Using Environmental DNA as a Plant Health Surveillance Tool in Forests
Abstract
1. Introduction
2. Materials and Methods
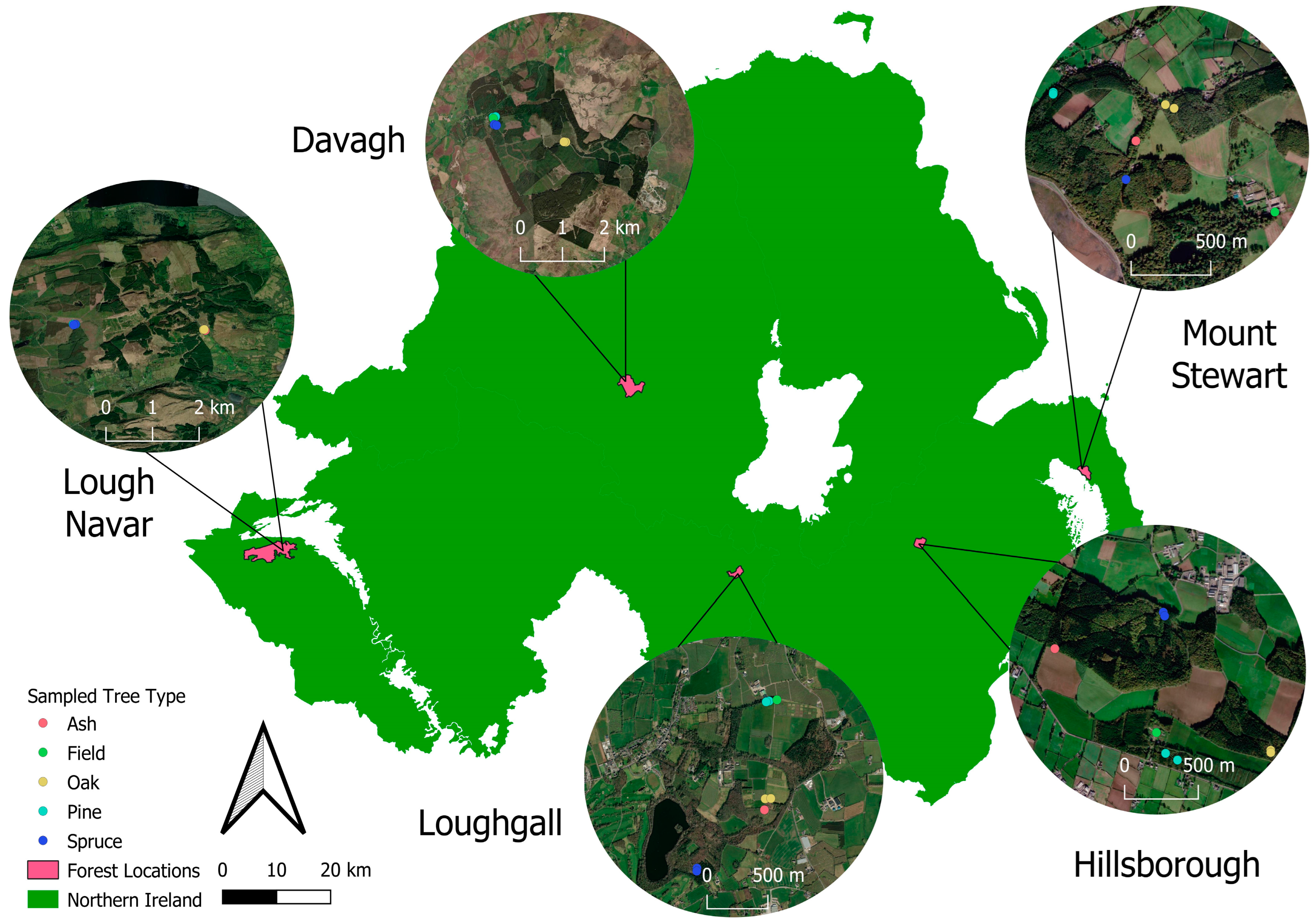
2.1. Information on Sites
2.2. Rainwater Trap Placement
2.3. Rainwater Collection and Filtration
2.4. DNA Extraction and Next-Generation Sequencing
2.5. Bioinformatics
2.6. Detection and Diversity Metrics and Visualisation
2.7. Generalised Linear Mixed Model (GLMM)
2.8. Analysis of Compositon of Microbiomes with Bias Correction (ANCOM-BC)
2.9. Data Submission and Availability
3. Results
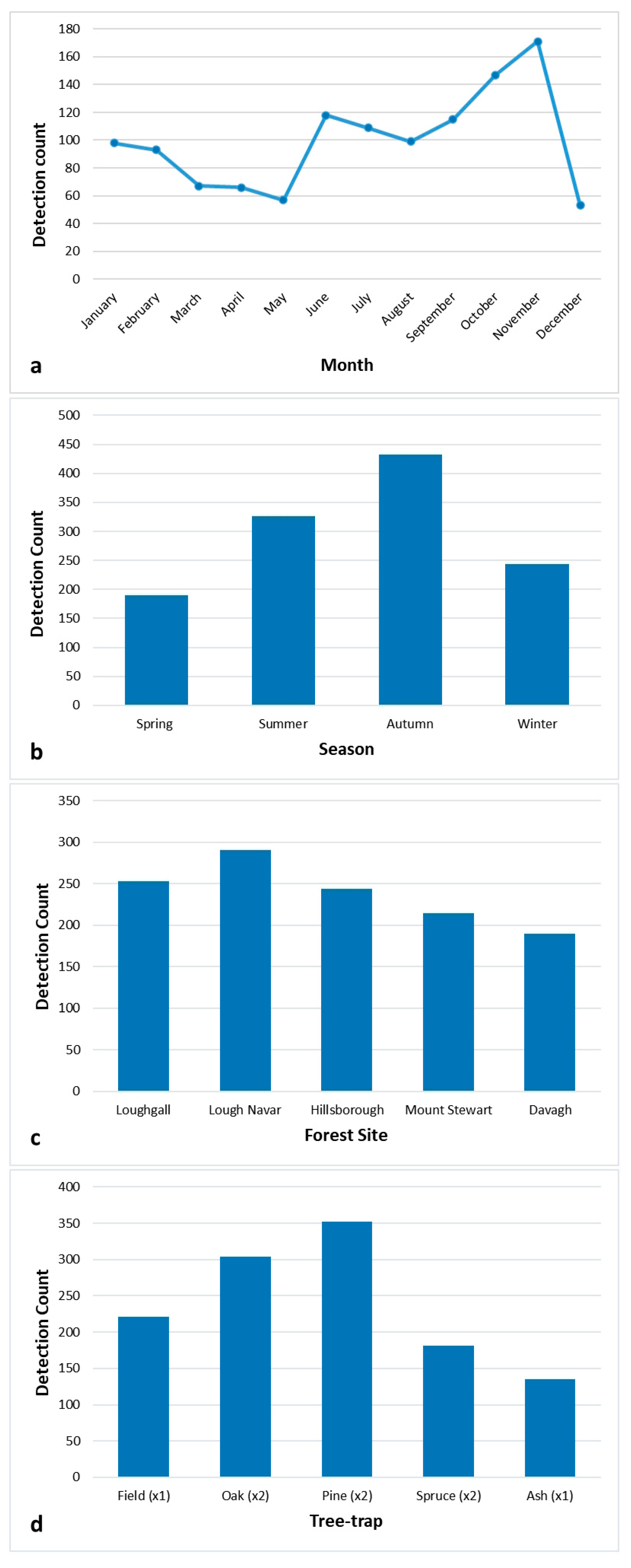
3.1. Trends in the Detection of Fungal and Oomycete Pests
