Genome-Wide Identification, Evolution and Expression Analysis of the U-Box E3 Ubiquitin Ligases Gene Family in Poplar (Populus alba × P. tremula var. glandulosa)
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome-Wide Identification of PUB Gene Family in Poplar
2.2. Multiple Sequence Alignment and Phylogenetic Analysis
2.3. Gene Structure, Conserved Domain, and Motif Analysis
2.4. Cis-Acting Element Analysis of PUB Gene Promoters
2.5. Collinearity Analysis of PUB Genes
2.6. Ka and Ks Analysis
2.7. Expression Pattern Analysis of PUB Gene Family
3. Results
3.1. Whole-Genome Identification of PUB Genes in Poplar
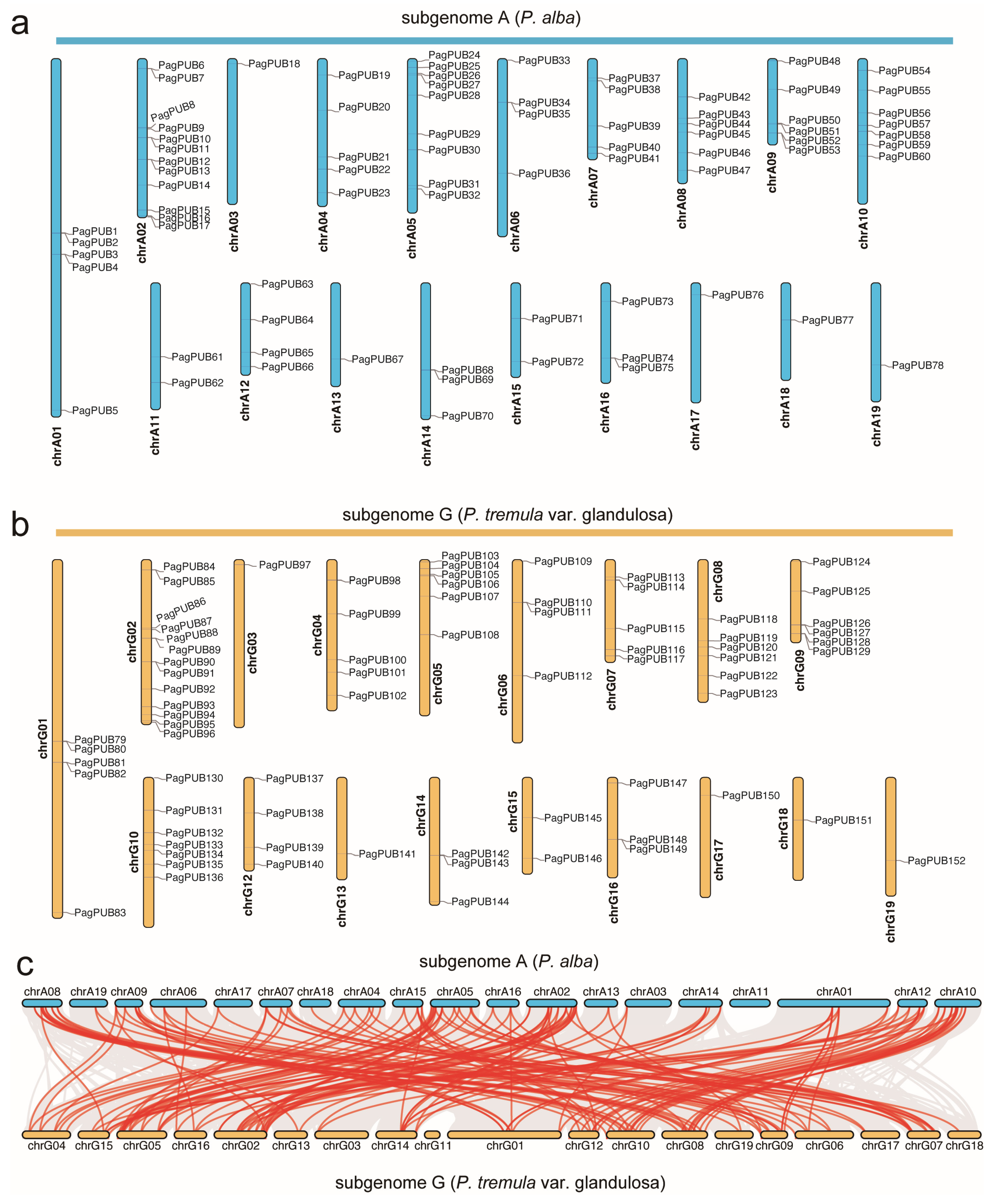
3.2. The Chromosomal Distribution Pattern of PUB Genes in the Poplar Genome
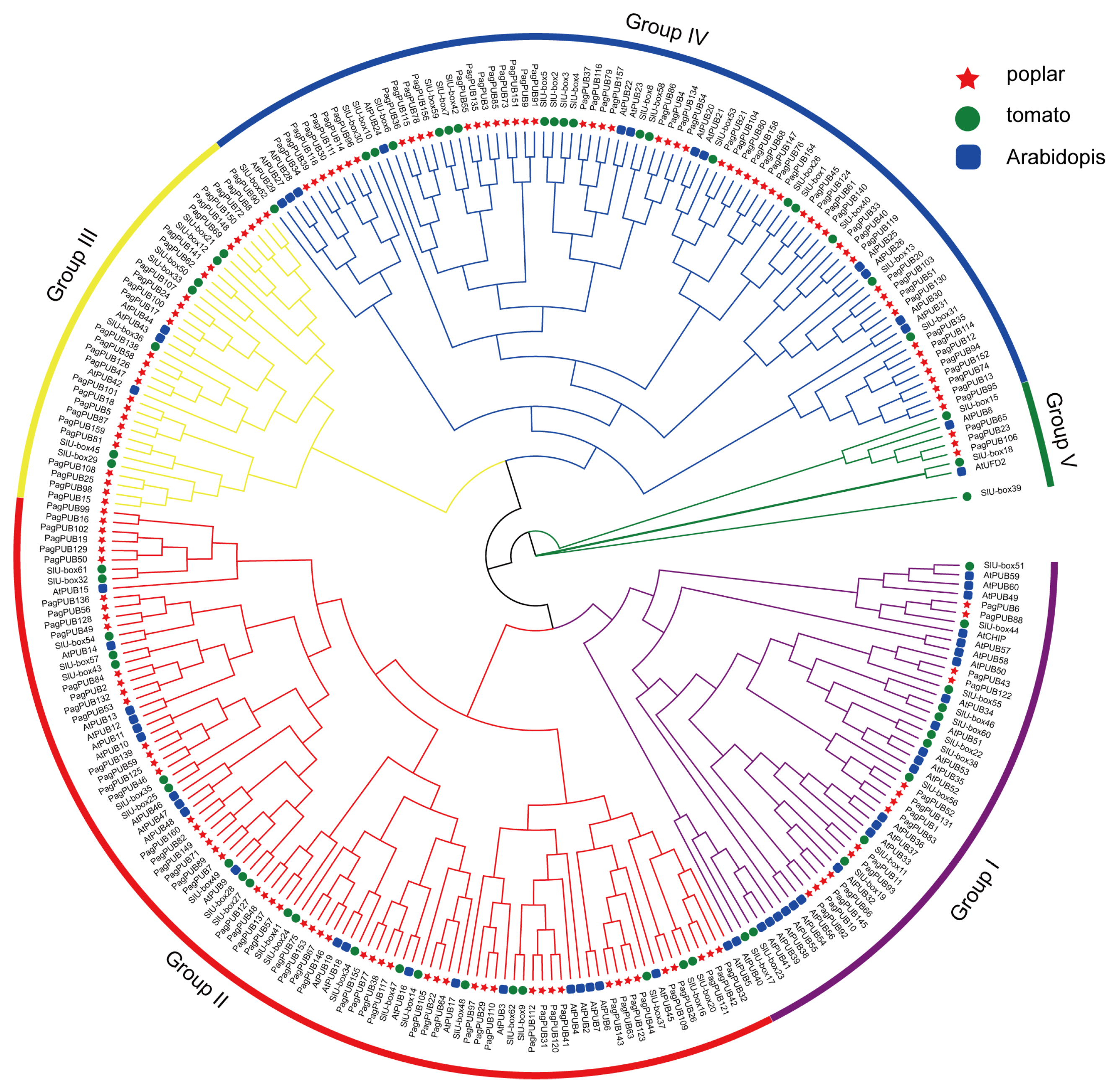
3.3. Phylogenetic Analysis of PUB Genes in Poplar, Arabidopsis and Tomato
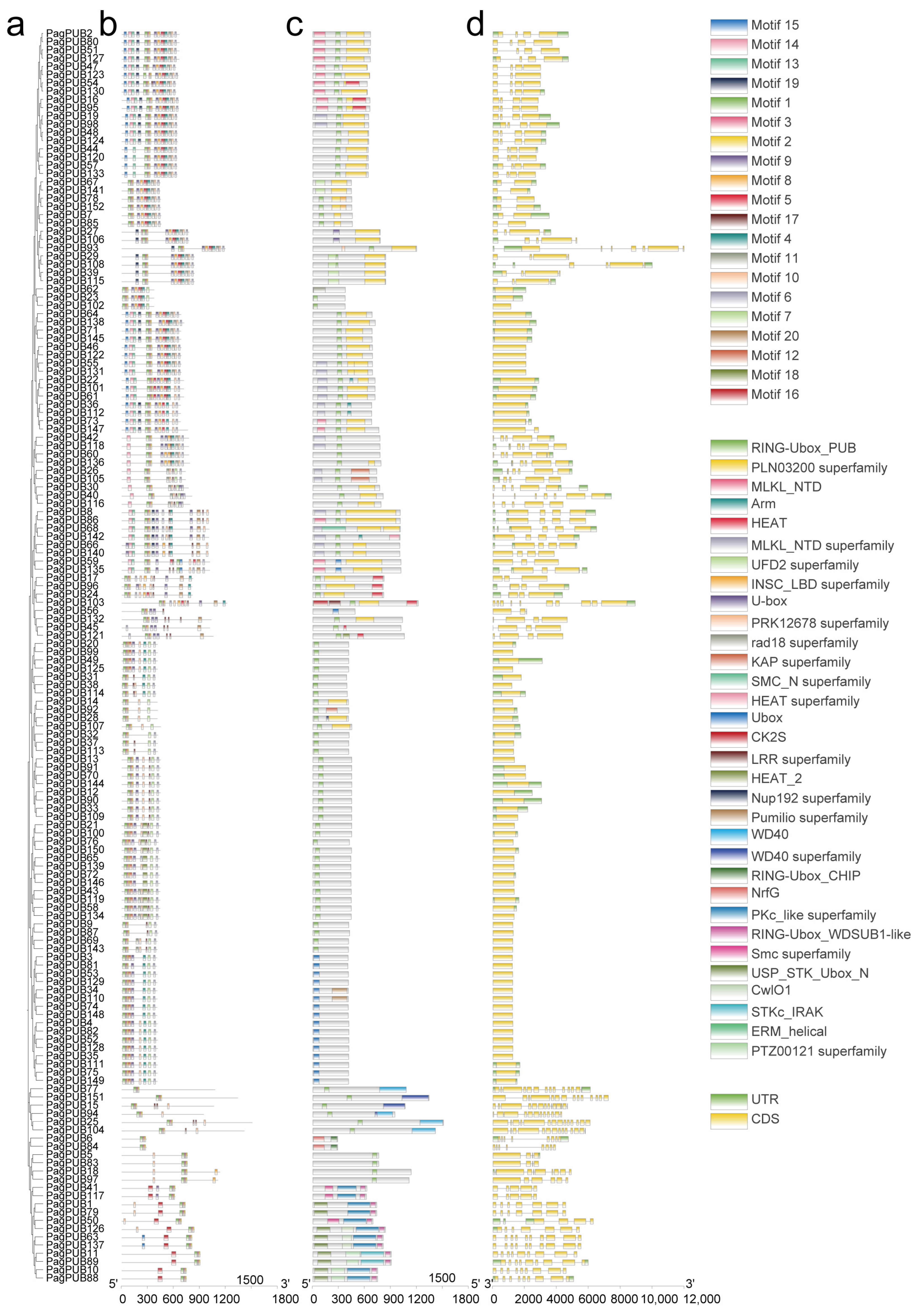
3.4. Gene Structure, Conserved Domain, and Motif Analysis of PagPUB Geness
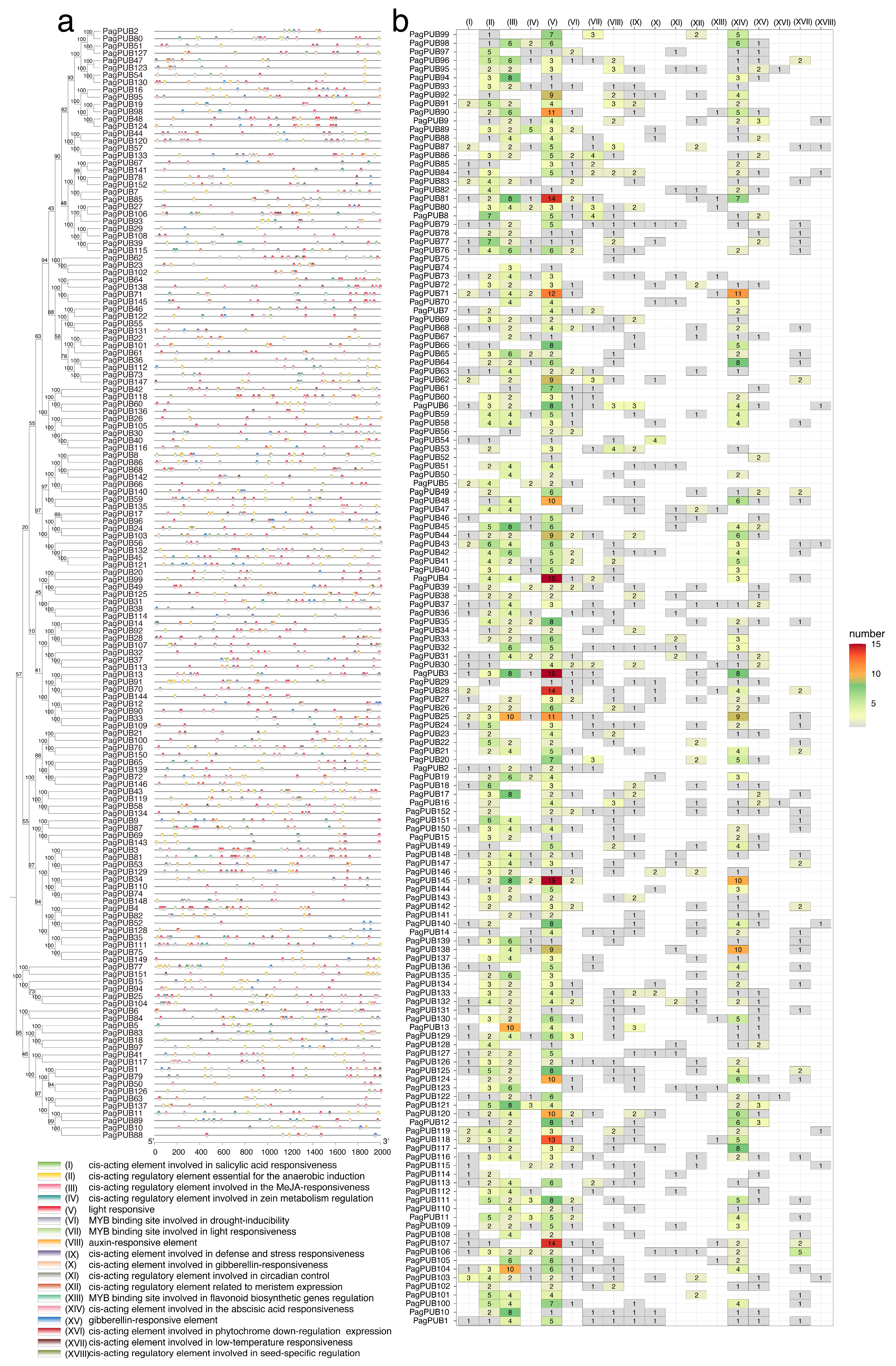
3.5. Cis-Acting Element Predication of PagPUB Genes
3.6. Synteny and Duplication Event Analyses Reveal Expansion of the PUB Gene Family
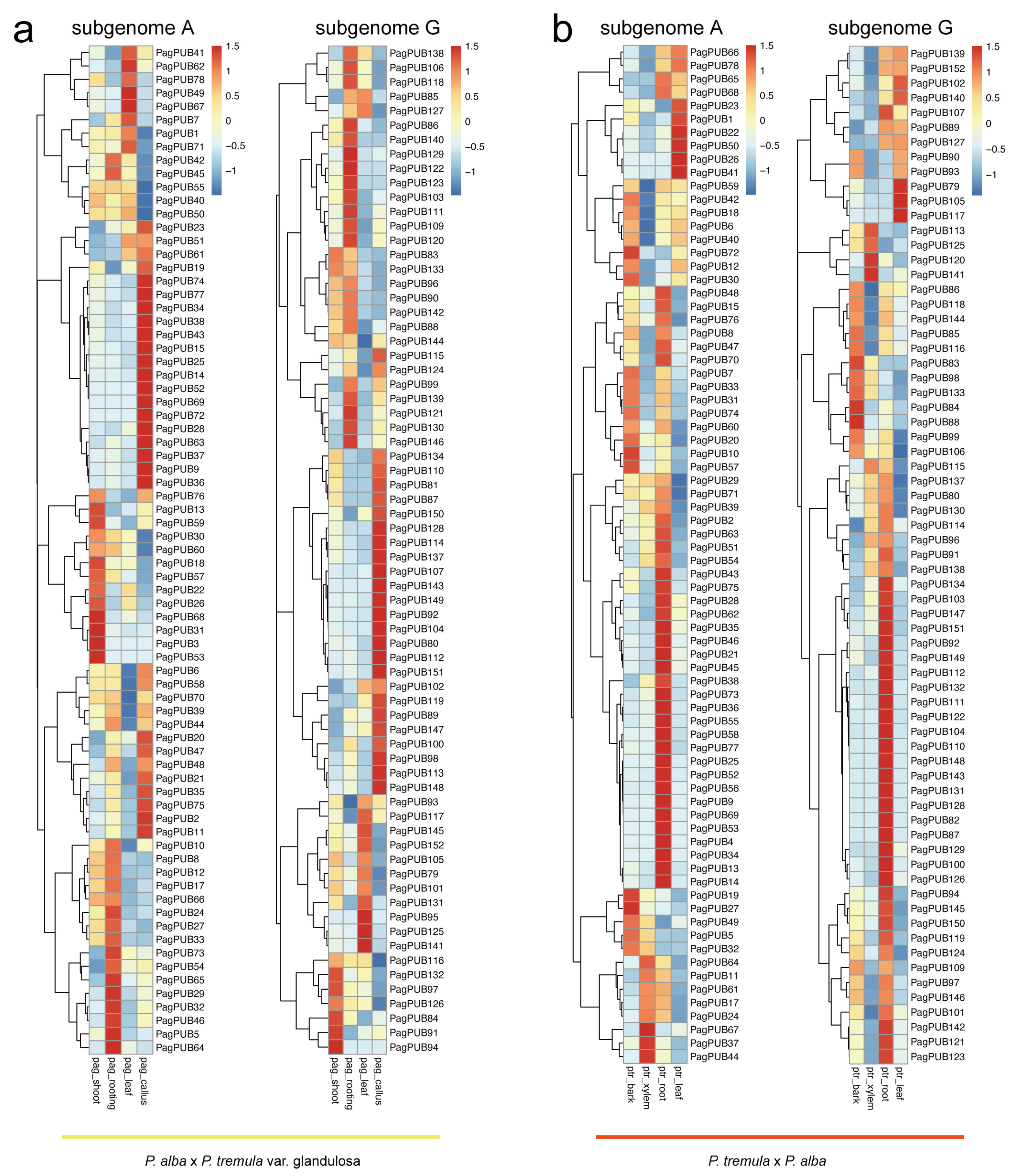
3.7. Expression Patterns of PUB Genes in Different Tissues of Poplar
3.8. Expression Patterns of PUB Genes Under Pathogen Stress
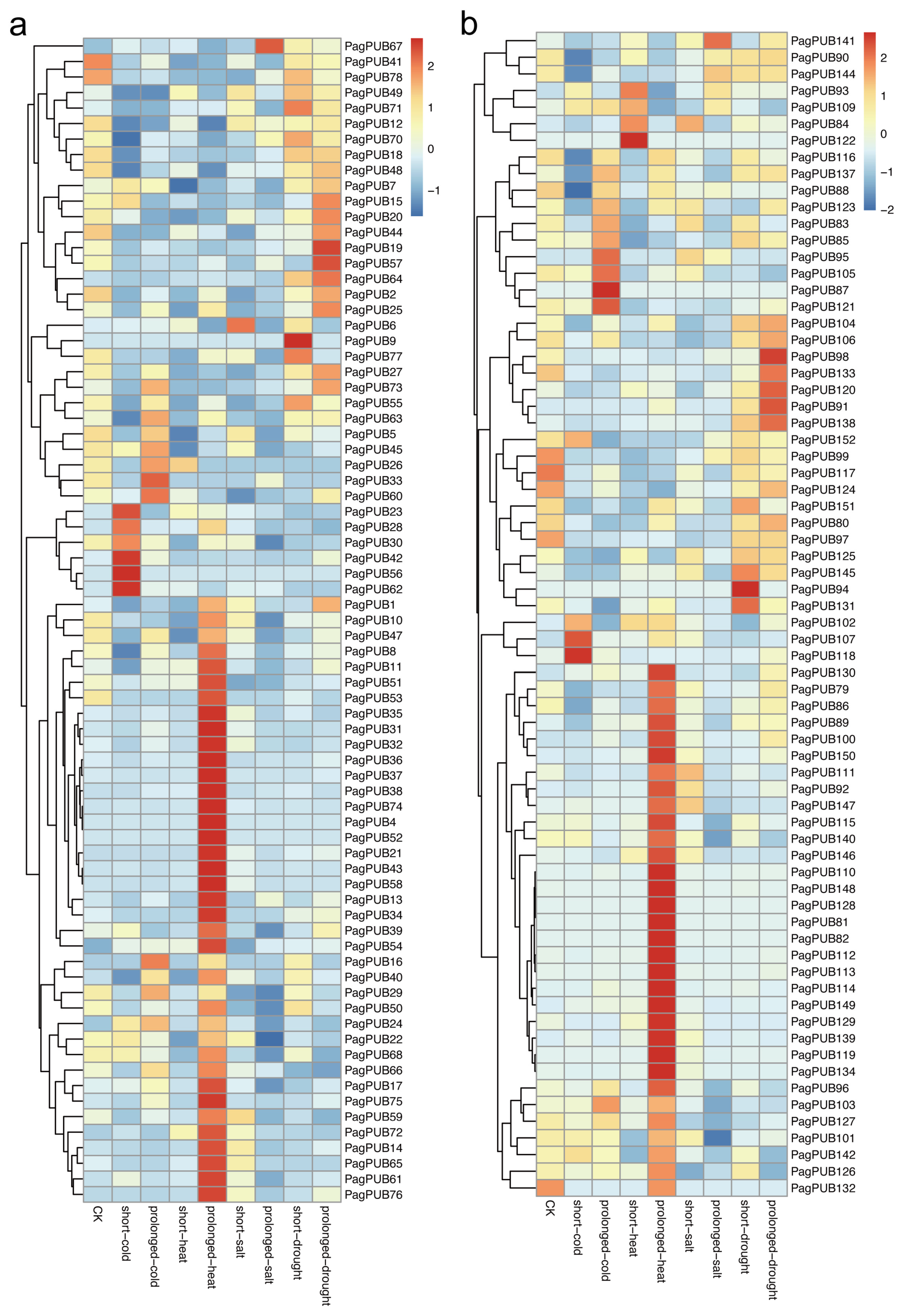
3.9. Expression Patterns of PUB Genes Under Abiotic Stress Conditions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| pI | Theoretical isoelectric points |
| WGD | Whole-genome duplication |
| HMM | Hidden Markov Model |
| pI | Theoretical isoelectric points |
| ABA | Abscisic acid |
| FPKM | Fragments per kilobase of exon per million mapped reads |
| WGD | Whole-genome duplication |
| TD | Tandem duplication |
| PD | Proximal duplication |
| DD | Dispersed duplication |
| SD | Singleton duplication |
| PUB | Plant U-box E3 ubiquitin ligases |
| PTM | Post-translational modification |
| TE | Transposable element |
References
