Using Citizen Science Data as Pre-Training for Semantic Segmentation of High-Resolution UAV Images for Natural Forests Post-Disturbance Assessment
Abstract
1. Introduction
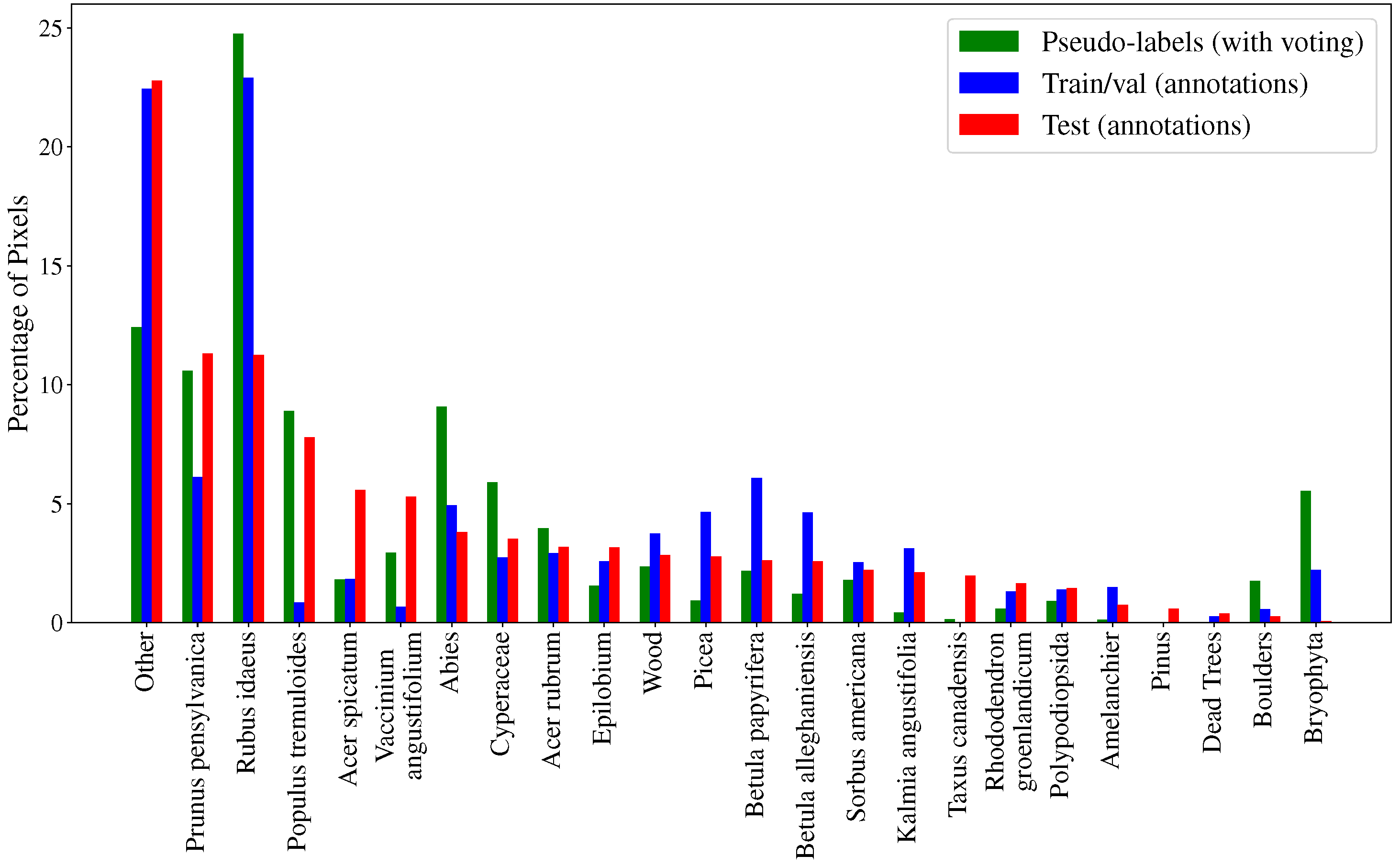
- A dataset of 11,269 full-size UAV images, called WilDReF-Q, taken at very low altitude and speed, with an average GSD of around , collected over around of natural regrowth environments;
- Accompanying ground truth for 153 cropped images, hand-labeled over 24 classes;
- Improving the quality of a previous sliding-window pseudo-labeling approach [9], notably by a thorough hyperparameter search and voting over multiple predictions;
- Demonstrating that when employed at scale, this pseudo-labeling framework surpasses the use of labeled UAV images.
2. Related Work
2.1. Impact of GSD on UAV Plant Species Mapping
2.2. Leveraging Citizen Science Contributions for Species Identification
2.3. Training Semantic Segmentation Networks Based on Pseudo-Labels
3. Materials and Methods
3.1. Areas and Species of Interest
- Fir/White Birch domain: Located in the southern part of the boreal vegetation zone, this bioclimatic domain is characterized by a dominant presence of fir and white birch trees. The sites in this domain are Montmorency, ZEC Des Passes, and Chic-Chocs.
- Fir/Yellow Birch domain: Situated in the northern temperate zone’s mixed forest sub-zone, this ecotone marks the transition between the northern temperate and boreal zones. The sites included in this domain are ZEC Wessoneau, ZEC Batiscan, and ZEC Chapais.
