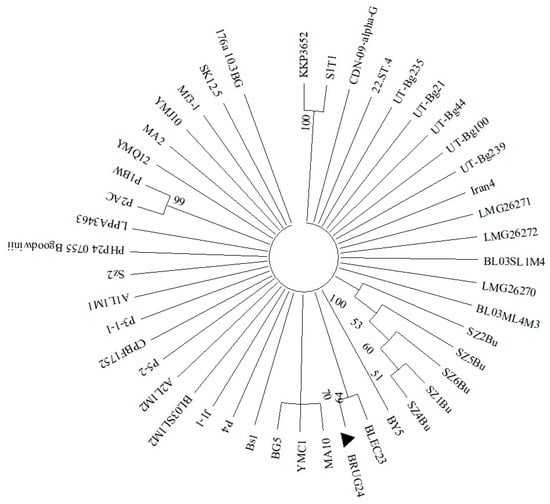
Figure 4.
Phylogenetic tree based on 16S gene sequencing of B. goodwinii isolate BRUG24 (▲). The strains KKP 3652 and S1T1 of S. fonticola represent the outlier and are genetically the farthest strain; all other strains reported in the phylogenetic tree refer to B. goodwinii. For the phylogenetic tree, maximum likelihood based on the Tamura–Nei model was used.
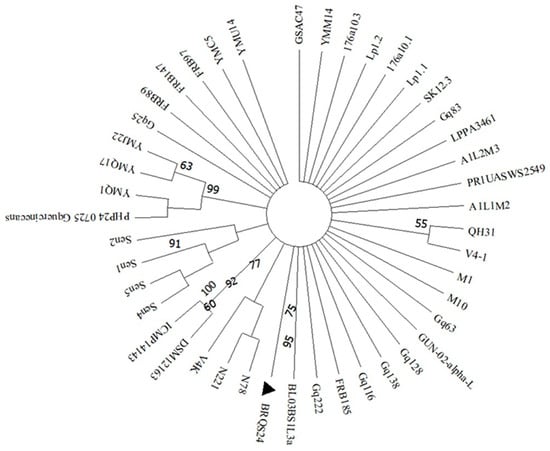
Figure 5.
Phylogenetic tree based on 16S gene sequencing of G. quercinecans isolate BRQS24 (▲). The strains DSM 12163 and ICMP 14143 of E. pyrifoliae represent the outlier and are the strain genetically most far; all other strains reported in the phylogenetic tree refer to G. quercinecans. For the phylogenetic tree, maximum likelihood based on the Tamura–Nei model was used.
3.3. Pathogenicity Assessments
The bacteria isolated from kermes oak and identified as B. goodwinii and G. quercinecans, one month after being inoculated on branches excised from healthy oaks, developed necrosis in the xylem and inner bark at all inoculation points as well as exudates external to the bark (Figure 6); the overall results revealed that 100% of the inoculation points showed necrosis from which the respective bacteria could be isolated. The symptoms observed on the branches during the pathogenicity tests entirely resembled those observed in nature. Furthermore, no necrosis was found on the mock-inoculated branch.

Figure 6.
The picture shows the state of the twigs one month after inoculation. (A) The branch on which the mock inoculation was carried out showed no signs of wood necrosis; (B) the branch inoculated with B. goodwinii showed extensive wood necrosis, and (C) the bark showed a moist brown patch; (D) the branch inoculated with G. quercinecans also showed necrosis of the wood extending beyond the inoculum point; (E) also in this case, the outer bark showed an extensive brown, moist patch with a brownish exudate. As a reference, the bar in the bottom-left-hand corner equals 1 cm.