- Martín-Cardoso, H.; San Segundo, B. Impact of Nutrient Stress on Plant Disease Resistance. Int. J. Mol. Sci. 2025, 26, 1780. [Google Scholar] [CrossRef]
- Sachdev, S.; Ansari, S.A.; Ansari, M.I.; Fujita, M.; Hasanuzzaman, M. Abiotic Stress and Reactive Oxygen Species: Generation, Signaling, and Defense Mechanisms. Antioxidants 2021, 10, 277. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Xiong, L.; Shi, H.; Yang, S.; Herrera-Estrella, L.R.; Xu, G.; Chao, D.-Y.; Li, J.; Wang, P.-Y.; Qin, F. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [PubMed]
- Hirayama, T.; Shinozaki, K. Research on plant abiotic stress responses in the post-genome era: Past, present and future. Plant J. 2010, 61, 1041–1052. [Google Scholar] [CrossRef] [PubMed]
- Smalle, J.; Vierstra, R.D. The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 2004, 55, 555–590. [Google Scholar] [CrossRef]
- McClellan, A.J.; Tam, S.; Kaganovich, D.; Frydman, J. Protein quality control: Chaperones culling corrupt conformations. Nat. Cell Biol. 2005, 7, 736–741. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, J.; Liu, H.; Chong, K.; Xu, Y. Roles of ubiquitination-mediated protein degradation in plant responses to abiotic stresses. Environ. Exp. Bot. 2015, 114, 92–103. [Google Scholar] [CrossRef]
- Jansen, A.H.; Reits, E.A.; Hol, E.M. The ubiquitin proteasome system in glia and its role in neurodegenerative diseases. Front. Mol. Neurosci. 2014, 7, 73. [Google Scholar] [CrossRef]
- Santner, A.; Estelle, M. The ubiquitin-proteasome system regulates plant hormone signaling. Plant J. 2010, 61, 1029–1040. [Google Scholar] [CrossRef]
- Song, J.; Mo, X.; Yang, H.; Yue, L.; Song, J.; Mo, B. The U-box family genes in Medicago truncatula: Key elements in response to salt, cold, and drought stresses. PLoS ONE 2017, 12, e0182402. [Google Scholar] [CrossRef]
- Azevedo, C.; Santos-Rosa, M.J.; Shirasu, K. The U-box protein family in plants. Trends Plant Sci. 2001, 6, 354–358. [Google Scholar] [CrossRef] [PubMed]