- Maple/Basswood domain: Found in the northern temperate zone’s deciduous forest sub-zone, this domain contains a diverse flora, with many species reaching their northern distribution limits here. The Windsor site represents this domain.
- Division: Bryophyta (Moss).
- Class: Polypodiopsida (Fern).
- Family: Cyperaceae (Sedge).
- Genus: Abies (Fir), Amelanchier (Serviceberry), Epilobium (Willowherb), Picea (Spruce), Pinus (Pine).
- Species: Acer rubrum (Red Maple), Acer spicatum (Mountain Maple), Betula alleghaniensis (Yellow Birch), Betula papyrifera (Paper Birch), Kalmia angustifolia (Sheep Laurel), Populus tremuloides (Trembling Aspen), Prunus pensylvanica (Fire Cherry), Rhododendron groenlandicum (Bog Labrador Tea), Rubus idaeus (Red Raspberry), Sorbus americana (American Mountain-Ash), Taxus canadensis (Canadian Yew), Vaccinium angustifolium (Lowbush Blueberry).
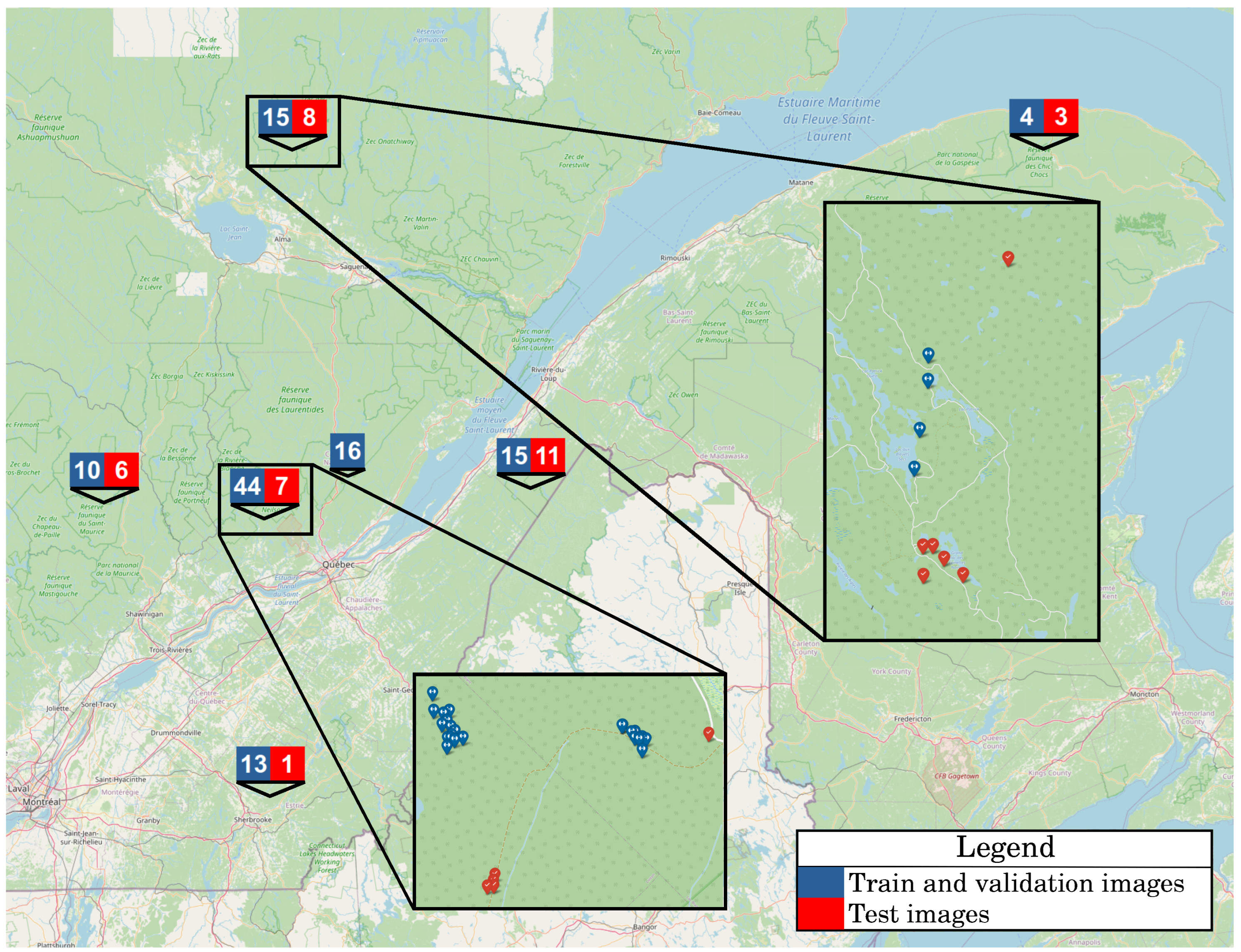
3.2. UAV Image Acquisition
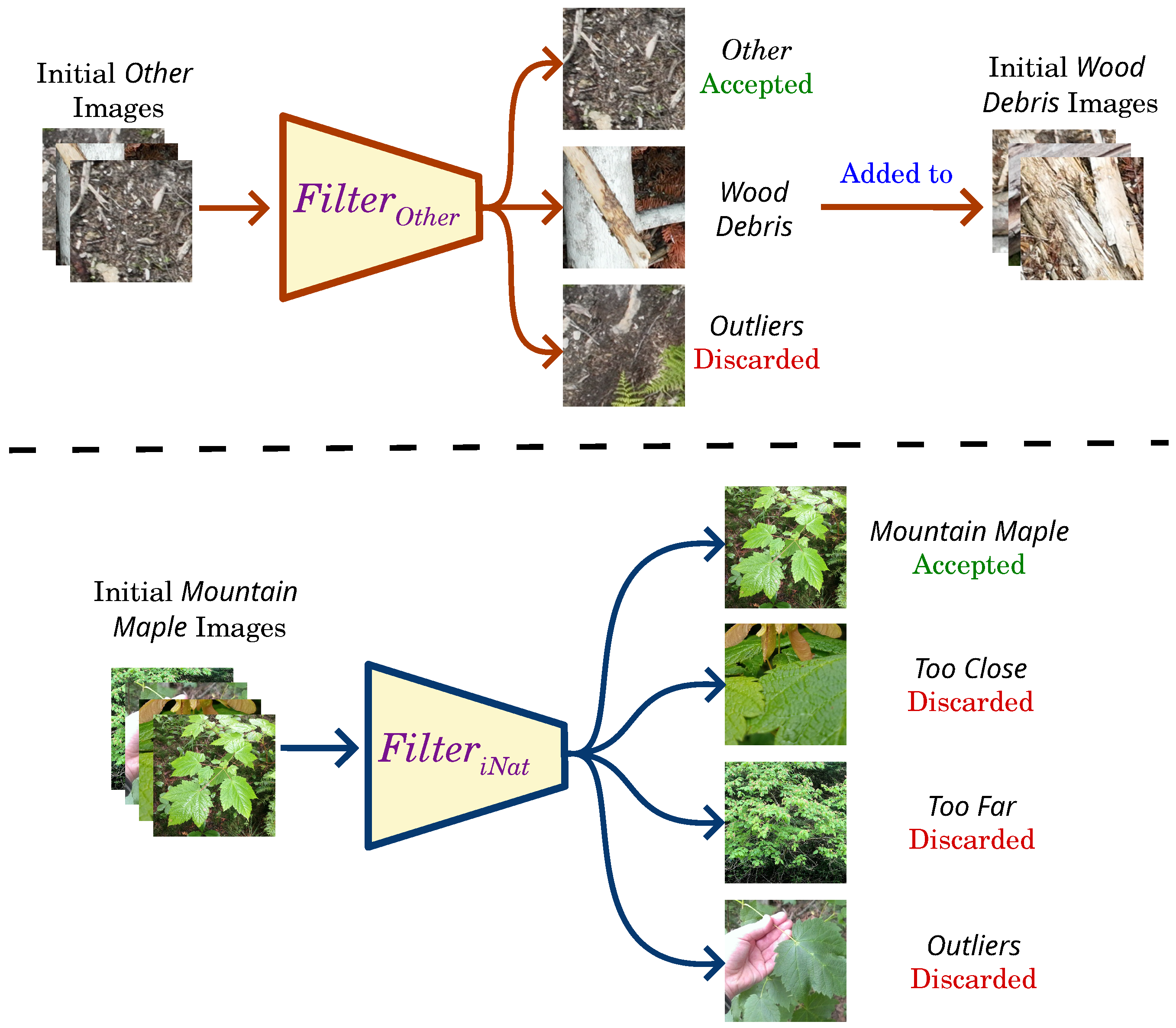
3.3. Training Data for Image Classifier
3.4. Training of Image Classifier
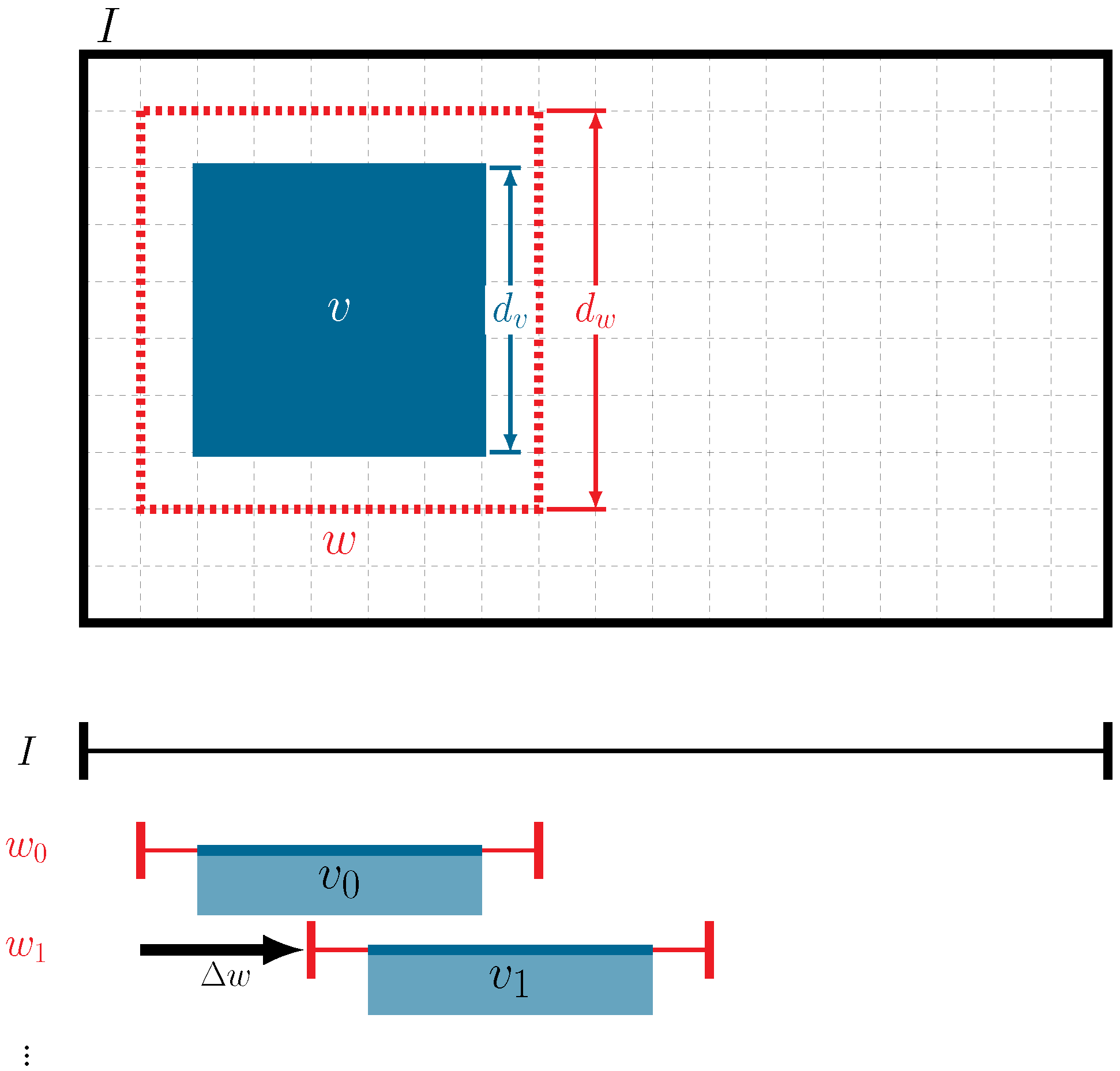
3.5. Generating Pseudo-Labels with a Moving-Window () Approach for Pre-Training Data
3.6. Training a Segmentation Model
4. Results
4.1. Image Classifier
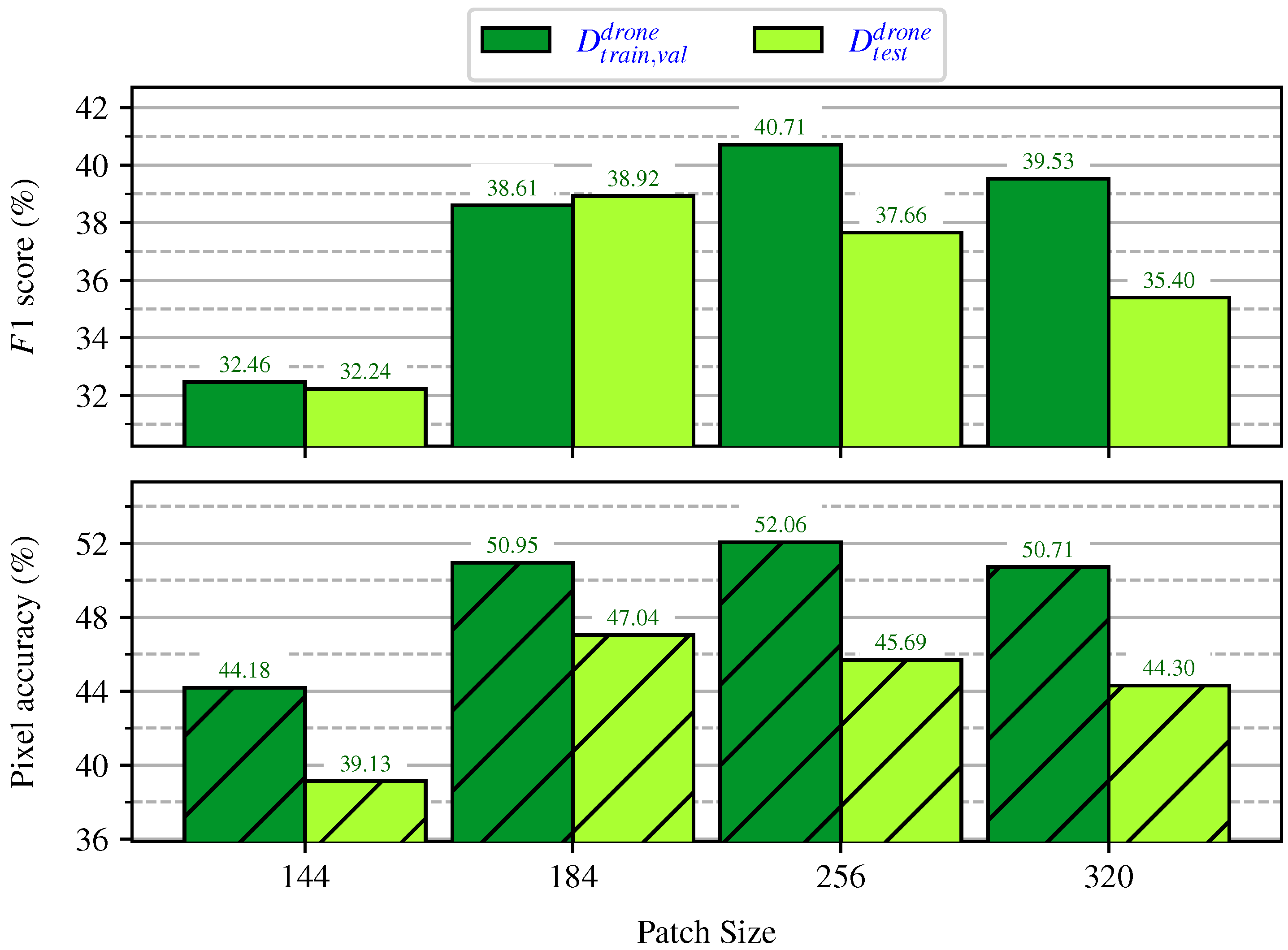
Evaluating the Impact of Patch Sizes on Inference
