Abstract
Pinus yunnanensis Franch. is a common woodland species in the southwest of China. Its trunk frequently twists during growth, affecting timber quality. The explanation for the twisted-trunk formation is unknown. In this work, we examined the variety of endophytes and metabolites by comparing the straight and twisted trunk types of P. yunnanensis. The results showed that the twisted trunk had a distinct endosymbiont composition compared to the straight trunk. Pseudomonas and Craurococcus bacteria, as well as the fungus taxa Phaeosphaeria and Epichloë, spread significantly more in the twisted trunk compared to the straight trunk. However, there was less Dechloromonas in the twisted trunk. Metabolomic analysis revealed differences in metabolites in the straight and twisted trunks, which were associated with four metabolic pathways: beta-alanine metabolism, metabolism of Jasmonic acid and trans-Zeatin metabolism, linoleic acid metabolism, and pentose phosphate pathway. The compounds were linked to certain endophyte bacteria species. Our findings suggested that the twisted trunk was subjected to more stress than the straight trunk because of endosymbionts. Moreover, we speculated that hormone signal transduction and the absorption, transport, and utilization of phosphorus elements and their interaction with microorganisms may be closely connected to the formation of twists. This is the first study to characterize the microbiome and metabolome in the twisted trunks of P. yunnanensis, and the results enhance our understanding of the underlying causes of twisted-trunk formation in P. yunnanensis.
1. Introduction
Pinus yunnanensis Franch. is a kind of pine tree native to southwestern China, which is the main forest vegetation type in Yunnan Province and the pioneer species of Yunnan afforestation, with a distribution area accounting for 29.98% of the national land area of Yunnan Province, 52% of the province’s forested land area, and 32% of the area of the stored forest land [1,2,3]. P. yunnanensis wood has beautiful grain, stable and pressure-resistant properties, strong workability. and is a good material for making handicrafts and high-quality furniture, as well as an important accessory for Chinese folk architectural structures, and is especially used in house frames, floors, roof beams, doors and windows, and other wooden structural components. In addition, P. yunnanensis has also been made into a landscape tree species and is widely used in the construction of protective forests, which is better at wind and sand resistance, soil consolidation and ecological restoration.
However, due to the extensive use of rough afforestation of P. yunnanensis, mixed forests, and serious degradation, a large number of twisted forms (trunks that are both twisted and curved) are found in more than two-thirds of P. yunnanensis stands in planted secondary forests or pure forests in the central Yunnan region, especially [4,5,6]. The occurrence of twisting seriously affects forest success, directly leading to a sharp decrease in forestry timber output and a decline in the annual production of usable timber, which seriously restricts the production of construction, good furniture, and timber and greatly affects people’s lives. Improving the trunk characteristics of P. yunnanensis has been the focus of tree improvement in southwest China for decades [7,8]. However, nothing is known about the causes of P. yunnanensis trunk twisting.
Plants do not grow in isolation; rather, they are constantly influenced by abiotic and biotic stresses, directly impacting productivity and health [9,10]. During long-term evolution, complicated interactions have gradually formed between plants and their commensal microbiome [11,12]. In recent years, their interplay has emerged as a hotspot for research [8,13,14,15,16]. Current research on plant–microbe interactions is moving toward the idea that the plant’s characteristics are controlled by more than just its genes [14], also modulated by plant–microbe interactions [17,18,19]. For example, microbes modify the host’s primary and secondary metabolism to affect the plant responses [12,20]. Scherling et al. (2009) have studied specific plant–microbe interactions utilizing poplar plants grown in vitro [21]. The inoculation of Paenibacillus sp. strain P22 to endophyte-free plants cultivated in vitro induced significant modifications in the metabolome of the inoculated plants. In addition, it was reported that the interplay between Zea mays and the fungus Colletotrichum graminicola triggered systemic resistance, which was associated with elevated levels of salicylic acid and abscisic acid [22]. Several pieces of evidence indicated that endophytic fungi enhanced the uptake and accumulation of Ni in the plants by upregulation of several genes [23]. Moreover, the effects of the microbiome on the edible quality of sugarcane through the modulation of its secondary metabolitess have been determined [18].
The most direct indicator of an organism’s phenotype is its metabolome, which is the technologies to assess plant–microbe interactions and is located downstream of the transcriptome and proteome [17]. By identifying and measuring tiny molecules (often represent the end product of regulatory circuits), this method offers indirect insight into how the microbiome affects the phenotype of the host plant [24,25,26]. This field is still in its infancy and details on how diverse microbiomes may influence the metabolic profile of plants are currently insufficient and not well understood [17,18,27]. However, it is nevertheless a useful tool for studying plant phenotypes.