3.2. Statisitcal Analysis of Fungal and Oomycete Pest Detection
3.3. Trends in the Diversity of Fungal and Oomycetes Pests
3.4. Statistical Analysis Diversity of Fungal and Oomycete Pest Diversity
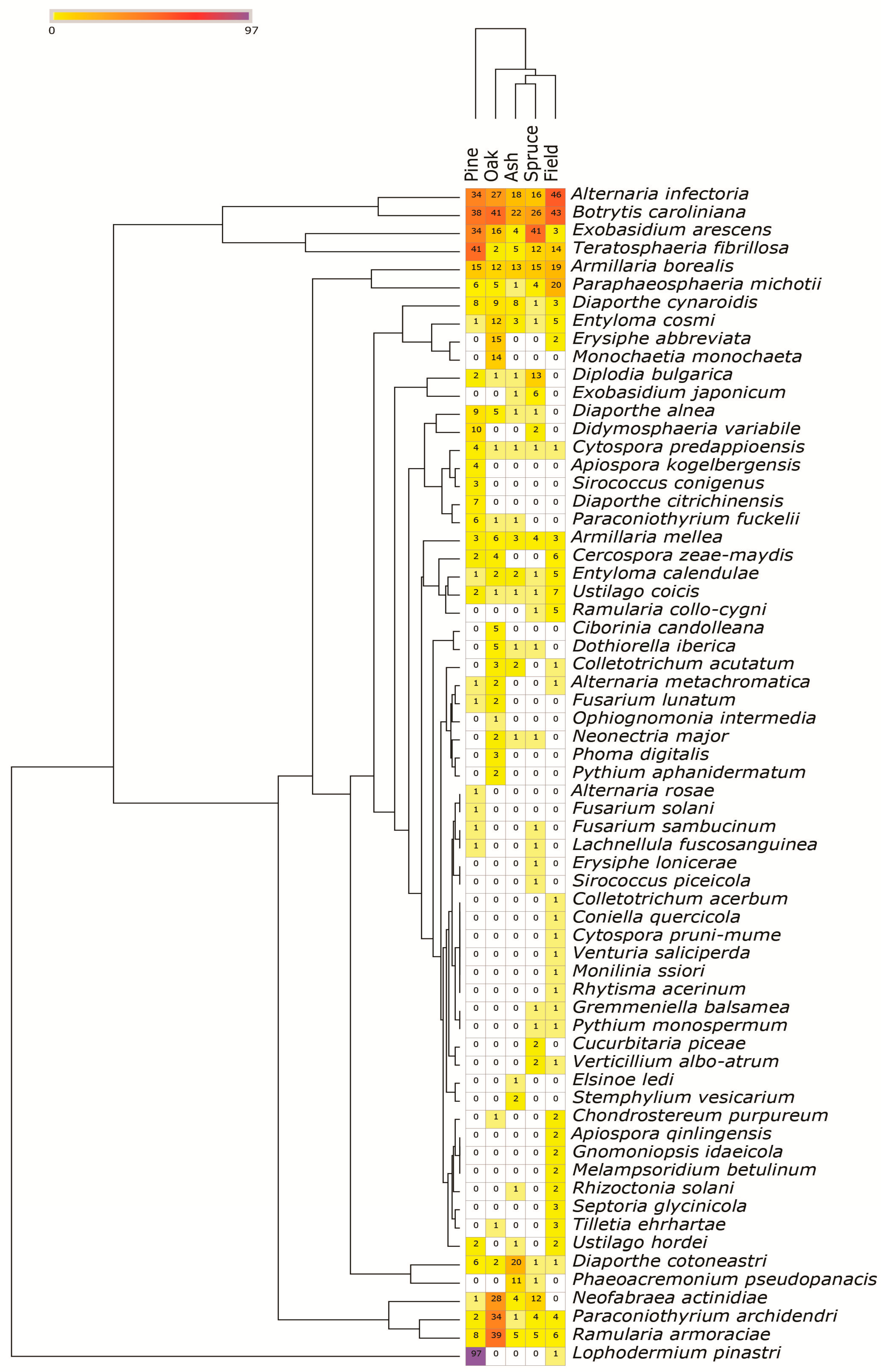
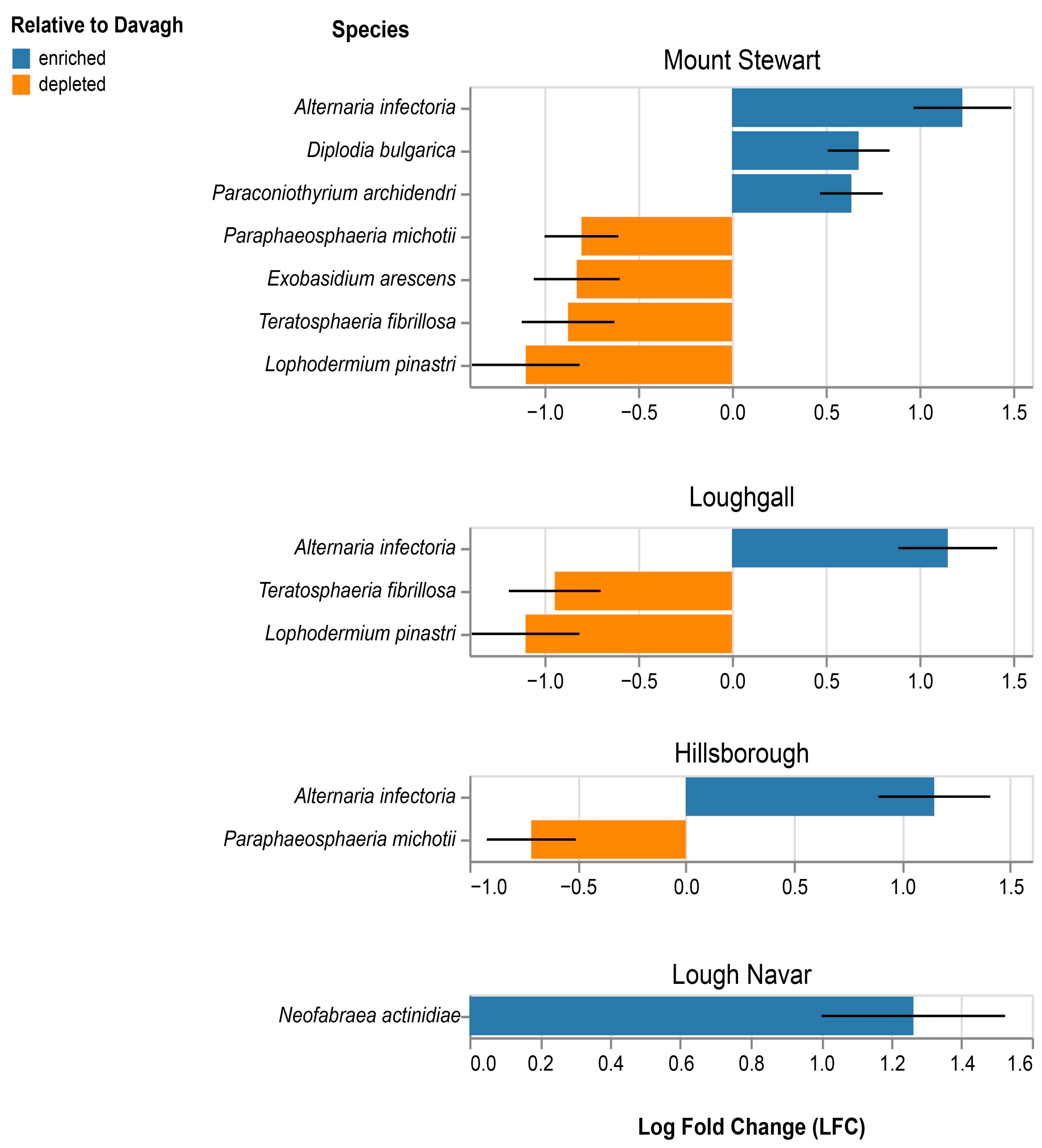
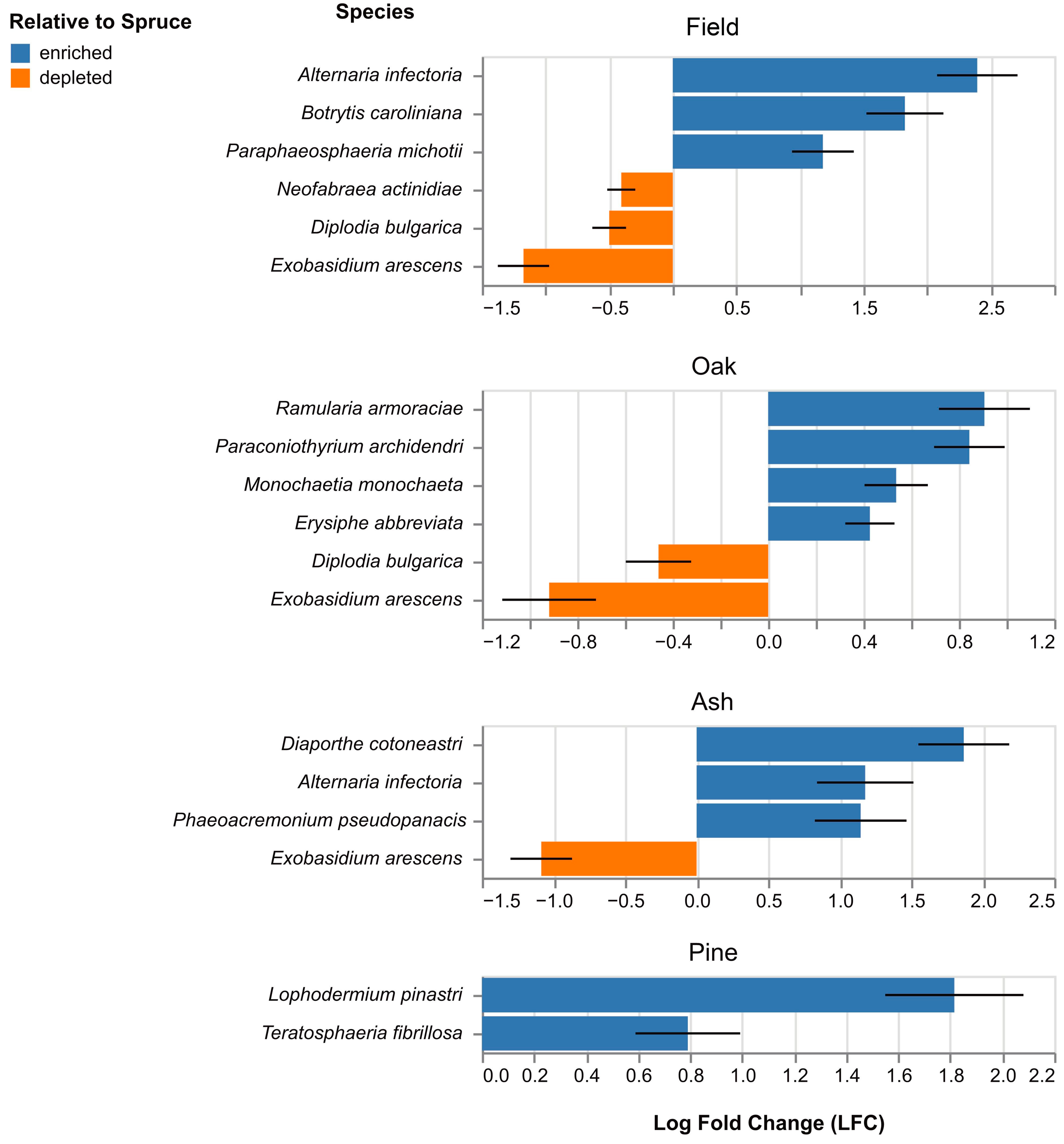
3.5. Composition of Fungal and Oomycete Pests
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- IPPC Secretariat. Scientific Review of the Impact of Climate Change on Plant Pests—A Global Challenge to Prevent and Mitigate Plant Pest Risks in Agriculture, Forestry and Ecosystems, 1st ed.; Food