4. Discussion
In Europe, bacteria associated with AOD have been identified predominantly on plants of the Quercus genus, so given that the affected oak species are found in diverse pedoclimatic environments, a comparative analysis of symptom types, severity, and eventual decline recovery across different oak species could provide essential data on the interaction between host and pathogens. Also, since AOD-related bacteria exhibit necrogenic [33,34] activity and bacteria of the same genus are capable of degrading cell walls [35,36,37], it would be beneficial to investigate the response of different oak species to the infection, as different oaks have different cell wall composition, for example, in terms of chemistry and ultrastructure [38,39]. Thus, understanding these responses and possible differences could provide important insights into the species’ diverse resilience and susceptibility to the disease and shed light on specific factors such as lesion dimensions, lesion count, and the bacteria’s ability to spread within the plant. These findings support and expand existing hypotheses regarding host range and species specificity of AOD-associated bacteria. Although Quercus coccifera is ecologically distinct and highly tolerant to drought and poor soils [6], it appears equally vulnerable to B. goodwinii and G. quercinecans. This suggests that host susceptibility may not solely depend on environmental adaptation but possibly on specific host–pathogen interactions, as hypothesized in previous studies [33,34]. Furthermore, the confirmation of the necrogenic activity of these bacteria on Q. coccifera supports the idea that oak species across various ecological niches can act as viable hosts, which aligns with earlier reports from different European regions on species such as Q. robur, Q. ilex, Q. cerris, and Q. suber [9,10,11,12,13,20,21]. Such information can be beneficial for several reasons: (a) guiding reforestation programs by selecting oak species that are less susceptible to AOD; (b) informing plant breeding and screening programs aimed at preserving the biodiversity of oak species and maintaining the integrity of European forest ecosystems [40]; and (c) enhancing our understanding of the abiotic factors involved in the development and spread of AOD (for instance, as there is a correlation between AOD symptoms and drought) [41]. Thus, it would be valuable to compare the susceptibility to AOD between oak species adapted to drier environments, such as Q. coccifera [6], and those typical of temperate forests, such as Q. robur, which, by contrast, have a lifestyle more dependent on water availability [42].
However, since, outside of Europe, B. goodwinii and G. quercinecans are reported to infect not only exotic oaks, such as Q. castaneifolia and Q. brantii [24], but also other species, such as Elaeagnus angustifolia [43], Juglans regia [44], or Carpinus betulus [24], there exists the possibility that these microorganisms may not be strictly oak pathogens and might be a potential vector in Europe. Furthermore, in Europe, such bacteria have been reported on other genera, such as Tilia sp. [11], Malus sp. ((NCBI) [online]., 2021), Ulmus sp., and Acer sp. ((NCBI) [online], 2019; (NCBI) [online], 2023); no further details about those interactions are available, given the few reports.
Although its role in AOD is not well understood and its role as a proven vector has not been established, A. biguttatus, a beetle found throughout Europe, is suspected to be involved in AOD [45]. The literature frequently describes it as an aggressive pest for oaks, especially the species reported in the literature with AOD symptoms, such as Q. robur, Q. pertaea, Q. ilex, and Q. cerris, and occasionally other Fagaceae [46]. Alongside, there are other European beetles of the same genus, like A. sulcicollis, A. angustulus, and related buprestid, such as Coraebus florentinus or C. undatus, whose lifestyles are closely associated with oak trees that, in the past, have been reported for damaging oaks [46,47], may act as a vector for AOD bacteria, and explain the association between B. goodwinii, G. quercinecans, and oaks in Europe. Lastly, our findings contribute to the ongoing discussion on the role of insect vectors, particularly xylophagous beetles such as Agrilus biguttatus, whose galleries were observed in association with bacterial exudates. While a definitive vector–pathogen relationship remains to be proven, this aligns with previous reports proposing a vector-mediated facilitation of AOD symptoms [45,46,47].
In addition, the results corroborate previous hypotheses suggesting that AOD pathogens exhibit a broader host range than initially assumed, potentially influenced by bacterial capacity for cell wall degradation [35,36,37] and the anatomical or chemical features of different oak species [38,39]. The identification of typical symptoms (exudates, necrosis, crown dieback) and the successful re-isolation of the pathogens following inoculation further reinforce the consistency with disease expression patterns described in other Quercus hosts [14,15,16,17,18].
While this study offers valuable insights into host–pathogen interactions in AOD across different oak species, certain aspects remain to be further explored. The diversity of environmental conditions among study sites as well as the limited temporal scope may have influenced the interpretation of symptom development and disease progression. Moreover, the complexity of microbial interactions and the potential influence of other biotic or abiotic stressors were beyond the scope of the current analysis.
Future studies could build on these findings by incorporating longer-term monitoring, more detailed analyses of host responses at cellular and molecular levels, and controlled experiments to better isolate species-specific patterns. A deeper investigation into the microbial communities involved may also help clarify the factors contributing to the varying susceptibility and resilience observed among oak species. Such research would complement the current results and contribute to a more comprehensive understanding of AOD dynamics.
5. Conclusions
In this study, symptomatic infection of Q. coccifera by B. goodwinii and G. quercinecans was reported, representing, to our knowledge, the first observation of such a host–pathogen relationship. After detecting kermes oaks with symptoms attributable to AOD, the infection was initially diagnosed by qPCR on symptomatic trees and subsequently confirmed by sequencing the 16S rRNA gene with the construction of phylogenetic trees, where isolates of B.goodwinii and G. quercinecans clustered with the respective strains. Finally, pathogenicity tests demonstrated that the two bacteria isolated from infected tissue are indeed capable of causing wood necrosis, as inoculation of branch segments of Q. coccifera showed that wood necrosis and exudates developed in the presence of the bacteria and the two pathogens were again isolated from necrotic tissue.