- Wiborg, J.; O’Shea, C.; Skriver, K. Biochemical function of typical and variant Arabidopsis thaliana U-box E3 ubiquitin-protein ligases. Biochem. J. 2008, 413, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.-R.; Park, C.H.; Venu, R.C.; Gough, J.; Wang, G.-L. Classification, Expression Pattern, and E3 Ligase Activity Assay of Rice U-Box-Containing Proteins. Mol. Plant 2008, 1, 800–815. [Google Scholar] [CrossRef]
- Sharma, B.; Taganna, J. Genome-wide analysis of the U-box E3 ubiquitin ligase enzyme gene family in tomato. Sci. Rep. 2020, 10, 9581. [Google Scholar] [CrossRef]
- Lu, X.; Shu, N.; Wang, D.; Wang, J.; Chen, X.; Zhang, B.; Wang, S.; Guo, L.; Chen, C.; Ye, W. Genome-wide identification and expression analysis of PUB genes in cotton. BMC Genom. 2020, 21, 213. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Dong, C.; Sun, D.; Hu, Y.; Xie, J. Genome-Wide Identification and Analysis of U-Box E3 Ubiquitin-Protein Ligase Gene Family in Banana. Int. J. Mol. Sci. 2018, 19, 3874. [Google Scholar] [CrossRef]
- Wang, C.; Duan, W.; Riquicho, A.R.; Jing, Z.; Liu, T.; Hou, X.; Li, Y. Genome-wide survey and expression analysis of the PUB family in Chinese cabbage (Brassica rapa ssp. pekinesis). Mol. Genet. Genom. 2015, 290, 2241–2260. [Google Scholar] [CrossRef]
- Seo, D.H.; Ryu, M.Y.; Jammes, F.; Hwang, J.H.; Turek, M.; Kang, B.G.; Kwak, J.M.; Kim, W.T. Roles of Four Arabidopsis U-Box E3 Ubiquitin Ligases in Negative Regulation of Abscisic Acid-Mediated Drought Stress Responses. Plant Physiol. 2012, 160, 556–568. [Google Scholar] [CrossRef]
- Wang, J.; Qu, B.; Dou, S.; Li, L.; Yin, D.; Pang, Z.; Zhou, Z.; Tian, M.; Liu, G.; Xie, Q.; et al. The E3 ligase OsPUB15 interacts with the receptor-like kinase PID2 and regulates plant cell death and innate immunity. BMC Plant Biol. 2015, 15, 49. [Google Scholar] [CrossRef]