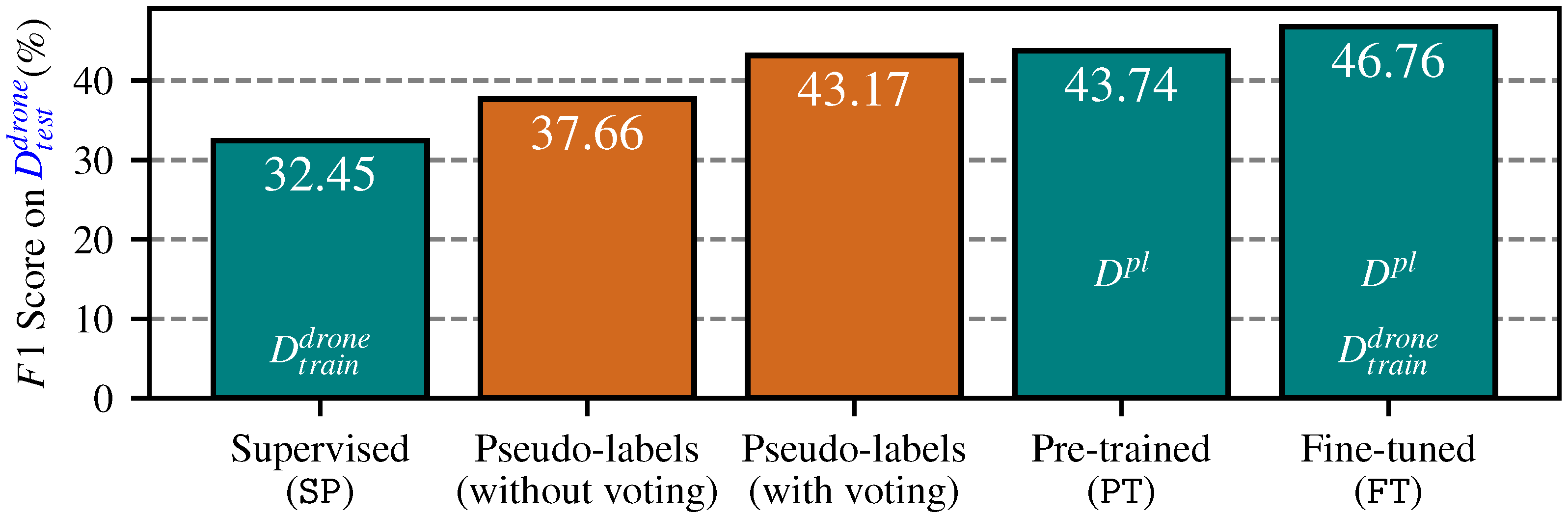
4.2. Pseudo-Label Generation
4.2.1. Impact of Different Prediction Assignments
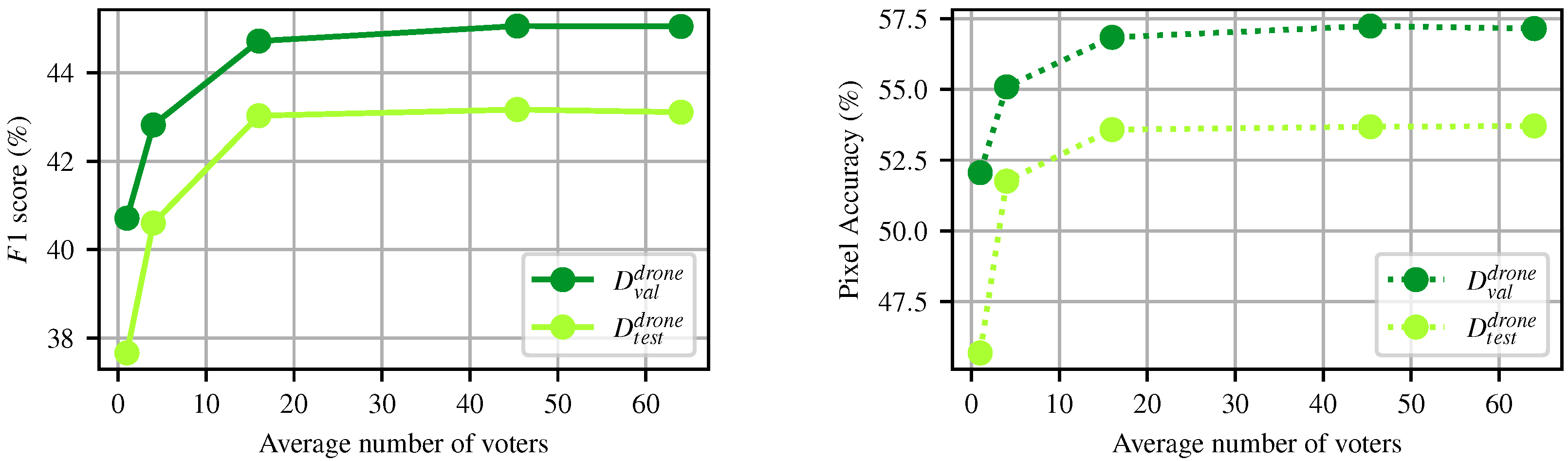
4.2.2. Impact of the Number of Votes
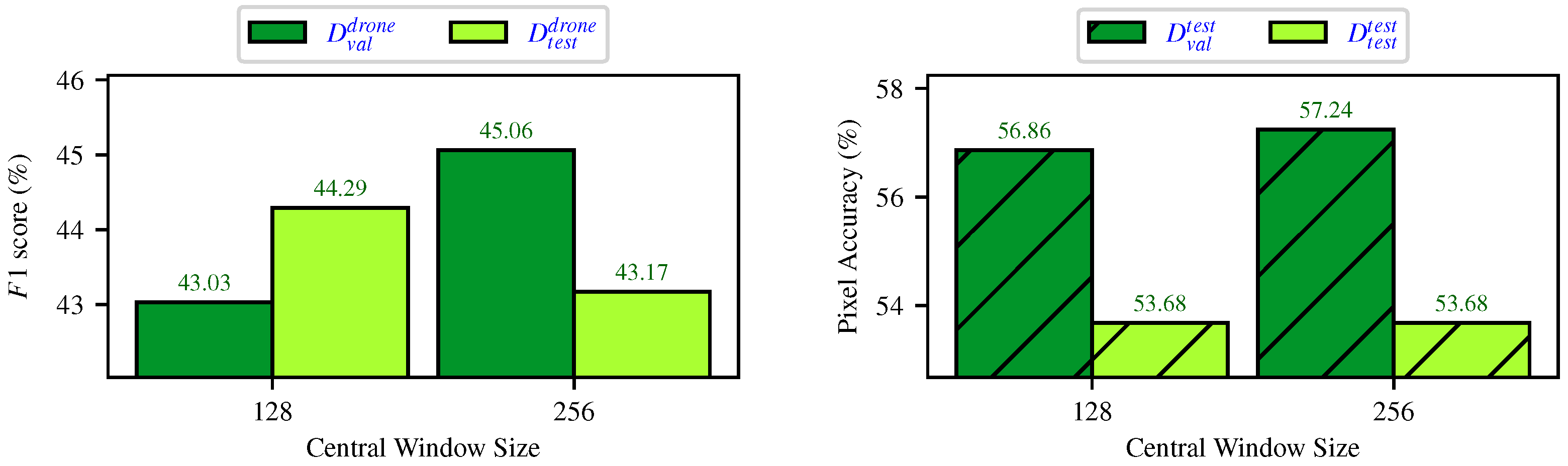
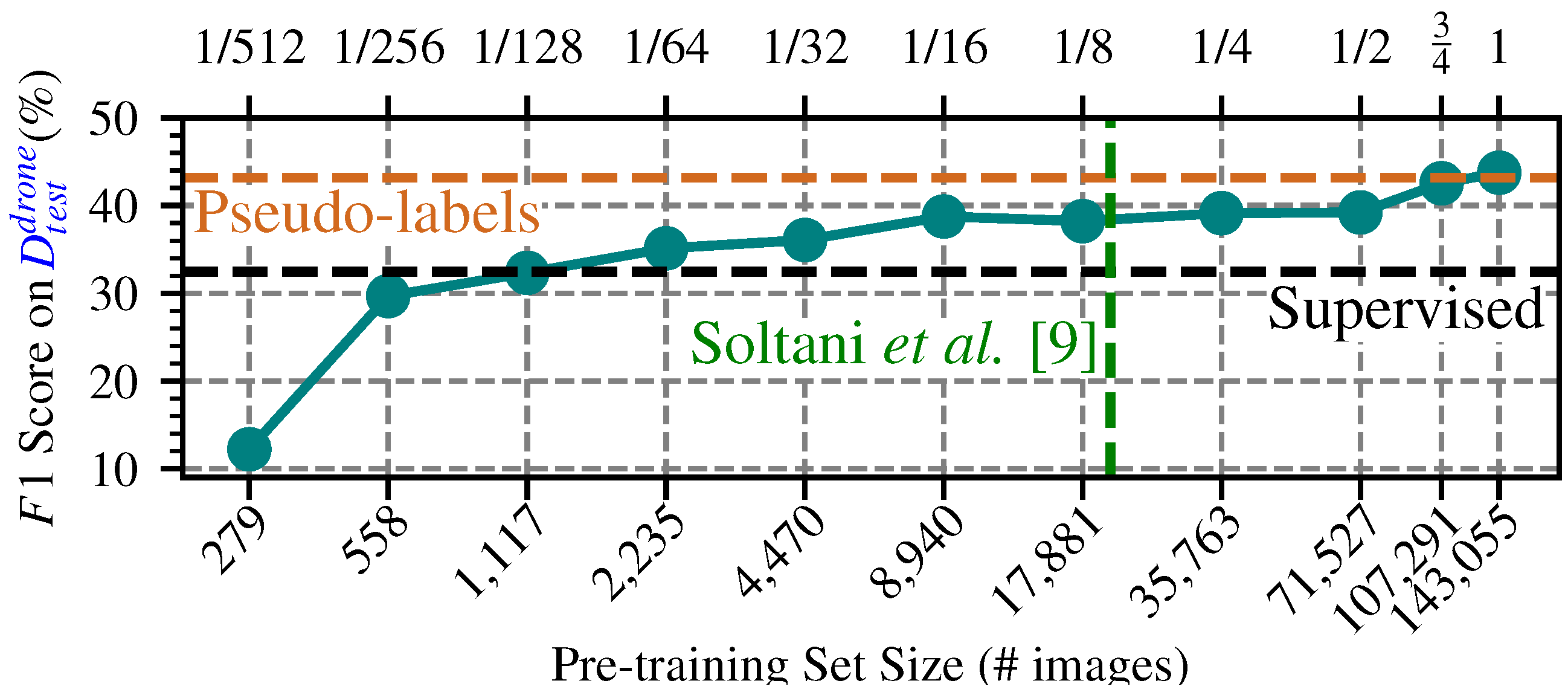
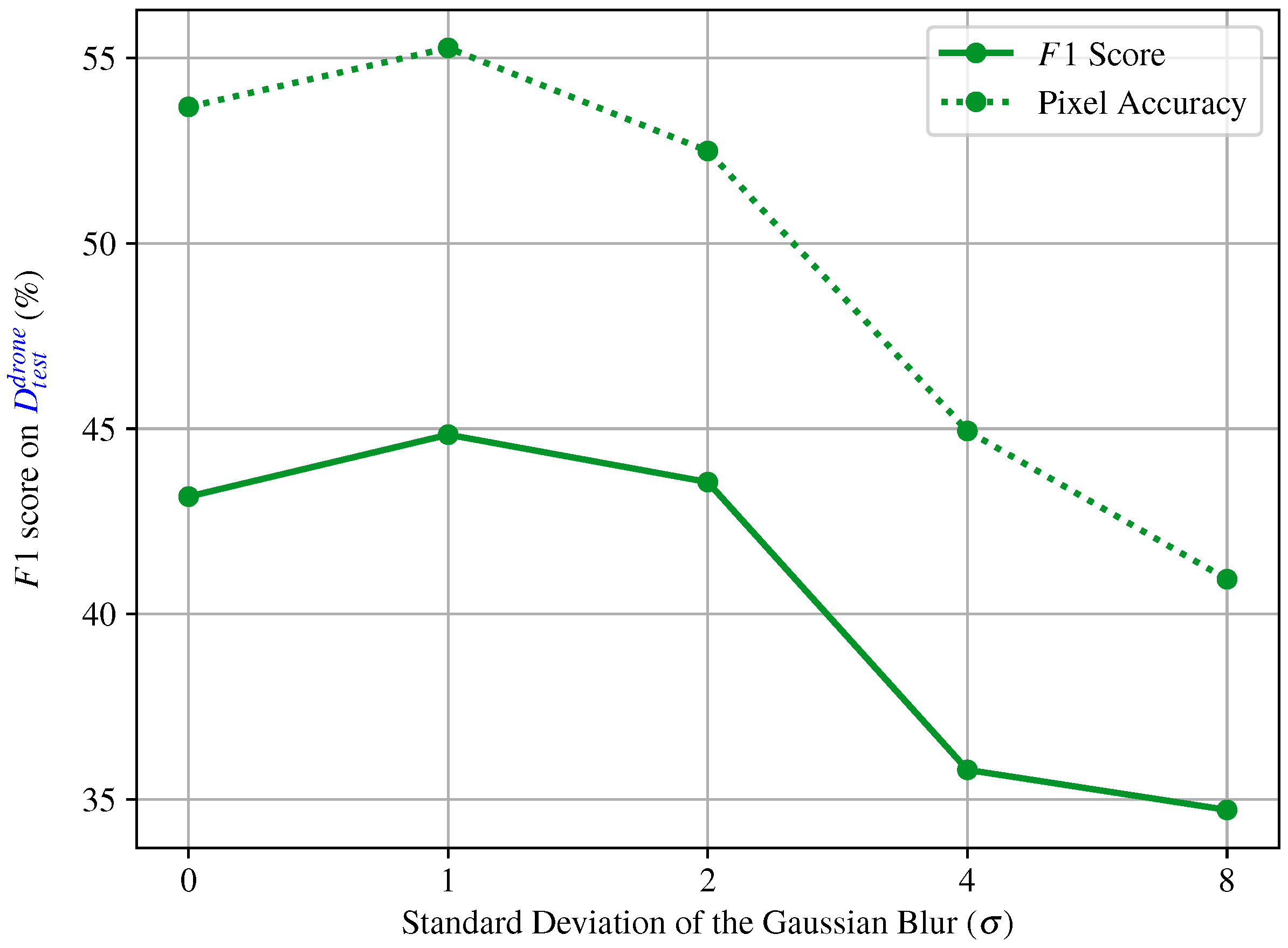
4.3. End-to-End Segmentation with
5. Discussion
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Oliver, T.H.; Isaac, N.J.; August, T.A.; Woodcock, B.A.; Roy, D.B.; Bullock, J.M. Declining resilience of ecosystem functions under biodiversity loss. Nat. Commun. 2015, 6, 10122. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Latifi, H.; Stereńczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.T.; Straub, C.; Ghosh, A. Review of studies on tree species classification from remotely sensed data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Van den Broeck, W.A.J.; Terryn, L.; Cherlet, W.; Cooper, Z.T.; Calders, K. Three-Dimensional Deep Learning for Leaf-Wood Segmentation of Tropical Tree Point Clouds. Int. Arch. Photogramm. Remote. Sens. Spat. Inf. Sci. 2023, XLVIII-1/W2-2023, 765–770. [Google Scholar] [CrossRef]
- Sylvain, J.D.; Drolet, G.; Thiffault, É.; Anctil, F. High-resolution mapping of tree species and associated uncertainty by combining aerial remote sensing data and convolutional neural networks ensemble. Int. J. Appl. Earth Obs. Geoinf. 2024, 131, 103960. [Google Scholar] [CrossRef]