This finding opens the way to several lines of research concerning the study of a possible species specificity of AOD-related bacteria and potential vectors, leading to oaks being the main affected plants. It is also important to determine which species are most impacted as well as whether there is a range of symptom intensity or the plant’s ability to recover.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/f16050789/s1, Table S1: Phenotypic descriptors collected for each oak used to evaluate the decline type and severity.
Author Contributions
Conceptualization, G.C., M.V. and A.L.; Data curation, G.C. and M.V.; Formal analysis, G.C., M.V. and L.P.; Investigation, G.C. and A.D.D.; Methodology, G.C., M.D.P., and A.B.; Software, G.C. and M.D.P.; Supervision, L.D.B. and A.L.; Validation, G.C., M.V. and A.L.; Writing—original draft, G.C.; Writing—review and editing, M.V., L.D.B. and A.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Martínez-Ferri, E.; Balaguer, L.; Valladares, F.; Chico, J.M.; Manrique, E. Energy Dissipation in Drought-Avoiding and Drought-Tolerant Tree Species at Midday during the Mediterranean Summer. Tree Physiol. 2000, 20, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Cañellas, I.; San Miguel, A. Litter Fall and Nutrient Turnover in Kermes Oak (Quercus coccifera L.) Shrublands in Valencia (Eastern Spain). Ann. Sci. For. 1998, 55, 589–597. [Google Scholar] [CrossRef]
- Jasprica, N.; Škvorc, Ž.; Dolina, K.; Ruščić, M.; Kovačić, S.; Franjić, J. Composition and Ecology of the Quercus coccifera L. Communities along the Eastern Adriatic Coast (NE Mediterranean). Plant Biosyst. 2016, 150, 1140–1155. [Google Scholar] [CrossRef]
- Rodà, F.; Retana, J.; Gracia, C.A.; Bellot, J. Ecology of Mediterranean Evergreen Oak Forests; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2021. [Google Scholar]
- Tsiouvaras, C.N. Ecology and Management of Kermes Oak (Quercus coccifera L.) Shrublands in Greece: A Review. J. Range Manag. 1987, 40, 542. [Google Scholar] [CrossRef]
- FO.RE.S.T.A.S. Sardegna Foreste–Quercia Spinosa. 2024. Available online: https://www.sardegnaforeste.it/flora/quercia-spinosa (accessed on 15 November 2024).
- Caudullo, G.; Welk, E.; San-Miguel-Ayanz, J. Chorological Maps for the Main European Woody Species. Data Br. 2017, 12, 662–666. [Google Scholar] [CrossRef]
- EFI. EUFORGEN- Quercus Coccifera. 2020. Available online: https://www.euforgen.org/species/quercus-coccifera (accessed on 26 November 2024).
- Ruffner, B.; Schneider, S.; Meyer, J.B.; Queloz, V.; Rigling, D. First Report of Acute Oak Decline Disease of Native and Non-native Oaks in Switzerland. New Dis. Rep. 2020, 41, 18. [Google Scholar] [CrossRef]
- Tkaczyk, M.; Celma, L.; Ruņģis, D.E.; Bokuma, G. First Report of Brenneria Goodwinii and Gibbsiella Quercinecans Bacteria, Detected on Weaken Oak Trees in Poland. Balt. For. 2021, 27, 166–169. [Google Scholar] [CrossRef]
- Tkaczyk, M.; Sikora, K.; Plewa, R. Dieback of Small-Leaved Lime Trees (Tilia Cordata Mill.) Caused by Gibsiella quercinecans in Urban Areas in Poland. For. Pathol. 2024, 54, e12861. [Google Scholar] [CrossRef]
- Fernandes, C.; Duarte, L.; Naves, P.; Sousa, E.; Cruz, L. First Report of Brenneria goodwinii Causing Acute Oak Decline on Quercus suber in Portugal. J. Plant Pathol. 2022, 104, 837–838. [Google Scholar] [CrossRef]
- Carluccio, G.; Sabella, E.; Greco, D.; Vergine, M.; Delle Donne, A.G.; Nutricati, E.; Aprile, A.; De Bellis, L.; Luvisi, A. Acute and Chronic Oak Decline in Urban and Forest Ecosystems in Southern Italy. For. Int. J. For. Res. 2024, 97, 739–749. [Google Scholar] [CrossRef]