The phloem, a crucial component of the plant vascular system, plays an essential role in transporting photosynthates and various molecules, such as sugars, lipids, amino acids, nucleic acids, proteins, and phytohormones, from source tissues like mature leaves to sink tissues, including buds, flowers, seeds, and roots [28,29,30,31]. Moreover, the phloem is closely related to the radial growth of the plant [32,33]. Plant-related microbial communities depend on the microenvironment provided by plant tissue [10]. The phloem, as the tissue that transports nutrients from the plant, is responsible for transporting photosynthetically produced organic matter downward to other organs of the plant, and many insects and pathogens draw their nutrients from the phloem tissue deep in the temporal part of the plant. The phloem is so nutrient-rich that numerous pathogens and insects have developed specialized mechanisms to extract nutrients from it, with some of the most destructive among them feeding on or residing within the phloem itself [31]. We consider that exploring the diversity and correlations of microorganisms and metabolites in the phloem might provide new insights into the formation of twisted-trunk P. yunnanensis. In this study, we employed a comprehensive approach that combines gene sequencing with non-targeted ultra-high-performance liquid chromatography coupled with quadrupole time-of-flight high-resolution mass spectrometry (UHPLC-QTOF-MS) metabolomics to assess microbial diversity and analyze the abundance of metabolites In summary, this study aims to use metabolomics and microbiome technologies to compare the metabolites and microorganisms in the phloem of twisted and straight trunks of P. yunnanensis. The study will analyze the differential microorganisms and metabolites obtained, and attempt to elucidate the relationship between specific microorganisms and metabolites and the morphological development of twisted trunks in P. yunnanensis.
2. Materials and Methods
2.1. Sample Collection and Processing
In this study, the phloem of P. yunnanensis was collected in August 2020 from three sampling sites in Yunnan Province, China: Kunming, Chuxiong and Dali (Supplementary Table S1). Specifically, we selected ten P. yunnanensis trees, around 25 years old, with five exhibiting straight trunks and five with twisted trunks. These trees were chosen for their comparable height and diameter at breast height, ensuring uniformity across the sample plants at each location A sterile scalpel was used to scrape the thick bark surface at a height of 1.5 m, and exposed an area 100 mm long and 100 mm wide and 3 samples were taken from one tree. The phloem samples, each approximately 1 mm thick, were carefully extracted and placed into freezing tubes. These samples were then transported to the laboratory using liquid nitrogen for preservation and stored in a −80 °C freezer for subsequent analysis. Individuals from these samples were then divided into two sections for endophyte and metabolome analysis, respectively.
2.2. DNA Extraction and HIGH-Throughput Sequencing
DNA from both bacterial and fungal sources was extracted from the phloem samples of two trunk types of P. yunnanensis using the MN NucleoSpin 96 Soil kit (Macherey-Nagel, Düren, Germany). The concentration of the DNA was measured using a spectrophotometer at A260/280 (NanoDrop 1000; Thermo Scientific, Wilmington, DE, USA), while its integrity was evaluated using an Agilent 2100 system (Agilent Technologies, Santa Clara, CA, USA). Bacterial communities were barcoded and identified on the basis of the V3-V4 hypervariable regions of 16S rRNA gene. Fungal communities were identified and barcoded by analyzing internal transcribed spacer 1 (ITS1) DNA sequences from the rRNA gene regions. Polymerase chain reaction (PCR) amplification was carried out using specific primer combinations of 338F (5′-ACTCCTACGGGAGGCAGCA-3′)/806R (5′-GGACTACHVGGGTWTCTAAT-3′) and ITS3F (GCATCGATGAAGAACGCAGC)/ITS4 (TCCTCCGCTTATTGATATGC), respectively. High-throughput sequencing analyses of bacterial and fungal genes were performed on the Illumina HiSeq 2500 platform (2 × 250 paired ends) (Illumina, San Diego, CA, USA) at Biomarker Technologies Corporation (Beijing, China).
2.3. Statistical Analysis of Microbiota
The paired-ends reads obtained from sequencing were first merged, filtered, and removed chimera using software FLASH v1.2.7 [34], Trimmomatic v0.33 [35], and UCHIME v4.2 [36], respectively, to obtain optimized data. Search software [37] was used to perform taxonomic annotation for OTUs (operational taxonomic units) on the basis of Silva (Bacteria) [38] and UNITE (fungi) [39] taxonomic database. The OTUs were assigned from sequences at ≥97% similarity threshold. Alpha diversity reflects the species complexity of individual samples and species diversity. Alpha diversity indices of samples, including Chao1 (species richness) and Shannon (species diversity), were evaluated by Mothur v1.30 software. Moreover, the linear discriminant analysis (LDA) effect size (LEfSe) [40] method was used to detect significantly different taxa (LDA score > 3.0) with differential abundance. The functions of the bacterial communities were predicted using PICRUSt2 software (v 2.3.0) [41] based on the KEGG database. The functional classification (guild) of fungal OTUs was determined through the use of FUNGuild software (v 1.0) [42]. To assess the microbial community differences between the two groups, Student’s t-test and the two-tailed Wilcoxon rank-sum test were employed for statistical analysis.