- Kaur, H.; Salvi, P.; Chaurasia, A.; Mondal, T.K. PUB63, a novel early heat responsive ubiquitin E3 ligase from rice implicates heat stress tolerance. Plant Stress 2023, 10, 100291. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Madden, T. The BLAST Sequence Analysis Tool. In The NCBI Handbook, 2nd ed.; National Center for Biotechnology Information (NCBI): Bethesda, MD, USA, 2013; Volume 2, pp. 425–436. [Google Scholar]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Zhang, Z.; Xiao, J.; Wu, J.; Zhang, H.; Liu, G.; Wang, X.; Dai, L. ParaAT: A parallel tool for constructing multiple protein-coding DNA alignments. Biochem. Biophys. Res. Commun. 2012, 419, 779–781. [Google Scholar] [CrossRef] [PubMed]
- Harding, S.A.; Hu, H.; Nyamdari, B.; Xue, L.-J.; Naran, R.; Tsai, C.-J. Tubulins, rhythms and cell walls in poplar leaves: It’s all in the timing. Tree Physiol. 2018, 38, 397–408. [Google Scholar] [CrossRef]
- Xue, L.-J.; Frost, C.J.; Tsai, C.-J.; Harding, S.A. Drought response transcriptomes are altered in poplar with reduced tonoplast sucrose transporter expression. Sci. Rep. 2016, 6, 33655. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, J.; Yang, Z.; Matsui, A.; Seki, M.; Li, S.; Yan, X.; Kohnen, M.V.; Gu, L.; Prasad, K. PtWOX11 acts as master regulator conducting the expression of key transcription factors to induce de novo shoot organogenesis in poplar. Plant Mol. Biol. 2018, 98, 389–406. [Google Scholar] [CrossRef]
- Shu, W.; Zhou, H.; Jiang, C.; Zhao, S.; Wang, L.; Li, Q.; Yang, Z.; Groover, A.; Lu, M.Z. The auxin receptor TIR 1 homolog (Pag FBL 1) regulates adventitious rooting through interactions with Aux/IAA 28 in Populus. Plant Biotechnol. J. 2019, 17, 338–349. [Google Scholar] [CrossRef]
- Yang, G.; Wang, S.; Long, L.; Yu, X.; Cai, H.; Chen, P.; Gu, L.; Yang, M. Genome-wide identification and expression analysis of PtJAZ gene family in poplar (Populus trichocarpa). BMC Genom. Data 2023, 24, 55. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Subgroup, G.P.D.P. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Kolde, R. Package ‘pheatmap’. Available online: https://cran.r-project.org/web/packages/pheatmap/index.html (accessed on 22 April 2025).