- Brady, C.; Denman, S.; Kirk, S.; Venter, S.; Rodríguez-Palenzuela, P.; Coutinho, T. Description of Gibbsiella quercinecans Gen. Nov., Sp. Nov., Associated with Acute Oak Decline. Syst. Appl. Microbiol. 2010, 33, 444–450. [Google Scholar] [CrossRef] [PubMed]
- Denman, S.; Brady, C.; Kirk, S.; Cleenwerck, I.; Venter, S.; Coutinho, T.; De Vos, P. Brenneria goodwinii Sp. Nov., Associated with Acute Oak Decline in the UK. Int. J. Syst. Evol. Microbiol. 2012, 62, 2451–2456. [Google Scholar] [CrossRef] [PubMed]
- Brady, C.; Arnold, D.; McDonald, J.; Denman, S. Taxonomy and Identification of Bacteria Associated with Acute Oak Decline. World J. Microbiol. Biotechnol. 2017, 33, 143. [Google Scholar] [CrossRef]
- Denman, S.; Kirk, S.; Webber, J. Managing Acute Oak Decline; FC Practice Note 15; FC: Edinburgh, Scotland, 2010; p. 6. [Google Scholar]
- Denman, S.; Brown, N.; Kirk, S.; Jeger, M.; Webber, J. A Description of the Symptoms of Acute Oak Decline in Britain and a Comparative Review on Causes of Similar Disorders on Oak in Europe. Forestry 2014, 87, 535–551. [Google Scholar] [CrossRef]
- Broberg, M.; Doonan, J.; Mundt, F.; Denman, S.; McDonald, J.E. Integrated Multi-Omic Analysis of Host-Microbiota Interactions in Acute Oak Decline. Microbiome 2018, 6, 21. [Google Scholar] [CrossRef]
- González, A.J.; Ciordia, M. Brenneria Goodwinii and Gibbsiella Quercinecans Isolated from Weeping Cankers on Quercus robur L. in Spain. Eur. J. Plant Pathol. 2020, 156, 965–969. [Google Scholar] [CrossRef]
- Zalkalns, O.; Celma, L. The Distribution of Bacteria Gibbsiella quercinecans and Brenneria goodwinii in Oak (Quercus robur L.) Stands in Latvia. IOP Conf. Ser. Earth Environ. Sci. 2021, 875, 012033. [Google Scholar] [CrossRef]
- Finch, J.P.; Brown, N.; Beckmann, M.; Denman, S.; Draper, J. Index Measures for Oak Decline Severity Using Phenotypic Descriptors. For. Ecol. Manag. 2021, 485, 118948. [Google Scholar] [CrossRef]
- Crampton, B.G.; Plummer, S.J.; Kaczmarek, M.; McDonald, J.E.; Denman, S. A Multiplex Real-time PCR Assay Enables Simultaneous Rapid Detection and Quantification of Bacteria Associated with Acute Oak Decline. Plant Pathol. 2020, 69, 1301–1310. [Google Scholar] [CrossRef]
- Moradi-Amirabad, Y.; Rahimian, H.; Babaeizad, V.; Denman, S. Brenneria Spp. and Rahnella victoriana Associated with Acute Oak Decline Symptoms on Oak and Hornbeam in Iran. For. Pathol. 2019, 49, e12535. [Google Scholar] [CrossRef]
- Pettifor, B. Survival of Brenneria Goodwinii and Gibbsiella quercinecans, Associated with Lesion Formation in Acute Oak Decline, in Rainwater and Forest Soil. Master’s Thesis, Bangor University, Gwynedd, UK, 2019; pp. 1–90. [Google Scholar]
- Lane, D.J. 16S/23S rRNA Sequencing. In Nucleic Acid Techniques in Bacterial Systematic; Wiley: Chichester, England, 1991; pp. 115–175. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J Mol Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the Number of Nucleotide Substitutions in the Control Region of Mitochondrial DNA in Humans and Chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Eichenlaub, L.; Denman, S.; Brady, C.; Maddock, D.; Robledo-Garcia, F.; Aubert, A.; Husson, C.; Robin, C. First Report of Brenneria goodwinii, Gibbsiella quercinecans and Rahnella victoriana in Declining Oaks in France. New Dis. Rep. 2024, 49, 2023–2024. [Google Scholar] [CrossRef]