and Agricultural Organization of the United Nation: (FAO): Rome, Itlay, 2021; p. 88. [Google Scholar]
- IPBES. Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services. Summary for Policy Makers of the Global Assessment Report on Biodiversity and Ecosystem Services of the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services; IPBES: Bonn, Germany, 2019. [Google Scholar]
- Bellard, C.; Cassey, P.; Blackburn, T.M. Alien species as a driver of recent extinctions. Biol. Lett. 2016, 12, 20150623. [Google Scholar] [CrossRef] [PubMed]
- Quirion, B.R.; Domke, G.M.; Walters, B.F.; Lovett, G.M.; Fargione, J.E.; Greenwood, L.; Serbesoff-King, K.; Randall, J.M.; Fei, S. Insect and disease disturbances correlate with reduced carbon sequestration in forests of the contiguous United States. Front. For. Glob. Change 2021, 4, 716582. [Google Scholar] [CrossRef]
- Keller, R.P.; Perrings, C. International Policy Options for Reducing the Environmental Impacts of Invasive Species. BioScience 2011, 61, 1005–1012. [Google Scholar]
- IPPC. Approved Specifications for Adopted ISPMs. 2024. Available online: https://www.ippc.int/en/core-activities/standards-setting/approved-specifications/approved-specifications-for-adopted-ispms/ (accessed on 4 December 2024).