- Bharath, P.; Gahir, S.; Raghavendra, A.S. Abscisic Acid-Induced Stomatal Closure: An Important Component of Plant Defense Against Abiotic and Biotic Stress. Front. Plant Sci. 2021, 12, 615114. [Google Scholar] [CrossRef] [PubMed]
- Yee, D.; Goring, D.R. The diversity of plant U-box E3 ubiquitin ligases: From upstream activators to downstream target substrates. J. Exp. Bot. 2009, 60, 1109–1121. [Google Scholar] [CrossRef]
- Zeng, L.-R.; Qu, S.; Bordeos, A.; Yang, C.; Baraoidan, M.; Yan, H.; Xie, Q.; Nahm, B.H.; Leung, H.; Wang, G.-L. Spotted leaf11, a Negative Regulator of Plant Cell Death and Defense, Encodes a U-Box/Armadillo Repeat Protein Endowed with E3 Ubiquitin Ligase Activity. Plant Cell 2004, 16, 2795–2808. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.-W.; González-Lamothe, R.; Ewan, R.A.; Rowland, O.; Yoshioka, H.; Shenton, M.; Ye, H.; O’Donnell, E.; Jones, J.D.G.; Sadanandom, A. The E3 Ubiquitin Ligase Activity of Arabidopsis PLANT U-BOX17 and Its Functional Tobacco Homolog ACRE276 Are Required for Cell Death and Defense. Plant Cell 2006, 18, 1084–1098. [Google Scholar] [CrossRef]
- Orosa, B.; He, Q.; Mesmar, J.; Gilroy, E.M.; McLellan, H.; Yang, C.; Craig, A.; Bailey, M.; Zhang, C.; Moore, J.D.; et al. BTB-BACK Domain Protein POB1 Suppresses Immune Cell Death by Targeting Ubiquitin E3 ligase PUB17 for Degradation. PLoS Genet. 2017, 13, e1006540. [Google Scholar] [CrossRef]
- Shu, K.; Yang, W. E3 Ubiquitin Ligases: Ubiquitous Actors in Plant Development and Abiotic Stress Responses. Plant Cell Physiol. 2017, 58, 1461–1476. [Google Scholar] [CrossRef]
- Wang, C.; Song, B.; Dai, Y.; Zhang, S.; Huang, X. Genome-wide identification and functional analysis of U-box E3 ubiquitin ligases gene family related to drought stress response in Chinese white pear (Pyrus bretschneideri). BMC Plant Biol. 2021, 21, 235. [Google Scholar] [CrossRef]
- Cho, S.K.; Ryu, M.Y.; Song, C.; Kwak, J.M.; Kim, W.T. Arabidopsis PUB22 and PUB23 are homologous U-Box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant Cell 2008, 20, 1899–1914. [Google Scholar] [CrossRef] [PubMed]
- Adler, G.; Konrad, Z.; Zamir, L.; Mishra, A.K.; Raveh, D.; Bar-Zvi, D. The Arabidopsis paralogs, PUB46 and PUB48, encoding U-box E3 ubiquitin ligases, are essential for plant response to drought stress. BMC Plant Biol. 2017, 17, 8. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Zhang, X.; Hou, Y.; Jia, C.; Dan, X.; Zhang, Y.; Jiang, Y.; Lai, Q.; Feng, J.; Feng, J.; et al. The super-pangenome of Populus unveils genomic facets for its adaptation and diversification in widespread forest trees. Mol. Plant 2024, 17, 725–746. [Google Scholar] [CrossRef] [PubMed]
- Machado, T.B.; Picorelli, A.C.R.; Azevedo, B.L.d.; Aquino, I.L.M.d.; Queiroz, V.F.; Rodrigues, R.A.L.; Araújo, J.P.; Ullmann, L.S.; Santos, T.M.d.; Marques, R.E.; et al. Gene duplication as a major force driving the genome expansion in some giant viruses. J. Virol. 2023, 97, e01309–e01323. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.-H.; Niu, M.-X.; Liu, X.; Bao, Y.; Liu, S.; Liu, M.; He, F.; Han, S.; Liu, C.; Wang, H.-L.; et al. Genome-Wide Analysis of the FBA Subfamily of the Poplar F-Box Gene Family and Its Role under Drought Stress. Int. J. Mol. Sci. 2023, 24, 4823. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, Y.; Zhong, H.; Chen, S.; Wong, K.-B.; Xia, Y. Arabidopsis PUB2 and PUB4 connect signaling components of pattern-triggered immunity. New Phytol. 2022, 233, 2249–2265. [Google Scholar] [CrossRef]


| Gene Name | Sequence ID | Chromosome | Start | End | Strand | Number of Amino Acid | Molecular Weight | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity | Prediction of Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PagPUB1 | Pop_A01G056850.T1 | chrA01 | 23,796,664 | 23,801,216 | − | 736 | 82,944 | 7 | 49 | 93 | −0.289 | nuclear |
| PagPUB2 | Pop_A01G056845.T1 | chrA01 | 23,858,439 | 23,863,158 | + | 665 | 72,007 | 6 | 38 | 100 | −0.17 | plasma membrane |
| PagPUB3 | Pop_A01G054584.T1 | chrA01 | 26,716,220 | 26,717,443 | + | 407 | 46,123 | 9 | 42 | 111 | 0 | chloroplast |
| PagPUB4 | Pop_A01G054588.T1 | chrA01 | 26,742,954 | 26,744,201 | − | 415 | 46,597 | 9 | 51 | 111 | 0 | cytoplasmic |
| PagPUB5 | Pop_A01G029509.T1 | chrA01 | 48,026,055 | 48,028,998 | − | 760 | 84,059 | 7 | 45 | 68 | −0.725 | nuclear |
| PagPUB6 | Pop_A02G032997.T1 | chrA02 | 1,345,295 | 1,350,018 | + | 287 | 32,751 | 5 | 48 | 87 | −0.549 | nuclear |
| PagPUB7 | Pop_A02G032998.T1 | chrA02 | 1,358,038 | 1,361,559 | − | 456 | 51,113 | 9 | 39 | 103 | −0.227 | nuclear |
| PagPUB8 | Pop_A02G012420.T1 | chrA02 | 9,419,791 | 9,426,215 | − | 1008 | 111,443 | 5 | 49 | 104 | −0.077 | nuclear |
| PagPUB9 | Pop_A02G012404.T1 | chrA02 | 9,601,590 | 9,602,834 | − | 414 | 46,147 | 8 | 41 | 110 | 0 | nuclear |
| PagPUB10 | Pop_A02G012262.T1 | chrA02 | 10,765,439 | 10,770,018 | − | 748 | 84,833 | 5 | 47 | 88 | −0.39 | nuclear |
| PagPUB11 | Pop_A02G012261.T1 | chrA02 | 10,777,449 | 10,782,710 | − | 906 | 101,393 | 6 | 55 | 80 | −0.479 | nuclear |
| PagPUB12 | Pop_A02G066389.T1 | chrA02 | 13,769,391 | 13,771,851 | + | 447 | 48,565 | 7 | 50 | 119 | 0 | cytoplasmic |
| PagPUB13 | Pop_A02G066388.T1 | chrA02 | 13,777,487 | 13,778,836 | + | 449 | 48,995 | 6 | 39 | 112 | 0 | cytoplasmic |
| PagPUB14 | Pop_A02G065553.T1 | chrA02 | 17,254,384 | 17,255,619 | − | 411 | 44,849 | 6 | 43 | 115 | 0 | cytoplasmic |
| PagPUB15 | Pop_A02G004950.T1 | chrA02 | 20,682,911 | 20,687,581 | − | 1063 | 117,967 | 9 | 46 | 91 | −0.139 | nuclear |
| PagPUB16 | Pop_A02G005103.T1 | chrA02 | 21,466,965 | 21,469,799 | + | 658 | 73,401 | 6 | 41 | 103 | −0.232 | nuclear |
| PagPUB17 | Pop_A02G026918.T1 | chrA02 | 21,609,395 | 21,612,784 | + | 822 | 90,116 | 5 | 42 | 110 | 0 | cytoplasmic |
| PagPUB18 | Pop_A03G034675.T1 | chrA03 | 666,747 | 671,647 | + | 1133 | 126,732 | 6 | 54 | 86 | −0.478 | cytoplasmic |