- Carluccio, G.; Vergine, M.; Vita, F.; Sabella, E.; Delle Donne, A.; De Bellis, L.; Luvisi, A. Long-Distance Finding of AOD-Related Bacteria in the Natural Environment: Risks to Quercus ilex (L.) in Italy. Forests 2024, 15, 2055. [Google Scholar] [CrossRef]
- Denman, S.; Doonan, J.; Ransom-Jones, E.; Broberg, M.; Plummer, S.; Kirk, S.; Scarlett, K.; Griffiths, A.R.; Kaczmarek, M.; Forster, J.; et al. Microbiome and Infectivity Studies Reveal Complex Polyspecies Tree Disease in Acute Oak Decline. ISME J. 2018, 12, 386–399. [Google Scholar] [CrossRef]
- Maddock, D.; Brady, C.; Denman, S.; Arnold, D. Bacteria Associated with Acute Oak Decline: Where Did They Come From? We Know Where They Go. Microorganisms 2023, 11, 2789. [Google Scholar] [CrossRef]
- Wang, L.; Li, C.; Wang, X.; Wang, G.; Shang, S.; Dou, Z.; Luo, Y. Gut Lignocellulose Activity and Microbiota in Asian Longhorned Beetle and Their Predicted Contribution to Larval Nutrition. Front. Microbiol. 2022, 13, 899865. [Google Scholar] [CrossRef]
- Wang, X.; Wang, H.; Zeng, J.; Cui, Z.; Geng, S.; Song, X.; Zhang, F.; Su, X.; Li, H. Distinct Gut Bacterial Composition in Anoplophora Glabripennis Reared on Two Host Plants. Front. Microbiol. 2023, 14, 1199994. [Google Scholar] [CrossRef]
- Zhang, H. Abiotic Stress Responses in Plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Le Floch, A.; Jourdes, M.; Teissedre, P.L. Polysaccharides and Lignin from Oak Wood Used in Cooperage: Composition, Interest, Assays: A Review. Carbohydr. Res. 2015, 417, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Daniel, G. Variations in Cell Wall Ultrastructure and Chemistry in Cell Types of Earlywood and Latewood in English Oak (Quercus robur). IAWA J. 2016, 37, 383–401. [Google Scholar] [CrossRef]
- Martínez, M.T.; Cuenca, B.; Mosteiro, F.; Piñeiro, P.; Pérez, F.; Solla, A.; Corredoira, E. Screening of Cork Oak for Resistance to Phytophthora cinnamomi and Micropropagation of Tolerant Seedlings. Horticulturae 2023, 9, 692. [Google Scholar] [CrossRef]
- Brown, N.; Jeger, M.; Kirk, S.; Xu, X.; Denman, S. Spatial and Temporal Patterns in Symptom Expression within Eight Woodlands Affected by Acute Oak Decline. For. Ecol. Manag. 2016, 360, 97–109. [Google Scholar] [CrossRef]
- Ducousso, A.; Bordacs, S. EUFORGEN Technical Guidelines for Genetic Conservation and Use for Pedunculate and Sessile Oaks (Quercus robur and Q. petraea); The International Plant Genetic Resources Institute: Maccarese, Italy, 2004; pp. 1–6. [Google Scholar]
- Basavand, E.; Khodaygan, P.; Doonan, J.M.; Rahimian, H. Gibbsiella quercinecans as New Pathogen Involved in Bacterial Canker of Russian Olive. 3 Biotech 2021, 11, 286. [Google Scholar] [CrossRef]
- Allahverdipour, T.; Shahryari, F.; FalahiCharkhabi, N. Gibbsiella quercinecans and Brenneria roseae subsp. roseae Associated to the Canker Disease of Walnut Trees in Northwestern Iran. Eur. J. Plant Pathol. 2021, 161, 783–797. [Google Scholar] [CrossRef]
- Brown, N.; Inward, D.J.G.; Jeger, M.; Denman, S. A Review of A Review of Agrilus biguttatus in UK Forests and Its Relationship with Acute Oak Decline. Forestry 2015, 88, 53–63. [Google Scholar] [CrossRef]
- Moraal, L.G.; Hilszczanski, J. The Oak Buprestid Beetle, Agrilus biguttatus (F.) (Col., Buprestidae), a Recent Factor in Oak Decline in Europe. Anz. Schädlingskunde 2000, 73, 134–138. [Google Scholar] [CrossRef]
- Sallé, A. Native buprestid and longhorn beetles in the Mediterranean Basin. In Insects and Diseases of Mediterranean Forest Systems; Springer International Publishing: Cham, Germany, 2016; pp. 199–210. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).