- APHIS. Animal and Plant Health Inspection Service, U.S. Department of Agriculture, Laws and Regulations. 2025. Available online: https://www.aphis.usda.gov/laws-regs (accessed on 4 December 2024).
- ANON. EU Plant Health Regulation 2016/2031. 2016: EUR-Lex. Available online: https://eur-lex.europa.eu/eli/reg/2016/2031/oj/eng (accessed on 19 February 2025).
- Sosnowski, M.R.; Fletcher, J.D.; Daly, A.M.; Rodoni, B.C.; Viljanen-Rollinson, S.L.H. Techniques for the treatment, removal and disposal of host material during programmes for plant pathogen eradication. Plant Pathol. 2009, 58, 621–635. [Google Scholar] [CrossRef]
- Straw, N.A.; Fielding, N.J.; Tilbury, C.; Williams, D.T.; Cull, T. History and development of an isolated outbreak of Asian longhorn beetle Anoplophora glabripennis (Coleoptera: Cerambycidae) in southern England. Agric. For. Entomol. 2016, 18, 280–293. [Google Scholar] [CrossRef]
- Forest and Wood Products of Australia, Exotic Pest Risk Real and Rising in Australia. 2017. Available online: https://fwpa.com.au/news/exotic-pest-risk-real-and-rising-in-australia-2/ (accessed on 4 December 2024).
- Schneider, K.; van der Werf, W.; Cendoya, M.; Mourits, M.; Navas-Cortés, J.A.; Vicent, A.; Oude Lansink, A. Impact of Xylella fastidiosa subspecies pauca in European olives. Proc. Natl. Acad. Sci. USA 2020, 117, 9250–9259. [Google Scholar] [CrossRef] [PubMed]
- Hill, L.; Hemery, G.; Hector, A.; Brown, N.; Fajardo, A. Maintaining ecosystem properties after loss of ash in Great Britain. J. Appl. Ecol. 2018, 56, 282–293. [Google Scholar] [CrossRef]
- Pluess, T.; Jarosik, V.; Pysek, P.; Cannon, R.; Pergl, J.; Breukers, A.; Bacher, S. Which factors affect the success or failure of eradication campaigns against alien species? PLoS ONE 2012, 7, e48157. [Google Scholar] [CrossRef]
- Venbrux, M.; Crauwels, S.; Rediers, H. Current and emerging trends in techniques for plant pathogen detection. Front. Plant Sci. 2023, 14, 1120968. [Google Scholar] [CrossRef]
- Green, S.; Dehnen-Schmutz, K.; Drakulic, J.; Eschen, R.; Orazio, C.; Douma, J.C.; Lundén, K.; Colombari, F.; Jactel, H. Awareness, detection and management of new and emerging tree pests and pathogens in Europe: Stakeholders’ perspectives. NeoBiota 2023, 8, 9–40. [Google Scholar] [CrossRef]
- Mizumoto, H.; Kishida, O.; Takai, K.; Matsuura, N.; Araki, H. Utilizing environmental DNA for wide-range distributions of reproductive area of an invasive terrestrial toad in Ishikari river basin in Japan. Biol. Invasions 2021, 24, 1199–1211. [Google Scholar] [CrossRef]
- Ruiz-Ramos, D.V.; Meyer, R.S.; Toews, D.; Stephens, M.; Kolster, M.K.; Sexton, J.P. Environmental DNA (eDNA) detects temporal and habitat effects on community composition and endangered species in ephemeral ecosystems: A case study in vernal pools. Environ. DNA 2022, 5, 85–101. [Google Scholar] [CrossRef]
- Català, S.; Pérez-Sierra, A.; Abad-Campos, P. The Use of Genus-Specific Amplicon Pyrosequencing to Assess Phytophthora Species Diversity Using eDNA from Soil and Water in Northern Spain. PLoS ONE 2015, 10, e0119311. [Google Scholar]
- Prigigallo, M.I.; Abdelfattah, A.; Cacciola, S.O.; Faedda, R.; Sanzani, S.M.; Cooke, D.E.L.; Schena, L. Metabarcoding Analysis of Phytophthora Diversity Using Genus-Specific Primers and 454 Pyrosequencing. Phytopathology 2016, 106, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Catalá, S.; Berbegal, M.; Pérez-Sierra, A.; Abad-Campos, P. Metabarcoding and development of new real-time specific assays reveal Phytophthora species diversity in holm oak forests in eastern Spain. Plant Pathol. 2017, 66, 115–123. [Google Scholar]
- Valentin, R.E.; Fonseca, D.M.; Gable, S.; Kyle, K.E.; Hamilton, G.C.; Nielsen, A.L.; Lockwood, J.L. Moving eDNA surveys onto land: Strategies for active eDNA aggregation to detect invasive forest insects. Mol. Ecol. Resour. 2020, 20, 746–755. [Google Scholar]
- Milian-Garcia, Y.; Young, R.; Madden, M.; Bullas-Appleton, E.; Hanner, R.H. Optimization and validation of a cost-effective protocol for biosurveillance of invasive alien species. Ecol. Evol. 2021, 11, 1999–2014. [Google Scholar] [CrossRef]