| PagPUB19 | Pop_A04G002612.T1 | chrA04 | 2,232,642 | 2,236,254 | − | 644 | 71,826 | 7 | 38 | 109 | −0.173 | cytoplasmic |
| PagPUB20 | Pop_A04G030625.T1 | chrA04 | 7,039,512 | 7,040,945 | + | 413 | 45,253 | 7 | 51 | 109 | 0 | plasma membrane |
| PagPUB21 | Pop_A04G029277.T1 | chrA04 | 13,405,290 | 13,406,636 | + | 448 | 50,282 | 8 | 39 | 102 | −0.076 | endoplasmic reticulum |
| PagPUB22 | Pop_A04G029140.T1 | chrA04 | 15,105,012 | 15,107,880 | − | 717 | 78,101 | 7 | 45 | 103 | 0 | chloroplast |
| PagPUB23 | Pop_A04G018204.T1 | chrA04 | 18,307,535 | 18,309,391 | − | 372 | 40,876 | 8 | 43 | 103 | −0.049 | cytoplasmic |
| PagPUB24 | Pop_A05G057115.T1 | chrA05 | 369,241 | 373,596 | − | 817 | 89,843 | 5 | 41 | 104 | −0.074 | cytoplasmic |
| PagPUB25 | Pop_A05G002002.T1 | chrA05 | 1,220,232 | 1,226,316 | + | 1506 | 167,956 | 6 | 47 | 91 | −0.21 | chloroplast |
| PagPUB26 | Pop_A05G001867.T1 | chrA05 | 2,028,105 | 2,033,060 | − | 736 | 81,596 | 6 | 48 | 100 | −0.091 | nuclear |
| PagPUB27 | Pop_A05G001837.T1 | chrA05 | 2,224,762 | 2,228,387 | − | 777 | 85,533 | 7 | 45 | 92 | −0.238 | chloroplast |
| PagPUB28 | Pop_A05G056236.T1 | chrA05 | 4,986,956 | 4,988,532 | + | 411 | 44,921 | 7 | 45 | 107 | 0 | cytoplasmic |
| PagPUB29 | Pop_A05G073436.T1 | chrA05 | 10,280,710 | 10,285,458 | − | 840 | 91,038 | 6 | 45 | 98 | −0.209 | peroxisome |
| PagPUB30 | Pop_A05G073075.T1 | chrA05 | 12,431,803 | 12,437,733 | + | 771 | 86,522 | 6 | 46 | 94 | −0.206 | nuclear |
| PagPUB31 | Pop_A05G071680.T1 | chrA05 | 17,332,961 | 17,334,731 | + | 390 | 43,718 | 6 | 53 | 106 | −0.051 | plasma membrane |
| PagPUB32 | Pop_A05G071756.T1 | chrA05 | 17,795,466 | 17,797,218 | − | 418 | 46,824 | 7 | 39 | 107 | 0 | chloroplast |
| PagPUB33 | Pop_A06G062990.T1 | chrA06 | 197,704 | 199,882 | + | 448 | 48,303 | 8 | 46 | 117 | 0 | nuclear |
| PagPUB34 | Pop_A06G053578.T1 | chrA06 | 5,979,373 | 5,980,602 | + | 409 | 45,910 | 9 | 47 | 101 | −0.017 | cytoplasmic |
| PagPUB35 | Pop_A06G053579.T1 | chrA06 | 5,994,456 | 5,995,697 | + | 413 | 45,627 | 8 | 44 | 110 | 0 | nuclear |
| PagPUB36 | Pop_A06G089340.T1 | chrA06 | 15,661,456 | 15,663,646 | − | 679 | 74,982 | 9 | 43 | 106 | 0 | chloroplast |
| PagPUB37 | Pop_A07G006696.T1 | chrA07 | 2,652,193 | 2,653,492 | + | 414 | 46,376 | 9 | 40 | 109 | 0 | cytoplasmic |
| PagPUB38 | Pop_A07G006636.T1 | chrA07 | 3,033,746 | 3,034,924 | − | 392 | 43,429 | 6 | 48 | 106 | −0.02 | extracellular |
| PagPUB39 | Pop_A07G011859.T1 | chrA07 | 9,187,531 | 9,191,737 | − | 839 | 91,065 | 6 | 48 | 96 | −0.241 | peroxisome |
| PagPUB40 | Pop_A07G022563.T1 | chrA07 | 12,074,623 | 12,082,041 | + | 811 | 90,727 | 6 | 47 | 93 | −0.254 | nuclear |
| PagPUB41 | Pop_A07G022689.T1 | chrA07 | 12,936,932 | 12,939,679 | + | 622 | 70,767 | 7 | 53 | 84 | −0.509 | cytoplasmic |
| PagPUB42 | Pop_A08G045991.T1 | chrA08 | 5,220,443 | 5,224,273 | − | 775 | 85,446 | 6 | 55 | 91 | −0.269 | nuclear |
| PagPUB43 | Pop_A08G086524.T1 | chrA08 | 8,137,824 | 8,139,155 | + | 443 | 49,421 | 8 | 40 | 103 | −0.032 | mitochondrial |
| PagPUB44 | Pop_A08G064119.T1 | chrA08 | 8,882,645 | 8,885,439 | − | 640 | 69,410 | 9 | 41 | 99 | −0.17 | plasma membrane |
| PagPUB45 | Pop_A08G063939.T1 | chrA08 | 10,037,789 | 10,042,023 | − | 1017 | 113,825 | 6 | 51 | 101 | −0.16 | nuclear |
| PagPUB46 | Pop_A08G057978.T1 | chrA08 | 12,841,028 | 12,843,094 | − | 688 | 75,652 | 8 | 45 | 107 | −0.007 | golgi apparatus |
| PagPUB47 | Pop_A08G002355.T1 | chrA08 | 15,252,593 | 15,255,576 | + | 627 | 68,486 | 6 | 39 | 101 | −0.248 | chloroplast |
| PagPUB48 | Pop_A09G077078.T1 | chrA09 | 300,369 | 303,683 | − | 644 | 71,635 | 8 | 33 | 109 | −0.178 | cytoplasmic |
| PagPUB49 | Pop_A09G026705.T1 | chrA09 | 4,212,205 | 4,215,293 | + | 413 | 45,154 | 7 | 49 | 109 | 0 | plasma membrane |
| PagPUB50 | Pop_A09G014477.T1 | chrA09 | 8,839,017 | 8,845,299 | − | 692 | 78,007 | 7 | 55 | 86 | −0.359 | extracellular |
| PagPUB51 | Pop_A09G059535.T1 | chrA09 | 8,931,456 | 8,935,612 | + | 663 | 72,050 | 6 | 38 | 98 | −0.254 | endoplasmic reticulum |
| PagPUB52 | Pop_A09G083679.T1 | chrA09 | 10,169,989 | 10,171,236 | + | 415 | 46,623 | 9 | 51 | 108 | −0.04 | cytoplasmic |
| PagPUB53 | Pop_A09G083546.T1 | chrA09 | 10,179,002 | 10,180,234 | − | 410 | 46,775 | 9 | 47 | 108 | −0.098 | cytoplasmic |
| PagPUB54 | Pop_A10G046735.T1 | chrA10 | 1,606,019 | 1,608,984 | − | 628 | 68,816 | 6 | 41 | 102 | −0.198 | plasma membrane |
| PagPUB55 | Pop_A10G069610.T1 | chrA10 | 4,319,997 | 4,322,063 | + | 688 | 75,339 | 8 | 43 | 105 | −0.025 | extracellular |
| PagPUB56 | Pop_A10G069115.T1 | chrA10 | 7,414,568 | 7,416,679 | + | 487 | 55,597 | 5 | 65 | 95 | −0.307 | nuclear |
| PagPUB57 | Pop_A10G001694.T1 | chrA10 | 9,124,333 | 9,127,627 | + | 640 | 69,309 | 9 | 40 | 101 | −0.165 | plasma membrane |
| PagPUB58 | Pop_A10G048475.T1 | chrA10 | 9,911,809 | 9,913,292 | − | 442 | 49,196 | 8 | 39 | 103 | −0.023 | mitochondrial |
| PagPUB59 | Pop_A10G047247.T1 | chrA10 | 11,735,560 | 11,739,663 | − | 1016 | 112,970 | 6 | 41 | 110 | −0.062 | chloroplast |
| PagPUB60 | Pop_A10G047474.T1 | chrA10 | 13,333,482 | 13,337,243 | + | 775 | 85,344 | 5 | 50 | 94 | −0.213 | nuclear |
| PagPUB61 | Pop_A11G056718.T1 | chrA11 | 10,089,153 | 10,091,828 | − | 717 | 78,288 | 7 | 45 | 100 | 0 | chloroplast |
| PagPUB62 | Pop_A11G056786.T1 | chrA11 | 13,612,918 | 13,614,976 | + | 374 | 40,804 | 8 | 42 | 105 | 0 | cytoplasmic |
| PagPUB63 | Pop_A12G008014.T1 | chrA12 | 131,338 | 136,861 | − | 812 | 92,531 | 6 | 46 | 98 | −0.254 | nuclear |
| PagPUB64 | Pop_A12G019268.T1 | chrA12 | 5,023,152 | 5,025,568 | − | 684 | 75,202 | 8 | 45 | 103 | 0 | nuclear |
| PagPUB65 | Pop_A12G073757.T1 | chrA12 | 9,494,982 | 9,496,301 | − | 439 | 49,360 | 7 | 48 | 99 | −0.097 | chloroplast |
| PagPUB66 | Pop_A12G081617.T1 | chrA12 | 11,425,998 | 11,431,252 | − | 1004 | 111,587 | 6 | 44 | 109 | −0.001 | chloroplast |
| PagPUB67 | Pop_A13G009822.T1 | chrA13 | 10,404,574 | 10,407,282 | − | 445 | 49,296 | 6 | 40 | 109 | −0.113 | cytoplasmic |