- LaRocque, A.; Phiri, C.; Leblon, B.; Pirotti, F.; Connor, K.; Hanson, A. Wetland Mapping with Landsat 8 OLI, Sentinel-1, ALOS-1 PALSAR, and LiDAR Data in Southern New Brunswick, Canada. Remote Sens. 2020, 12, 2095. [Google Scholar] [CrossRef]
- Pu, R. Mapping Tree Species Using Advanced Remote Sensing Technologies: A State-of-the-Art Review and Perspective. J. Remote Sens. 2021, 2021, 9812624. [Google Scholar] [CrossRef]
- Bolyn, C.; Lejeune, P.; Michez, A.; Latte, N. Mapping tree species proportions from satellite imagery using spectral–spatial deep learning. Remote Sens. Environ. 2022, 280, 113205. [Google Scholar] [CrossRef]
- Soltani, S.; Feilhauer, H.; Duker, R.; Kattenborn, T. Transfer learning from citizen science photographs enables plant species identification in UAV imagery. ISPRS Open J. Photogramm. Remote Sens. 2022, 5, 100016. [Google Scholar] [CrossRef]
- Soltani, S.; Ferlian, O.; Eisenhauer, N.; Feilhauer, H.; Kattenborn, T. From simple labels to semantic image segmentation: Leveraging citizen science plant photographs for tree species mapping in drone imagery. Biogeosciences 2024, 21, 2909–2935. [Google Scholar] [CrossRef]
- iNaturalist Contributors. iNaturalist Research-Grade Observations; iNaturalist Contributors: San Rafael, CA, USA, 2024. [Google Scholar] [CrossRef]
- iNaturalist Contributors. iNaturalist. Available online: www.inaturalist.org (accessed on 6 December 2024).