- Apangu, G.P.; Frisk, C.A.; Petch, G.M.; Muggia, L.; Pallavicini, A.; Hanson, M.; Skjøth, C.A. Environmental DNA reveals diversity and abundance of Alternaria species in neighbouring heterogeneous landscapes in Worcester, UK. Aerobiologia 2022, 38, 457–481. [Google Scholar] [CrossRef]
- Green, S.; Cooke, D.E.L.; Dunn, M.; Barwell, L.; Purse, B.; Chapman, D.S.; Valatin, G.; Schlenzig, A.; Barbrook, J.; Pettitt, T.; et al. PHYTO-THREATS: Addressing Threats to UK Forests and Woodlands from Phytophthora; Identifying Risks of Spread in Trade and Methods for Mitigation. Forests 2021, 12, 1617. [Google Scholar] [CrossRef]
- Green, S.; Cooke, D.E.L.; Barwell, L.; Purse, B.V.; Cock, P.; Frederickson-Matika, D.; Randall, E.; Keillor, B.; Pritchard, L.; Thorpe, P.; et al. The prevalence of Phytophthora in British plant nurseries; high-risk hosts and substrates and opportunities to implement best practice. Plant Pathol. 2025, 74, 696–717. [Google Scholar] [CrossRef]
- Ladin, Z.S.; Ferrell, B.; Dums, J.T.; Moore, R.M.; Levia, D.F.; Shriver, W.G.; D'Amico, V.; Trammell, T.L.E.; Setubal, J.C.; Wommack, K.E. Assessing the efficacy of eDNA metabarcoding for measuring microbial biodiversity within forest ecosystems. Sci. Rep. 2021, 11, 1629. [Google Scholar] [CrossRef]
- Macher, T.-H.; Schütz, R.; Hörren, T.; Beermann, A.J.; Leese, F. It’s raining species: Rainwash eDNA metabarcoding as a minimally invasive method to assess tree canopy invertebrate diversity. bioRxiv 2022. [Google Scholar] [CrossRef]
- Met Office, HadUK-Grid. 2025. Available online: https://www.metoffice.gov.uk/research/climate/maps-and-data/data/haduk-grid/haduk-grid (accessed on 21 January 2025).
- Boardman, L.; Marcelino, J.A.P.; Valentin, R.E.; Boncristiani, H.; Standley, J.M.; Ellis, J.D. Novel eDNA approaches to monitor Western honey bee (Apis mellifera L.) microbial and arthropod communities. Environ. DNA 2023, 6, e419. [Google Scholar] [CrossRef]
- Usyk, M.; Zolnik, C.P.; Patel, H.; Levi, M.H.; Burk, R.D. Novel ITS1 Fungal Primers for Characterization of the Mycobiome. Clin. Sci. Epidemiol. 2017, 2, e00488-17. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [PubMed]
- Li, P.-E.; Lo, C.-C.; Anderson, J.J.; Davenport, K.W.; Bishop-Lilly, K.A.; Xu, Y.; Ahmed, S.; Feng, S.; Mokashi, V.P.; Chain, P.S.G. Enabling the democratization of the genomics revolution with a fully integrated web-based bioinformatics platform. Nucleic Acids Res. 2016, 45, 67–80. [Google Scholar] [CrossRef]
- Lin, H.; Peddada, S.D. Analysis of compositions of microbiomes with bias correction. Nat. Commun. 2020, 11, 3514. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Hong, J.; Karaoz, U.; de Valpine, P.; Fithian, W. To rarefy or not to rarefy: Robustness and efficiency trade-offs of rarefying microbiome data. Bioinformatics 2022, 38, 2389–2396. [Google Scholar] [CrossRef]
- Abarenkov, K.; Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; May, T.W.; Frøslev, T.G.; Pawlowska, J.; Lindahl, B.; Põldmaa, K.; Truong, C.; et al. The UNITE database for molecular identification and taxonomic communication of fungi and other eukaryotes: Sequences, taxa and classifications reconsidered. Nucleic Acids Res. 2024, 52, D791–D797. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Broad Institute, Morpheus. 2025. Available online: https://software.broadinstitute.org/morpheus/ (accessed on 19 March 2025).
- Devkota, P.; Hammerschmidt, R. The infection process of Armillaria mellea and Armillaria solidipes. Physiol. Mol. Plant Pathol. 2020, 112, 101543. [Google Scholar] [CrossRef]
- DEFRA. UK Plant Health Risk Register. 2024. Available online: https://planthealthportal.defra.gov.uk/pests-and-diseases/uk-plant-health-risk-register/ (accessed on 16 October 2024).
- Hamberg, L.; Saksa, T.; Hantula, J. Role and function of Chondrostereum purpureum in biocontrol of trees. Appl. Microbiol. Biotechnol. 2020, 105, 431–440. [Google Scholar] [CrossRef] [PubMed]
- EPPO. EPPO Global Database. Available online: https://gd.eppo.int/ (accessed on 16 October 2024).