| PagPUB68 | Pop_A14G044946.T1 | chrA14 | 11,867,888 | 11,874,382 | + | 1010 | 111,633 | 5 | 49 | 106 | −0.072 | nuclear |
| PagPUB69 | Pop_A14G044935.T1 | chrA14 | 11,920,291 | 11,921,529 | − | 412 | 45,824 | 7 | 44 | 104 | 0 | cytoplasmic |
| PagPUB70 | Pop_A14G044906.T1 | chrA14 | 18,148,874 | 18,150,922 | + | 449 | 48,797 | 6 | 41 | 115 | 0 | plasma membrane |
| PagPUB71 | Pop_A15G017145.T1 | chrA15 | 4,862,048 | 4,864,485 | − | 684 | 75,348 | 8 | 47 | 103 | 0 | chloroplast |
| PagPUB72 | Pop_A15G064381.T1 | chrA15 | 10,751,345 | 10,752,762 | + | 440 | 49,341 | 7 | 44 | 101 | −0.069 | nuclear |
| PagPUB73 | Pop_A16G089936.T1 | chrA16 | 2,517,600 | 2,520,014 | − | 679 | 74,498 | 9 | 42 | 108 | 0 | chloroplast |
| PagPUB74 | Pop_A16G055207.T1 | chrA16 | 10,259,891 | 10,261,117 | + | 408 | 46,065 | 8 | 49 | 104 | 0 | cytoplasmic |
| PagPUB75 | Pop_A16G055218.T1 | chrA16 | 10,265,330 | 10,266,987 | + | 412 | 45,786 | 8 | 42 | 107 | 0 | nuclear |
| PagPUB76 | Pop_A17G075184.T1 | chrA17 | 1,624,650 | 1,625,918 | − | 422 | 46,912 | 8 | 43 | 103 | −0.012 | plasma membrane |
| PagPUB77 | Pop_A18G018919.T1 | chrA18 | 5,064,274 | 5,070,370 | + | 1077 | 118,630 | 6 | 47 | 97 | −0.096 | nuclear |
| PagPUB78 | Pop_A19G052378.T1 | chrA19 | 11,228,973 | 11,231,557 | − | 450 | 50,015 | 6 | 38 | 103 | −0.221 | chloroplast |
| PagPUB79 | Pop_G01G020781.T1 | chrG01 | 24,858,647 | 24,863,198 | − | 736 | 83,005 | 6 | 49 | 92 | −0.29 | nuclear |
| PagPUB80 | Pop_G01G020787.T1 | chrG01 | 24,917,617 | 24,921,318 | + | 665 | 71,971 | 6 | 36 | 99 | −0.165 | plasma membrane |
| PagPUB81 | Pop_G01G005854.T1 | chrG01 | 27,761,542 | 27,762,756 | + | 404 | 45,716 | 9 | 41 | 112 | 0 | chloroplast |
| PagPUB82 | Pop_G01G067834.T1 | chrG01 | 27,774,725 | 27,775,972 | − | 415 | 46,364 | 9 | 52 | 111 | 0 | cytoplasmic |
| PagPUB83 | Pop_G01G075640.T1 | chrG01 | 48,346,959 | 48,349,807 | − | 760 | 84,385 | 7 | 44 | 70 | −0.715 | nuclear |
| PagPUB84 | Pop_G02G015781.T1 | chrG02 | 1,403,385 | 1,407,298 | + | 287 | 32,729 | 5 | 50 | 86 | −0.563 | nuclear |
| PagPUB85 | Pop_G02G015782.T1 | chrG02 | 1,415,416 | 1,417,463 | − | 456 | 51,086 | 9 | 40 | 102 | −0.229 | cytoplasmic |
| PagPUB86 | Pop_G02G035213.T1 | chrG02 | 9,334,567 | 9,340,370 | − | 1008 | 111,437 | 6 | 49 | 104 | −0.089 | nuclear |
| PagPUB87 | Pop_G02G035233.T1 | chrG02 | 9,561,820 | 9,563,094 | − | 424 | 47,535 | 7 | 41 | 108 | 0 | nuclear |
| PagPUB88 | Pop_G02G031066.T1 | chrG02 | 10,740,805 | 10,745,856 | − | 748 | 84,891 | 5 | 47 | 89 | −0.38 | nuclear |
| PagPUB89 | Pop_G02G031070.T1 | chrG02 | 10,752,129 | 10,758,095 | − | 906 | 101,441 | 6 | 55 | 79 | −0.491 | nuclear |
| PagPUB90 | Pop_G02G005632.T1 | chrG02 | 13,967,116 | 13,970,164 | + | 447 | 48,509 | 7 | 50 | 118 | 0 | cytoplasmic |
| PagPUB91 | Pop_G02G005633.T1 | chrG02 | 13,975,351 | 13,977,393 | + | 449 | 48,996 | 6 | 39 | 111 | 0 | cytoplasmic |
| PagPUB92 | Pop_G02G005270.T1 | chrG02 | 17,688,838 | 17,690,348 | − | 415 | 45,443 | 7 | 41 | 114 | 0 | nuclear |
| PagPUB93 | Pop_G02G026854.T1 | chrG02 | 20,130,677 | 20,142,648 | + | 1197 | 131,795 | 7 | 48 | 83 | −0.458 | chloroplast |
| PagPUB94 | Pop_G02G065058.T1 | chrG02 | 21,231,438 | 21,235,743 | − | 946 | 105,425 | 8 | 47 | 85 | −0.319 | nuclear |
| PagPUB95 | Pop_G02G065388.T1 | chrG02 | 22,015,950 | 22,018,762 | + | 658 | 73,329 | 6 | 40 | 103 | −0.229 | nuclear |
| PagPUB96 | Pop_G02G065360.T1 | chrG02 | 22,255,103 | 22,259,863 | + | 813 | 89,295 | 5 | 42 | 109 | −0.009 | cytoplasmic |
| PagPUB97 | Pop_G03G076286.T1 | chrG03 | 692,443 | 697,128 | + | 1110 | 124,407 | 6 | 53 | 83 | −0.515 | nuclear |
| PagPUB98 | Pop_G04G059292.T1 | chrG04 | 2,808,865 | 2,813,024 | − | 644 | 71,765 | 7 | 39 | 109 | −0.173 | cytoplasmic |
| PagPUB99 | Pop_G04G028210.T1 | chrG04 | 7,399,400 | 7,400,641 | + | 413 | 45,267 | 7 | 52 | 111 | 0 | endoplasmic reticulum |
| PagPUB100 | Pop_G04G066587.T1 | chrG04 | 13,701,187 | 13,702,736 | + | 448 | 50,317 | 8 | 37 | 103 | −0.078 | endoplasmic reticulum |
| PagPUB101 | Pop_G04G066506.T1 | chrG04 | 15,411,436 | 15,414,188 | − | 717 | 78,215 | 8 | 45 | 102 | 0 | chloroplast |
| PagPUB102 | Pop_G04G023347.T1 | chrG04 | 18,596,595 | 18,597,713 | − | 372 | 40,846 | 8 | 43 | 103 | −0.042 | cytoplasmic |
| PagPUB103 | Pop_G05G008783.T1 | chrG05 | 333,289 | 342,205 | − | 1216 | 134,888 | 6 | 41 | 104 | −0.091 | nuclear |
| PagPUB104 | Pop_G05G057175.T1 | chrG05 | 1,236,419 | 1,242,211 | + | 1414 | 158,338 | 6 | 44 | 90 | −0.226 | chloroplast |
| PagPUB105 | Pop_G05G057312.T1 | chrG05 | 2,019,038 | 2,023,275 | − | 737 | 81,675 | 6 | 47 | 100 | −0.093 | nuclear |
| PagPUB106 | Pop_G05G057347.T1 | chrG05 | 2,218,107 | 2,223,363 | − | 778 | 85,635 | 7 | 45 | 93 | −0.235 | mitochondrial |
| PagPUB107 | Pop_G05G008350.T1 | chrG05 | 5,000,116 | 5,001,814 | + | 450 | 49,455 | 8 | 48 | 105 | 0 | chloroplast |
| PagPUB108 | Pop_G05G085107.T1 | chrG05 | 10,253,671 | 10,263,649 | − | 840 | 91,094 | 6 | 44 | 98 | −0.22 | peroxisome |
| PagPUB109 | Pop_G06G035460.T1 | chrG06 | 210,349 | 211,902 | + | 448 | 48,330 | 9 | 46 | 117 | 0 | nuclear |
| PagPUB110 | Pop_G06G003288.T1 | chrG06 | 5,823,057 | 5,824,286 | + | 409 | 45,852 | 8 | 49 | 103 | 0 | cytoplasmic |
| PagPUB111 | Pop_G06G003287.T1 | chrG06 | 5,842,794 | 5,844,482 | + | 413 | 45,588 | 8 | 44 | 110 | 0 | cytoplasmic |
| PagPUB112 | Pop_G06G051597.T1 | chrG06 | 15,896,001 | 15,898,264 | − | 679 | 74,861 | 9 | 43 | 106 | 0 | chloroplast |
| PagPUB113 | Pop_G07G061245.T1 | chrG07 | 2,390,486 | 2,391,781 | + | 414 | 46,455 | 9 | 41 | 107 | 0 | cytoplasmic |
| PagPUB114 | Pop_G07G061189.T1 | chrG07 | 2,790,289 | 2,792,326 | − | 397 | 43,978 | 6 | 45 | 104 | −0.038 | nuclear |
| PagPUB115 | Pop_G07G052040.T1 | chrG07 | 9,396,015 | 9,399,928 | − | 840 | 91,073 | 6 | 48 | 96 | −0.23 | peroxisome |