- Ferreira, M.P.; Almeida, D.R.A.d.; Papa, D.d.A.; Minervino, J.B.S.; Veras, H.F.P.; Formighieri, A.; Santos, C.A.N.; Ferreira, M.A.D.; Figueiredo, E.O.; Ferreira, E.J.L. Individual tree detection and species classification of Amazonian palms using UAV images and deep learning. For. Ecol. Manag. 2020, 475, 118397. [Google Scholar] [CrossRef]
- Cloutier, M.; Germain, M.; Laliberté, E. Influence of temperate forest autumn leaf phenology on segmentation of tree species from UAV imagery using deep learning. Remote Sens. Environ. 2024, 311, 114283. [Google Scholar] [CrossRef]
- Ecke, S.; Stehr, F.; Frey, J.; Tiede, D.; Dempewolf, J.; Klemmt, H.J.; Endres, E.; Seifert, T. Towards operational UAV-based forest health monitoring: Species identification and crown condition assessment by means of deep learning. Comput. Electron. Agric. 2024, 219, 108785. [Google Scholar] [CrossRef]
- Schiefer, F.; Kattenborn, T.; Frick, A.; Frey, J.; Schall, P.; Koch, B.; Schmidtlein, S. Mapping forest tree species in high resolution UAV-based RGB-imagery by means of convolutional neural networks. ISPRS J. Photogramm. Remote Sens. 2020, 170, 205–215. [Google Scholar] [CrossRef]
- Jozdani, S.; Chen, D.; Chen, W.; Leblanc, S.G.; Prévost, C.; Lovitt, J.; He, L.; Johnson, B.A. Leveraging Deep Neural Networks to Map Caribou Lichen in High-Resolution Satellite Images Based on a Small-Scale, Noisy UAV-Derived Map. Remote Sens. 2021, 13, 2658. [Google Scholar] [CrossRef]
- Hao, Z.; Lin, L.; Post, C.J.; Mikhailova, E.A.; Yu, K.; Fang, H.; Liu, J. The co-effect of image resolution and crown size on deep learning for individual tree detection and delineation. Int. J. Digit. Earth 2023, 16, 3753–3771. [Google Scholar] [CrossRef]
- Fromm, M.; Schubert, M.; Castilla, G.; Linke, J.; McDermid, G. Automated Detection of Conifer Seedlings in Drone Imagery Using Convolutional Neural Networks. Remote Sens. 2019, 11, 2585. [Google Scholar] [CrossRef]
- Gan, Y.; Wang, Q.; Iio, A. Tree Crown Detection and Delineation in a Temperate Deciduous Forest from UAV RGB Imagery Using Deep Learning Approaches: Effects of Spatial Resolution and Species Characteristics. Remote Sens. 2023, 15, 778. [Google Scholar] [CrossRef]
- Ball, J.G.C.; Hickman, S.H.M.; Jackson, T.D.; Koay, X.J.; Hirst, J.; Jay, W.; Archer, M.; Aubry-Kientz, M.; Vincent, G.; Coomes, D.A. Accurate delineation of individual tree crowns in tropical forests from aerial RGB imagery using Mask R-CNN. Remote Sens. Ecol. Conserv. 2023, 9, 641–655. [Google Scholar] [CrossRef]
- PlantNet. With the Pl@ntNet App, Identify One Plant from a Picture, and Be Part of a Citizen Science Project on Plant Biodiversity; PlantNet: Montpellier, France, 2024. [Google Scholar]
- Affouard, A.; Goëau, H.; Bonnet, P.; Lombardo, J.C.; Joly, A. Pl@ntNet app in the era of deep learning. In Proceedings of the International Conference on Learning Representations (ICLR), Toulon, France, 24–26 April 2017. [Google Scholar]
- Garcin, C.; Joly, A.; Bonnet, P.; Lombardo, J.C.; Affouard, A.; Chouet, M.; Servajean, M.; Lorieul, T.; Salmon, J. Pl@ntNet-300K: A plant image dataset with high label ambiguity and a long-tailed distribution. In Proceedings of the Conference on Neural Information Processing Systems (NeurIPS), Virtual, 6–14 December 2021. [Google Scholar] [CrossRef]
- Boone, M.E.; Basille, M. Using iNaturalist to Contribute Your Nature Observations to Science. EDIS 2019, 2019, 5. [Google Scholar] [CrossRef]
- Di Cecco, G.J.; Barve, V.; Belitz, M.W.; Stucky, B.J.; Guralnick, R.P.; Hurlbert, A.H. Observing the Observers: How Participants Contribute Data to iNaturalist and Implications for Biodiversity Science. BioScience 2021, 71, 1179–1188. [Google Scholar] [CrossRef]
- Konig, J.; Jenkins, M.D.; Mannion, M.; Barrie, P.; Morison, G. Weakly-Supervised Surface Crack Segmentation by Generating Pseudo-Labels Using Localization With a Classifier and Thresholding. IEEE Trans. Intell. Transp. Syst. 2022, 23, 24083–24094. [Google Scholar] [CrossRef]
- He, T.; Li, H.; Qian, Z.; Niu, C.; Huang, R. Research on Weakly Supervised Pavement Crack Segmentation Based on Defect Location by Generative Adversarial Network and Target Re-optimization. Constr. Build. Mater. 2024, 411, 134668. [Google Scholar] [CrossRef]
- Ferlian, O.; Cesarz, S.; Craven, D.; Hines, J.; Barry, K.E.; Bruelheide, H.; Buscot, F.; Haider, S.; Heklau, H.; Herrmann, S.; et al. Mycorrhiza in tree diversity–ecosystem function relationships: Conceptual framework and experimental implementation. Ecosphere 2018, 9, e02226. [Google Scholar] [CrossRef]
- Rolnick, D.; Veit, A.; Belongie, S.; Shavit, N. Deep Learning is Robust to Massive Label Noise. arXiv 2018, arXiv:1705.10694. [Google Scholar] [CrossRef]
- Cheng, B.; Misra, I.; Schwing, A.G.; Kirillov, A.; Girdhar, R. Masked-attention Mask Transformer for Universal Image Segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2022. [Google Scholar] [CrossRef]
- Ministère des Ressources Naturelles et des Forêts. Récolte et Autres Interventions Sylvicoles, [Jeu de Données]; Ministère des Ressources Naturelles et des Forêts: Québec, QC, Canada, 2017. [Google Scholar]
- Ministère des Ressources naturelles et de la Faune. Sustainable Management in the Boreal Forest: A Real Response to Environmental Challenges; Ministère des Ressources naturelles et de la Faune: Québec, QC, Canada, 2008. [Google Scholar]
- Zhu, F.; Ma, S.; Cheng, Z.; Zhang, X.Y.; Zhang, Z.; Liu, C.L. Open-world Machine Learning: A Review and New Outlooks. arXiv 2024, arXiv:2403.01759. [Google Scholar]
- Segments.ai Team. Segments.ai—The Training Data Platform for Computer Vision Engineers; Segments.ai Team: Leuven, Belgium, 2020; Available online: https://segments.ai (accessed on 15 August 2024).
- Carpentier, M.; Giguere, P.; Gaudreault, J. Tree Species Identification from Bark Images Using Convolutional Neural Networks. In Proceedings of the IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS), Madrid, Spain, 1–5 October 2018; pp. 1075–1081. [Google Scholar] [CrossRef]
- Standing Dead Tree Computer Vision Project. 2022. Available online: https://universe.roboflow.com/2905168025-qq-com/standing-dead-tree (accessed on 15 July 2024).