- Cline, W.O.; Brewer, M.T.; Brannen, P.M.; Scherm, H. Exobasidium leaf and fruit spot of blueberry in the southeastern United States. In Proceedings of the NABREW Conference, Atlantic City, NJ, USA, 25 June 2014. [Google Scholar]
- Stevanovic, M.; Ristic, D.; Zivkovic, S.; Aleksic, G.; Stankovic, I.; Krstic, B.; Bulajic, A. Characterization of Gnomoniopsis idaeicola, the Causal Agent of Canker and Wilting of Blackberry in Serbia. Plant Dis. 2019, 103, 249–258. [Google Scholar] [CrossRef]
- Yang, G. General Description of Rhizoctonia Species Complex; IntechOpen: London, UK, 2012. [Google Scholar]
- Molla, K.A.; Karmakar, S.; Molla, J.; Bajaj, P.; Varshney, R.K.; Datta, S.K.; Datta, K. Understanding sheath blight resistance in rice: The road behind and the road ahead. Plant Biotechnol. J. 2019, 18, 895–915. [Google Scholar] [CrossRef]
- Francis, A.; Ghosh, S.; Tyagi, K.; Prakasam, V.; Rani, M.; Singh, N.P.; Pradhan, A.; Sundaram, R.M.; Priyanka, C.; Laha, G.S.; et al. Evolution of pathogenicity-associated genes in Rhizoctonia solani AG1-IA by genome duplication and transposon-mediated gene function alterations. BMC Biol. 2023, 21, 15. [Google Scholar] [CrossRef]
- Pérez-Sierra, A.; Hardy, C.; Chitty, R.; Lewis, A.; McCartan, S. First report of Sirococcus piceicola associated with Sitka spruce seed in Britain. New Dis. Rep. 2021, 44, e12048. [Google Scholar] [CrossRef]
- White, R.; Marzano, M.; Fesenko, E.; Inman, A.; Jones, G.; Agstner, B.; Mumford, R. Technology development for the early detection of plant pests: A framework for assessing Technology Readiness Levels (TRLs) in environmental science. J. Plant Dis. Prot. 2022, 129, 1249–1261. [Google Scholar] [CrossRef]
- Reznik, Y.V.; Yermishev, O.V.; Palamarchuk, O.V.; Bobrovska, O.A.; Rodinkova, V.V. Features of the seasonal dynamics of airborne fungal spore concentrations in Ukraine. Biosyst. Divers. 2023, 31, 71–83. [Google Scholar] [CrossRef]
- Inderbitzin, P.; Bostock, R.M.; Davis, R.M.; Usami, T.; Platt, H.W.; Subbarao, K.V. Phylogenetics and taxonomy of the fungal vascular wilt pathogen Verticillium, with the descriptions of five new species. PLoS ONE 2011, 6, e28341. [Google Scholar]
- Li, X.; Fernández-Ortuño, D.; Chai, W.; Wang, F.; Schnabel, G. Identification and Prevalence of Botrytis spp. from Blackberry and Strawberry Fields of the Carolinas. Plant Dis. 2012, 96, 1634–1637. [Google Scholar] [CrossRef]
- Nannfeldt, J.A. Exobasidium, a Taxonomic Reassessment Applied to the European Species; Almqvist & Wiksell International: Stockholm, Sweden, 1981. [Google Scholar]
- Akulova, V.S.; Sharov, V.V.; Aksyonova, A.I.; Putintseva, Y.A.; Oreshkova, N.V.; Feranchuk, S.I.; Kuzmin, D.A.; Pavlov, I.N.; Litovka, Y.A.; Krutovsky, K.V. De novo sequencing, assembly and functional annotation of Armillaria borealis genome. BMC Genom. 2020, 21, 534. [Google Scholar] [CrossRef]
- Valentine, D.C.; Shaffer, B.T.; McGhee, G.C.; Bouska, C.; Stockwell, V.O. First Report of Neofabraea actinidiae Causing a Cranberry Fruit Rot in Oregon. Plant Dis. 2024, 108, 1405. [Google Scholar]
- Hudler, G.W. Unusual endemic of tar spot on Norway maple in Upstate New York. Plant Dis. 1987, 71, 65–68. [Google Scholar]
- Guo, Y.S.; Crous, P.W.; Bai, Q.; Fu, M.; Yang, M.M.; Wang, X.H.; Du, Y.M.; Hong, N.; Xu, W.X.; Wang, G.P. High diversity of Diaporthe species associated with pear shoot canker in China. Persoonia Mol. Phylogeny Evol. Fungi 2020, 45, 132–162. [Google Scholar]
- Poursafar, A.; Ghosta, Y.; Orina, A.S.; Gannibal, P.B.; Javan-Nikkhah, M.; Lawrence, D.P. Taxonomic study on Alternaria sections Infectoriae and Pseudoalternaria associated with black (sooty) head mold of wheat and barley in Iran. Mycol. Prog. 2017, 17, 343–356. [Google Scholar]
- McMullan, M.; Rafiqi, M.; Kaithakottil, G.; Clavijo, B.J.; Bilham, L.; Orton, E.; Percival-Alwyn, L.; Ward, B.J.; Edwards, A.; Saunders, D.G.O.; et al. The ash dieback invasion of Europe was founded by two genetically divergent individuals. Nat. Ecol. Evol. 2018, 2, 1000–1008. [Google Scholar]
- DAERA. Phytophthora Ramorum. 2025. Available online: https://www.daera-ni.gov.uk/articles/phytophthora-ramorum (accessed on 21 January 2025).