| PagPUB116 | Pop_G07G062480.T1 | chrG07 | 12,295,996 | 12,300,385 | + | 787 | 87,606 | 6 | 47 | 94 | −0.213 | nuclear |
| PagPUB117 | Pop_G07G062608.T1 | chrG07 | 13,162,944 | 13,165,676 | + | 617 | 70,151 | 7 | 50 | 84 | −0.483 | cytoplasmic |
| PagPUB118 | Pop_G08G082443.T1 | chrG08 | 8,100,134 | 8,104,736 | − | 775 | 85,400 | 6 | 53 | 91 | −0.257 | cytoplasmic |
| PagPUB119 | Pop_G08G046414.T1 | chrG08 | 11,101,251 | 11,102,877 | + | 443 | 49,334 | 8 | 39 | 104 | −0.02 | mitochondrial |
| PagPUB120 | Pop_G08G058547.T1 | chrG08 | 11,942,784 | 11,945,493 | − | 640 | 69,898 | 9 | 45 | 99 | −0.201 | endoplasmic reticulum |
| PagPUB121 | Pop_G08G058369.T1 | chrG08 | 13,137,618 | 13,141,997 | − | 1056 | 117,980 | 6 | 52 | 99 | −0.183 | chloroplast |
| PagPUB122 | Pop_G08G063491.T1 | chrG08 | 15,864,328 | 15,866,394 | − | 688 | 75,670 | 8 | 47 | 107 | −0.001 | golgi apparatus |
| PagPUB123 | Pop_G08G058063.T1 | chrG08 | 18,316,014 | 18,318,995 | + | 656 | 71,874 | 6 | 39 | 102 | −0.196 | plasma membrane |
| PagPUB124 | Pop_G09G011673.T1 | chrG09 | 282,639 | 285,957 | − | 644 | 71,655 | 8 | 32 | 108 | −0.186 | cytoplasmic |
| PagPUB125 | Pop_G09G077318.T1 | chrG09 | 4,280,957 | 4,282,198 | + | 413 | 45,154 | 6 | 48 | 109 | 0 | plasma membrane |
| PagPUB126 | Pop_G09G014114.T1 | chrG09 | 8,901,734 | 8,907,147 | − | 837 | 94,111 | 7 | 50 | 90 | −0.375 | nuclear |
| PagPUB127 | Pop_G09G014123.T1 | chrG09 | 8,954,631 | 8,959,350 | + | 663 | 71,909 | 6 | 39 | 98 | −0.232 | endoplasmic reticulum |
| PagPUB128 | Pop_G09G027806.T1 | chrG09 | 10,117,973 | 10,119,220 | + | 415 | 46,785 | 9 | 51 | 108 | −0.062 | cytoplasmic |
| PagPUB129 | Pop_G09G027805.T1 | chrG09 | 10,136,565 | 10,137,797 | − | 410 | 46,721 | 9 | 47 | 106 | −0.121 | chloroplast |
| PagPUB130 | Pop_G10G007781.T1 | chrG10 | 54,394 | 57,627 | − | 628 | 68,785 | 6 | 42 | 102 | −0.201 | plasma membrane |
| PagPUB131 | Pop_G10G047951.T1 | chrG10 | 4,501,468 | 4,503,534 | + | 688 | 75,089 | 8 | 42 | 107 | −0.015 | golgi apparatus |
| PagPUB132 | Pop_G10G048131.T1 | chrG10 | 7,567,131 | 7,571,788 | + | 1034 | 116,154 | 6 | 51 | 100 | −0.148 | chloroplast |
| PagPUB133 | Pop_G10G048375.T1 | chrG10 | 9,217,913 | 9,220,579 | + | 640 | 69,239 | 9 | 42 | 101 | −0.152 | plasma membrane |
| PagPUB134 | Pop_G10G000218.T1 | chrG10 | 9,983,796 | 9,985,124 | − | 442 | 49,164 | 8 | 41 | 104 | −0.011 | mitochondrial |
| PagPUB135 | Pop_G10G060717.T1 | chrG10 | 11,889,860 | 11,895,757 | − | 1016 | 112,857 | 6 | 40 | 109 | −0.057 | chloroplast |
| PagPUB136 | Pop_G10G020266.T1 | chrG10 | 13,673,677 | 13,678,664 | + | 786 | 86,555 | 5 | 50 | 92 | −0.225 | nuclear |
| PagPUB137 | Pop_G12G084750.T1 | chrG12 | 125,343 | 130,854 | + | 812 | 92,332 | 6 | 45 | 96 | −0.248 | nuclear |
| PagPUB138 | Pop_G12G068392.T1 | chrG12 | 4,872,982 | 4,875,690 | − | 720 | 79,050 | 9 | 42 | 103 | 0 | nuclear |
| PagPUB139 | Pop_G12G050752.T1 | chrG12 | 9,589,324 | 9,590,643 | − | 439 | 49,362 | 7 | 47 | 97 | −0.126 | chloroplast |
| PagPUB140 | Pop_G12G011606.T1 | chrG12 | 11,895,133 | 11,898,971 | − | 1004 | 111,624 | 6 | 44 | 109 | 0 | nuclear |
| PagPUB141 | Pop_G13G071258.T1 | chrG13 | 10,457,642 | 10,459,957 | − | 445 | 49,315 | 6 | 38 | 107 | −0.141 | cytoplasmic |
| PagPUB142 | Pop_G14G077975.T1 | chrG14 | 10,667,099 | 10,672,502 | + | 1006 | 111,101 | 6 | 49 | 106 | −0.081 | nuclear |
| PagPUB143 | Pop_G14G077964.T1 | chrG14 | 10,718,090 | 10,719,325 | − | 411 | 45,623 | 8 | 46 | 103 | 0 | cytoplasmic |
| PagPUB144 | Pop_G14G030545.T1 | chrG14 | 17,003,553 | 17,006,588 | + | 450 | 48,956 | 6 | 41 | 115 | 0 | endoplasmic reticulum |
| PagPUB145 | Pop_G15G048830.T1 | chrG15 | 5,528,163 | 5,530,599 | − | 684 | 75,479 | 8 | 47 | 103 | 0 | chloroplast |
| PagPUB146 | Pop_G15G074299.T1 | chrG15 | 11,090,167 | 11,091,489 | + | 440 | 49,338 | 6 | 44 | 99 | −0.085 | chloroplast |
| PagPUB147 | Pop_G16G068843.T1 | chrG16 | 757,514 | 760,365 | − | 762 | 83,266 | 9 | 43 | 107 | 0 | chloroplast |
| PagPUB148 | Pop_G16G057564.T1 | chrG16 | 8,502,944 | 8,504,170 | + | 408 | 46,119 | 8 | 47 | 104 | 0 | cytoplasmic |
| PagPUB149 | Pop_G16G057553.T1 | chrG16 | 8,508,310 | 8,509,821 | + | 412 | 45,818 | 8 | 42 | 107 | 0 | chloroplast |
| PagPUB150 | Pop_G17G033358.T1 | chrG17 | 2,438,610 | 2,440,217 | − | 446 | 49,883 | 8 | 44 | 100 | −0.094 | plasma membrane |
| PagPUB151 | Pop_G18G080275.T1 | chrG18 | 5,834,978 | 5,842,201 | + | 1340 | 148,348 | 6 | 48 | 100 | −0.094 | endoplasmic reticulum |
| PagPUB152 | Pop_G19G009310.T1 | chrG19 | 11,390,424 | 11,393,402 | − | 450 | 50,018 | 6 | 38 | 103 | −0.232 | chloroplast |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, B.; Wei, Q.; Liu, X.; Sun, K.; Liao, L.; Zu, A.; Wei, Y.; Liu, Q.; Fu, F.; Ming, M. Genome-Wide Identification, Evolution and Expression Analysis of the U-Box E3 Ubiquitin Ligases Gene Family in Poplar (Populus alba × P. tremula var. glandulosa). Forests 2025, 16, 749. https://doi.org/10.3390/f16050749
Song B, Wei Q, Liu X, Sun K, Liao L, Zu A, Wei Y, Liu Q, Fu F, Ming M. Genome-Wide Identification, Evolution and Expression Analysis of the U-Box E3 Ubiquitin Ligases Gene Family in Poplar (Populus alba × P. tremula var. glandulosa). Forests. 2025; 16(5):749. https://doi.org/10.3390/f16050749
Chicago/Turabian StyleSong, Bobo, Qixuan Wei, Xudong Liu, Kexin Sun, Lingdou Liao, Anning Zu, Yifan Wei, Qian Liu, Fangfang Fu, and Meiling Ming. 2025. "Genome-Wide Identification, Evolution and Expression Analysis of the U-Box E3 Ubiquitin Ligases Gene Family in Poplar (Populus alba × P. tremula var. glandulosa)" Forests 16, no. 5: 749. https://doi.org/10.3390/f16050749
APA StyleSong, B., Wei, Q., Liu, X., Sun, K., Liao, L., Zu, A., Wei, Y., Liu, Q., Fu, F., & Ming, M. (2025). Genome-Wide Identification, Evolution and Expression Analysis of the U-Box E3 Ubiquitin Ligases Gene Family in Poplar (Populus alba × P. tremula var. glandulosa). Forests, 16(5), 749. https://doi.org/10.3390/f16050749