- Oquab, M.; Darcet, T.; Moutakanni, T.; Vo, H.V.; Szafraniec, M.; Khalidov, V.; Fernandez, P.; HAZIZA, D.; Massa, F.; El-Nouby, A.; et al. DINOv2: Learning Robust Visual Features without Supervision. arXiv 2024, arXiv:2304.07193. [Google Scholar]
- Buslaev, A.; Iglovikov, V.I.; Khvedchenya, E.; Parinov, A.; Druzhinin, M.; Kalinin, A.A. Albumentations: Fast and Flexible Image Augmentations. Information 2020, 11, 125. [Google Scholar] [CrossRef]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical Vision Transformer using Shifted Windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 10–17 October 2021; pp. 9992–10002. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Springer International Publishing: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the International Conference on Learning Representations (ICLR), San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- MMSegmentation Contributors. OpenMMLab Semantic Segmentation Toolbox and Benchmark. Apache-2.0 License. 2020. Available online: https://github.com/open-mmlab/mmsegmentation (accessed on 15 November 2024).
- Zhai, X.; Kolesnikov, A.; Houlsby, N.; Beyer, L. Scaling Vision Transformers. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2022; pp. 1204–1213. [Google Scholar] [CrossRef]
- Burt, P.; Adelson, E. The Laplacian Pyramid as a Compact Image Code. IEEE Trans. Commun. 1983, 31, 532–540. [Google Scholar] [CrossRef]
- Wandell, B.A. Foundations of Vision; Sinauer Associates: Sunderland, MA, USA, 1995. [Google Scholar]
- Piccolo, R.L.; Warnken, J.; Chauvenet, A.L.M.; Castley, J.G. Location biases in ecological research on Australian terrestrial reptiles. Sci. Rep. 2020, 10, 9691. [Google Scholar] [CrossRef]
- Ouaknine, A.; Kattenborn, T.; Laliberté, E.; Rolnick, D. OpenForest: A data catalogue for machine learning in forest monitoring. arXiv 2024, arXiv:2311.00277. [Google Scholar] [CrossRef]
- Berrio, J.S.; Shan, M.; Worrall, S.; Nebot, E. Camera-Lidar Integration: Probabilistic sensor fusion for semantic mapping. arXiv 2020, arXiv:2007.05490. [Google Scholar]
- Valjarević, A.; Djekić, T.; Stevanović, V.; Ivanović, R.; Jandziković, B. GIS numerical and remote sensing analyses of forest changes in the Toplica region for the period of 1953–2013. Appl. Geogr. 2018, 92, 131–139. [Google Scholar] [CrossRef]
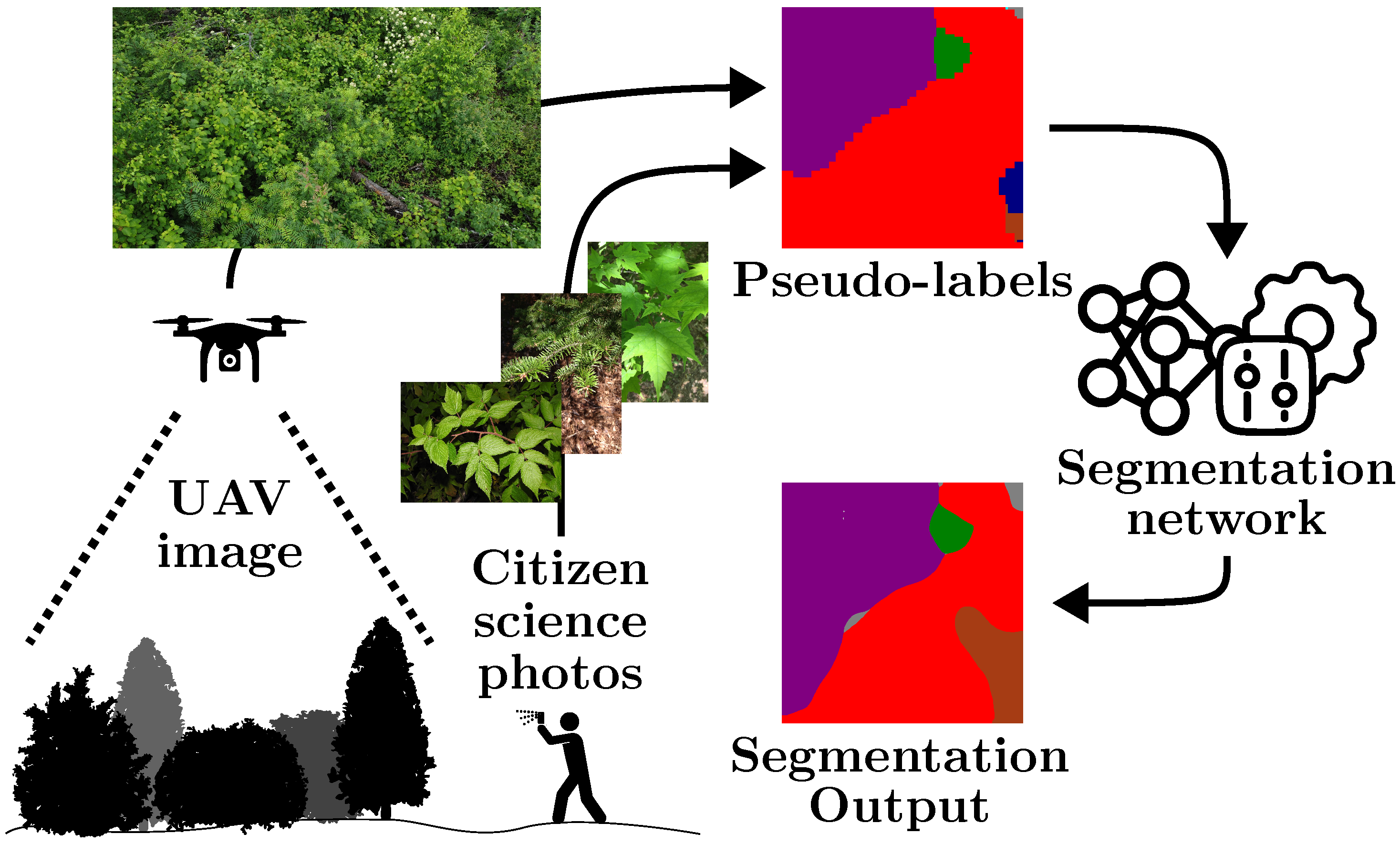
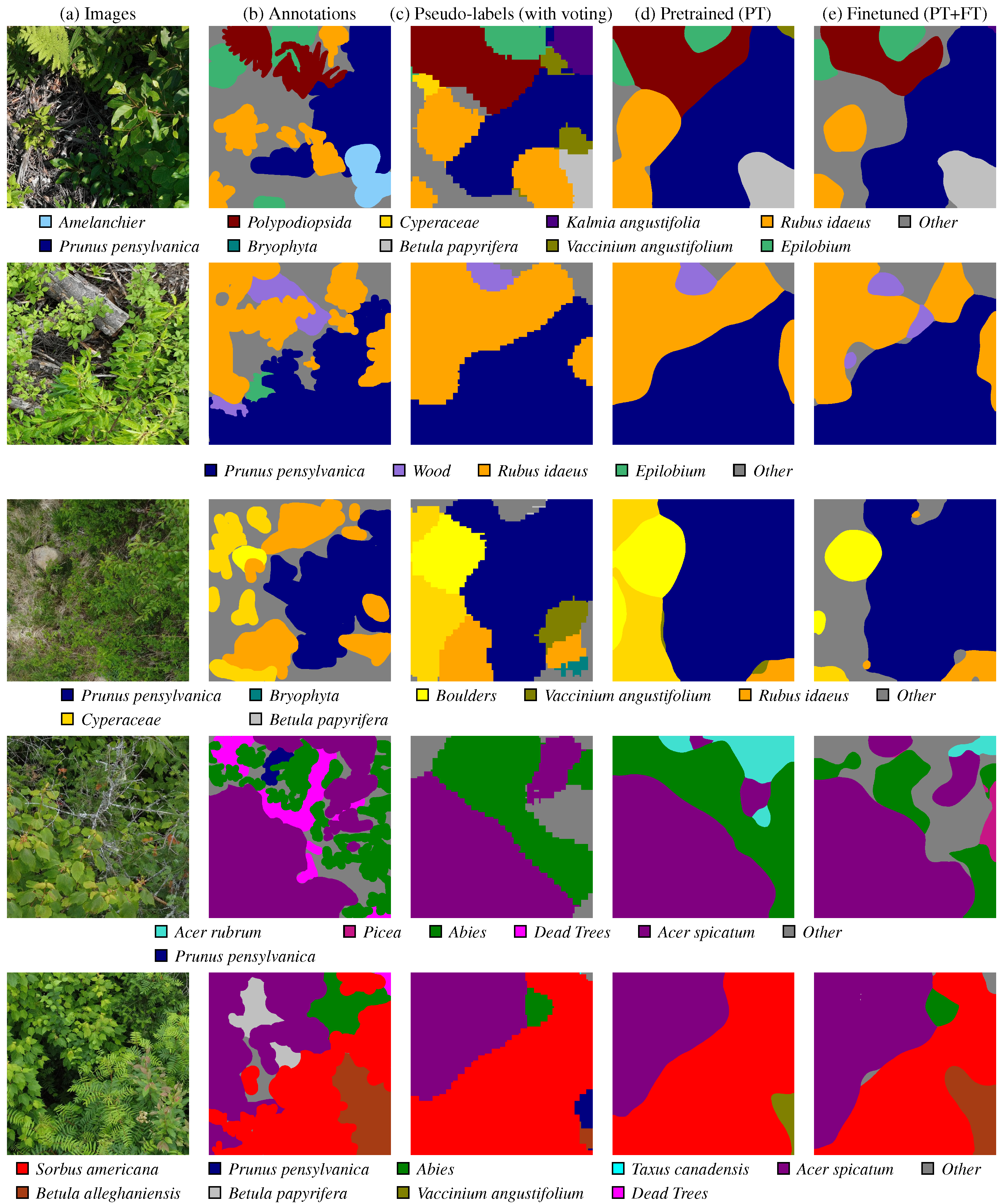
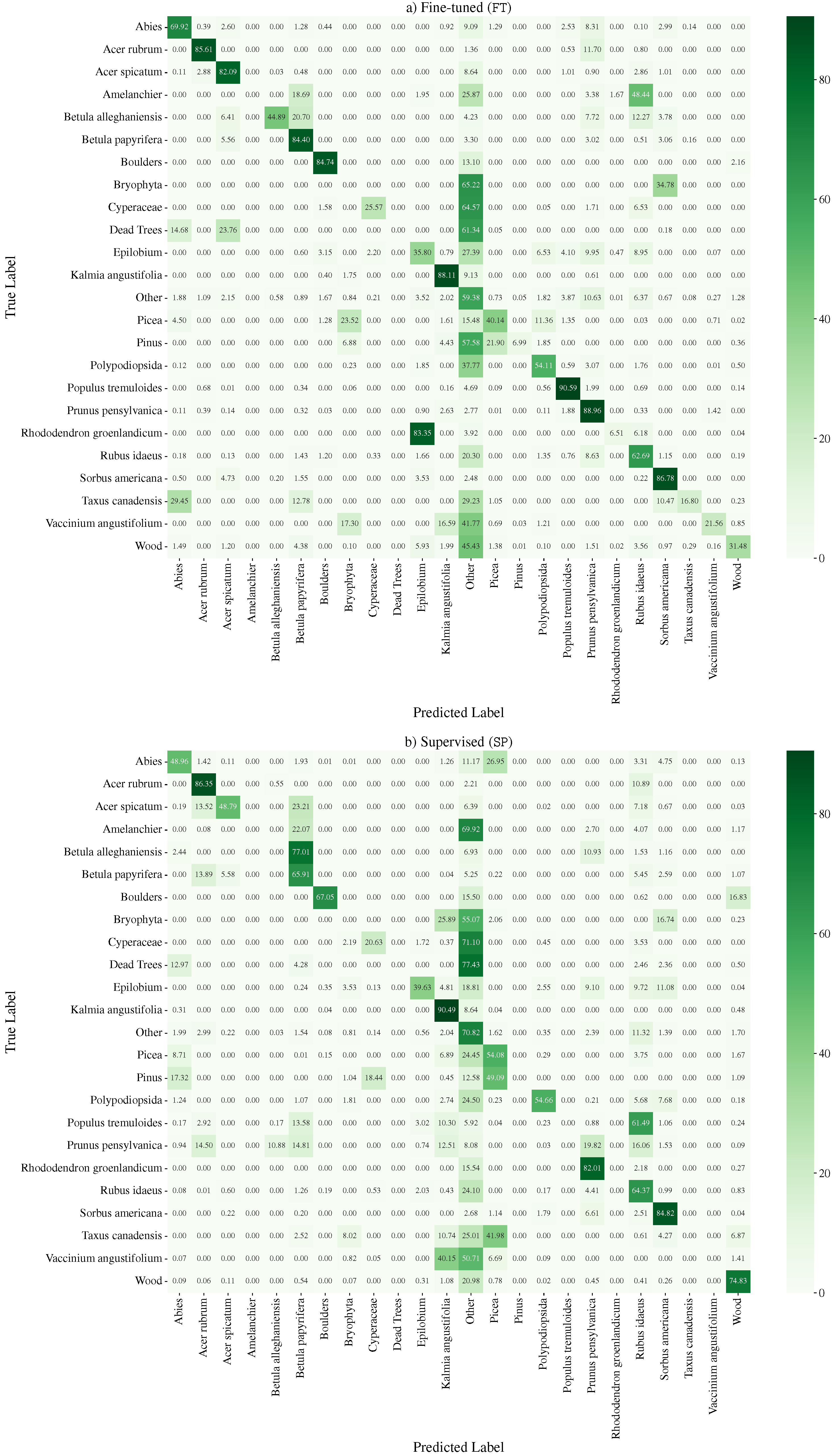
 Prunus pensylvanica is not present in the ground truth, and the
Prunus pensylvanica is not present in the ground truth, and the  Betula papyrifera is missing. However, the pre-training stage can mitigate some of these pseudo-label mistakes, due to an implicit averaging of labels. It also smooths out the granularity. Finally, fine-tuning () the semantic segmentation network on ground truth annotation provides some, but marginal, improvements. The iNaturalist images are sourced from iNaturalist [10] and the predictions are actual results from our method.