- Kroon, L.P.N.M.; Brouwer, H.; de Cock, A.W.A.M.; Govers, F. The Genus Phytophthora Anno 2012. Phytopathology 2012, 102, 348–364. [Google Scholar]
- Martin, K.J.; Rygiewicz, P.T. Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiol. 2005, 5, 28. [Google Scholar]
- Johnson, J.S.; Spakowicz, D.J.; Hong, B.Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M.; et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019, 10, 5029. [Google Scholar] [CrossRef] [PubMed]
- Olmos, A.; Boonham, N.; Candresse, T.; Gentit, P.; Giovani, B.; Kutnjak, D.; Liefting, L.; Maree, H.J.; Minafra, A.; Moreira, A.; et al. High-throughput sequencing technologies for plant pest diagnosis: Challenges and opportunities. EPPO Bull. 2018, 48, 219–224. [Google Scholar] [CrossRef]
- Massart, S.; Candresse, T.; Gil, J.; Lacomme, C.; Predajna, L.; Ravnikar, M.; Reynard, J.-S.; Rumbou, A.; Saldarelli, P.; Škorić, D.; et al. A Framework for the Evaluation of Biosecurity, Commercial, Regulatory, and Scientific Impacts of Plant Viruses and Viroids Identified by NGS Technologies. Front. Microbiol. 2017, 8, 45. [Google Scholar] [CrossRef] [PubMed]



| Species | Risk Register Status | Month | Site | Detections over the Year | Confidence Limit (CL) | Details |
|---|---|---|---|---|---|---|
| Armillaria mellea | Regulated non-quarantine pest | October and November | Davagh, Loughgall and Lough Navar | 16 | 1 | Highly virulent and is responsible for causing immense damage to fruit and nut trees, as well coniferous and broad leaf trees worldwide [41]. Some of the main hosts of A. mellea include birch, apple, pine, blackcurrant and ash [42] |
| Chondrostereum purpureum | Regulated non-quarantine pest | January | Davagh | 3 | 1 | Causes silver leaf disease [42,43]. C. purpureum has an extensive list of hosts including ash, maple, birch, pine, stone and pome fruit trees |
| Colletotrichum acutatum | Regulated non-quarantine pest (score 60) | June, September, October and November | Loughgall, Mount Stewart and Hillsborough | 6 | 0.84 | C. acutatum poses a risk to the strawberry industry [42]. Hosts include celery, lupine, pine, tomato, strawberry and olive [44] |
| Exobasidium japonicum | Regulated non-quarantine pest | January, February, September, October and November | Hillsborough and Lough Navar | 7 | 0.96 | Many species of the genus Exobasidium are reported pathogens of the Ericaceae family, causing galls on leaf and stems, spots and red shoots [45] |
| Gnomoniopsis idaeicola | Non-regulated (score 24) | September and October | Loughgall and Mount Stewart | 2 | 0.99 | The hosts of this species mostly belong to the genus Rubus and it is noted as being a risk to the Rubus industry, causing cane canker and wilting in blackberry [42,46] |
| Rhizoctonia solani | Regulated non-quarantine pest | June and July | Loughgall, Hillsborough and Mount Stewart | 3 | 0.78 | R. solani is a necrotrophic pathogen that infects a wide range of hosts, including economically important crops such as potato, rice, turf grass, barely and maize, and can persist in the soil for years without a specific host [47,48,49] |
| Sirococcus piceicola | Regulated non-quarantine pest (score 40) | July | Lough Navar | 1 | 1 | The first incidence of this pathogen in the UK was recorded in 2021 on Sitka spruce seeds from Wales [50]. This pathogen can negatively impact Sitka spruce production in nurseries or cause damage to established trees [50] |
| Stemphylium vesicarium | Non-regulated (score 36) | February and March | Mount Stewart | 2 | 0.99 | Described as a potential pathogen of pears [42,44]. Other hosts include onion, asparagus and tomato in which S. vesicarium can cause leaf spot or leaf blight [42,44] |
| Verticillium albo-atrum | Regulated non-quarantine pest (score 60) | February and March | Hillsbourgh and Mount Stewart | 3 | 0.97 | A pathogen affecting fruit and nut trees, with major hosts including European hazelnut, quince, strawberry, tomato, apple and pear [42] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McLaughlin, K.E.; Belaouni, H.A.; McClure, A.; McCullough, K.; Craig, D.; McKeown, J.; Stevenson, M.A.; Carmichael, E.; Dalzell, J.; O’Hanlon, R.; et al. Using Environmental DNA as a Plant Health Surveillance Tool in Forests. Forests 2025, 16, 632. https://doi.org/10.3390/f16040632
McLaughlin KE, Belaouni HA, McClure A, McCullough K, Craig D, McKeown J, Stevenson MA, Carmichael E, Dalzell J, O’Hanlon R, et al. Using Environmental DNA as a Plant Health Surveillance Tool in Forests. Forests. 2025; 16(4):632. https://doi.org/10.3390/f16040632
Chicago/Turabian StyleMcLaughlin, Kirsty Elizabeth, Hadj Ahmed Belaouni, Andrew McClure, Kelly McCullough, David Craig, Joanne McKeown, Michael Andrew Stevenson, Eugene Carmichael, Johnathan Dalzell, Richard O’Hanlon, and et al. 2025. "Using Environmental DNA as a Plant Health Surveillance Tool in Forests" Forests 16, no. 4: 632. https://doi.org/10.3390/f16040632
APA StyleMcLaughlin, K. E., Belaouni, H. A., McClure, A., McCullough, K., Craig, D., McKeown, J., Stevenson, M. A., Carmichael, E., Dalzell, J., O’Hanlon, R., Murchie, A. K., & Warnock, N. (2025). Using Environmental DNA as a Plant Health Surveillance Tool in Forests. Forests, 16(4), 632. https://doi.org/10.3390/f16040632