Betula papyrifera is missing. However, the pre-training stage can mitigate some of these pseudo-label mistakes, due to an implicit averaging of labels. It also smooths out the granularity. Finally, fine-tuning () the semantic segmentation network on ground truth annotation provides some, but marginal, improvements. The iNaturalist images are sourced from iNaturalist [10] and the predictions are actual results from our method.
 Prunus pensylvanica is not present in the ground truth, and the
Prunus pensylvanica is not present in the ground truth, and the  Betula papyrifera is missing. However, the pre-training stage can mitigate some of these pseudo-label mistakes, due to an implicit averaging of labels. It also smooths out the granularity. Finally, fine-tuning () the semantic segmentation network on ground truth annotation provides some, but marginal, improvements. The iNaturalist images are sourced from iNaturalist [10] and the predictions are actual results from our method.
Betula papyrifera is missing. However, the pre-training stage can mitigate some of these pseudo-label mistakes, due to an implicit averaging of labels. It also smooths out the granularity. Finally, fine-tuning () the semantic segmentation network on ground truth annotation provides some, but marginal, improvements. The iNaturalist images are sourced from iNaturalist [10] and the predictions are actual results from our method.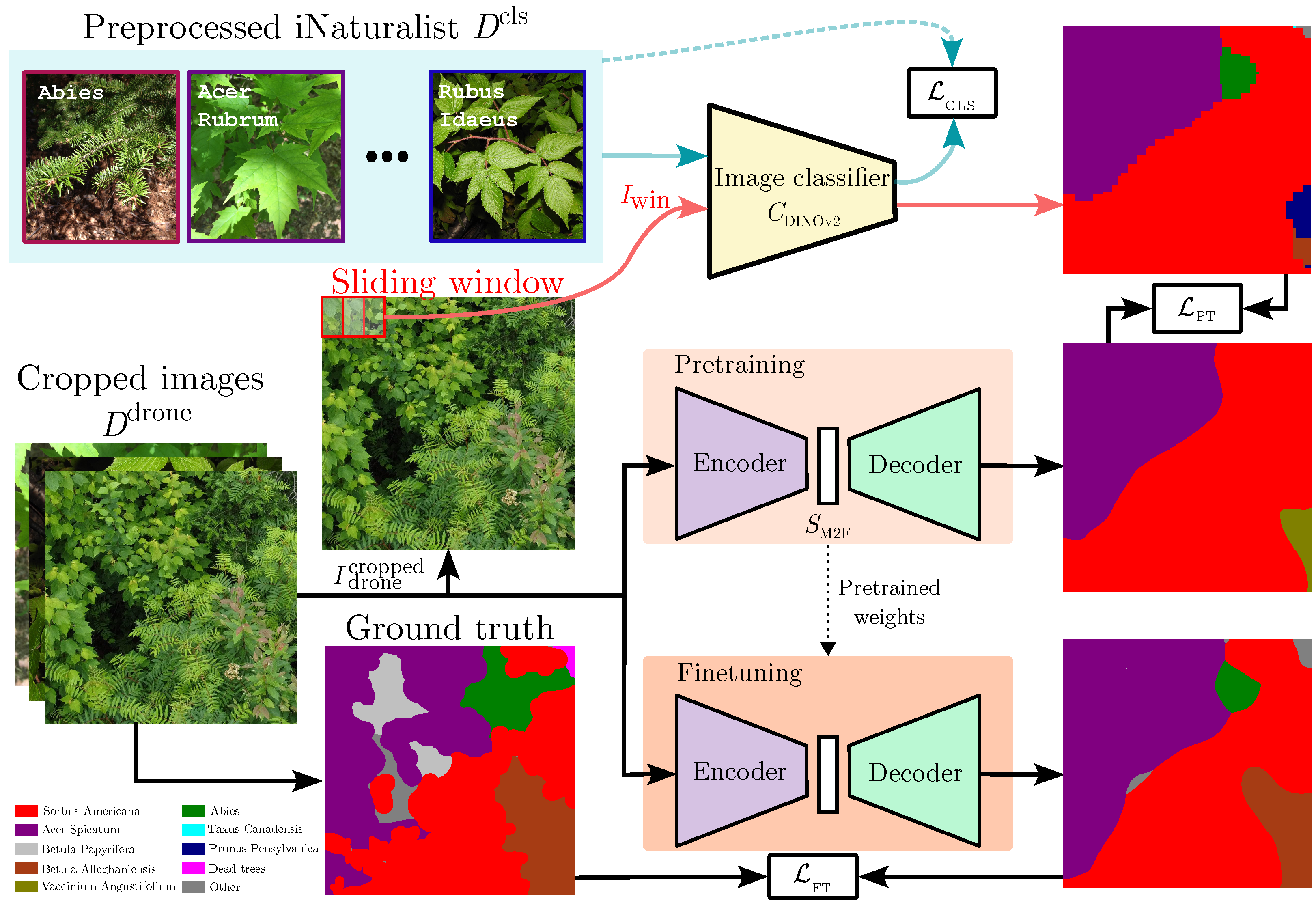
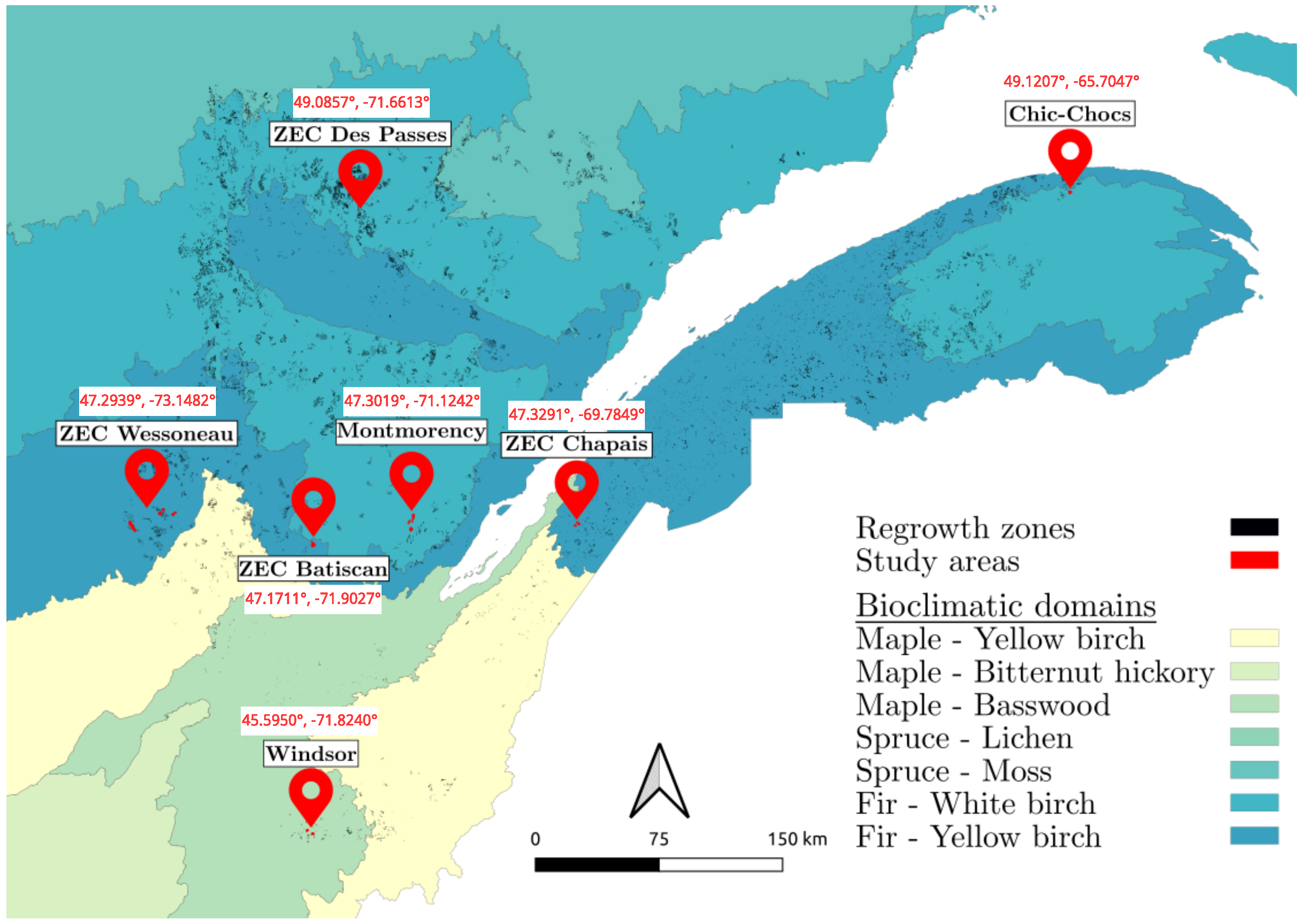





| Site | Vegetation Zone | Sub-Zone | Bioclimatic Domain | Elevation |
|---|---|---|---|---|
| ZEC Batiscan | Northern temperate | Mixed | Fir/Yellow Birch | 550 |
| ZEC Chapais | Northern temperate | Mixed | Fir/Yellow Birch | 350 |
| Chic-Chocs | Boreal | Continuous | Fir/White Birch | 600 |
| ZEC Des Passes | Boreal | Continuous | Fir/White Birch | 300 |
| Montmorency | Boreal | Continuous | Fir/White Birch | 700 |
| ZEC Wessoneau | Northern temperate | Mixed | Fir/Yellow Birch | 400 |
| Windsor | Northern temperate | Deciduous | Maple/Basswood | 250 |
| Dataset | Count | Image Size ( | Description |
|---|---|---|---|
| 318k | variable | Aggregated citizen science and other images to train | |
| 11,269 | 20 MPix or 9 MPix | Raw UAV images | |
| 143,208 | UAV image crops | ||
| 153 | Annotated UAV image crops |
| Augmentation | Experiments | ||||
|---|---|---|---|---|---|
| Aug 0 | Aug 1 | Aug 2 | Aug 3 | Aug 4 | |
| SmallestMaxSize | ● | ● | ● | ● | ● |
| RandomResizedCrop | ● | ● | ● | ● | ● |
| HorizontalFlip | ● | ● | ● | ● | ● |
| ColorJitter | ● | ● | ● | ● | ● |
| Blur | ● | ● | ● | ● | ● |
| ShiftScaleRotate | ○ | ● | ● | ● | ● |
| Perspective | ○ | ○ | ● | ● | ● |
| MotionBlur | ○ | ○ | ○ | ◐ | ◐ |
| MedianBlur | ○ | ○ | ○ | ◑ | ◑ |
| OpticalDistortion | ○ | ○ | ○ | ◓ | ◓ |
| GridDistortion | ○ | ○ | ○ | ◒ | ◒ |
| Defocus | ○ | ○ | ○ | ○ | ● |
| RandomFog | ○ | ○ | ○ | ○ | ● |
| Technique | Experiments | |||||
|---|---|---|---|---|---|---|
| Unfiltered | iNaturalist Filtered | Fully Filtered | Augmentations | Balance | Final | |
| ○ | ● | ● | ● | ● | ● | |
| ○ | ○ | ● | ● | ● | ● | |
| ImbalancedDatasetSampler | ○ | ○ | ○ | ○ | ● | ● |
| Experiment | () | () |
|---|---|---|
| Unfiltered baseline | 24.93% | 29.59% |
| Filtering technique | ||
| iNaturalist filtered | 24.37% ↓ −0.56 %pt | 27.26% ↓ −2.32 %pt |
| Fully filtered | 34.17% ↑ 9.24 %pt | 34.12% ↑ 4.53 %pt |
| Balancing technique | ||
| Balance | 34.23% ↑ 9.31 %pt | 34.04% ↑ 4.46 %pt |
| Augmentation technique | ||
| Aug 0 | 34.40% ↑ 9.48 %pt | 37.34% ↑ 7.76 %pt |
| Aug 1 | 35.12% ↑ 10.19 %pt | 36.53% ↑ 6.94 %pt |
| Aug 2 | 35.80% ↑ 10.87 %pt | 37.13% ↑ 7.54 %pt |
| Aug 3 | 36.29% ↑ 11.36 %pt | 37.83% ↑ 8.24 %pt |
| Aug 4 | 36.64% ↑ 11.71 %pt | 36.81% ↑ 7.22 %pt |
| Final (Balance + Aug 4) | 38.63% ↑ 13.70 %pt | 37.84% ↑ 8.25 %pt |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nasiri, K.; Guimont-Martin, W.; LaRocque, D.; Jeanson, G.; Bellemare-Vallières, H.; Grondin, V.; Bournival, P.; Lessard, J.; Drolet, G.; Sylvain, J.-D.; et al. Using Citizen Science Data as Pre-Training for Semantic Segmentation of High-Resolution UAV Images for Natural Forests Post-Disturbance Assessment. Forests 2025, 16, 616. https://doi.org/10.3390/f16040616
Nasiri K, Guimont-Martin W, LaRocque D, Jeanson G, Bellemare-Vallières H, Grondin V, Bournival P, Lessard J, Drolet G, Sylvain J-D, et al. Using Citizen Science Data as Pre-Training for Semantic Segmentation of High-Resolution UAV Images for Natural Forests Post-Disturbance Assessment. Forests. 2025; 16(4):616. https://doi.org/10.3390/f16040616
Chicago/Turabian StyleNasiri, Kamyar, William Guimont-Martin, Damien LaRocque, Gabriel Jeanson, Hugo Bellemare-Vallières, Vincent Grondin, Philippe Bournival, Julie Lessard, Guillaume Drolet, Jean-Daniel Sylvain, and et al. 2025. "Using Citizen Science Data as Pre-Training for Semantic Segmentation of High-Resolution UAV Images for Natural Forests Post-Disturbance Assessment" Forests 16, no. 4: 616. https://doi.org/10.3390/f16040616
APA StyleNasiri, K., Guimont-Martin, W., LaRocque, D., Jeanson, G., Bellemare-Vallières, H., Grondin, V., Bournival, P., Lessard, J., Drolet, G., Sylvain, J.-D., & Giguère, P. (2025). Using Citizen Science Data as Pre-Training for Semantic Segmentation of High-Resolution UAV Images for Natural Forests Post-Disturbance Assessment. Forests, 16(4), 616. https://doi.org/10.3390/f16040616