2.4. Extraction of Phloem Metabolites
Non-targeted LC-MS based metabolomics analysis was performed to examine the phloem metabolites from two distinct trunk types. In brief, 50 mg of sample was taken, and 1000 μL of the mixture was added (methanol: acetonitrile: water = 40:40:20 v/v/v, internal standard L-2-chlorophenylalanine 2 mg/L) and vortexed for 30 s. The mixture was ground and sonicated for 10 min each. After centrifugation and spin-drying, metabolites were resuspended with 160 μL of the extracting solution (acetonitrile: water = 50:50 v/v). Following centrifugation at 12,000 rpm for 15 min at 4 °C, 120 μL of the supernatant was carefully transferred into sample vials.
2.5. Metabolites Detection
Metabolite detection was carried out on the UHPLC-QTOF-MS analysis platform, comprising the Waters Acquity I-Class PLUS UHPLC and the Waters Xevo G2-XS QTof HRMS, at Biomarker Technologies Corporation (Beijing, China). Chromatographic separation was performed using an Acquity UPLC HSS T3 column (Waters, 100 mm × 2.1 mm, 1.8 μm). The mobile phases used for the analysis were A (water containing 0.1% formic acid) and B (acetonitrile with 0.1% formic acid). A gradient elution profile was employed with the following steps: 2% B from 0 to 0.25 min, a linear increase from 2% to 98% B between 0.25 and 10.0 min, 98% B maintained from 10.0 to 13.0 min, a return to 2% B from 13.0 to 13.1 min, and finally, 2% B held from 13.1 to 15.0 min. The injection volume was 1 μL, and the flow rate was set to 0.4 mL/min. Data acquisition occurred at a rate of 5 spectra per second. Metabolite detection was carried out using capillary voltages of 2.0 kV in positive ion mode and 1.5 kV in negative ion mode. The MS data acquisition parameters were configured as follows: cone voltage set to 30 V, ion source maintained at a temperature of 150 °C and desolvent gas heated to 500 °C. The backflush gas flow rate was set to 50 L/h, while the desolventizing gas flow was set to 800 L/h.
2.6. Statistical Analysis of Metabolomic Data
The raw data collection was performed with MassLynx v4.2 and processed using Progenesis QI software to perform peak alignment, peak picking, etc., for each metabolite. In August 2020, to confirm the accurate qualitative and relative quantitative results of specific metabolites, peaks were matched with the METLIN (http://metlin.scripps.edu/) database and self-compiled database (Biomarker Technologies Corporation, Beijing, China) to obtain the accurate qualitative and relative quantitative results. Meanwhile, theoretical fragment identification is carried out, and the mass deviation is within 100 ppm.
To confirm the identities, these metabolites were annotated using free online databases, including KEGG (https://www.kegg.jp/), HMDB (https://hmdb.ca), and Lipidmaps database (http://www.lipidmaps.org). Principal component analysis (PCA), carried out using R software (v3.3.2), was used to preliminarily understand the clustering trend for the multidimensional data and give a comprehensive view of the variation degree among samples within the group. Furthermore, a t-test was performed to calculate the statistical significance (p value) of selected compounds between straight and twisted trunks of P. yunnanensis according to their contents. Combining the p < 0.05 and |log2(FC)| ≥ 1.5 were identified as metabolites showing significant differences. To visualize and identify significant changes, volcano plots were employed to further provide visualization of up- and down-regulated differential metabolites. In the differential metabolites analyses, the R software (v3.3.2) was used to obtain the heatmap. Ultimately, the biological roles of these metabolites and their associated metabolic pathways were explored through the KEGG database.
2.7. Correlation Between Microbiota and Metabolome of P. yunnanensis
To further analyze the correlations between microorganisms and metabolites in the phloem, Spearman’s correlation analysis and visual presentation of their correlations were performed by psych and heat map package in R. A value of p < 0.05 was regarded as statistically significant.
3. Results
3.1. Sequencing and Phloem Microbial Diversity Analysis
We used a high-throughput sequencing technique to detect the phloem microbial diversity. After that, we analyze the differences in microbial species between straight and twisted trunks of P. yunnanensis. A total of 4,615,591 raw reads were obtained based on sequencing analysis of the V3-V4 region of the 16S rDNA gene in all 30 samples, and an average of 142,756 effective reads were obtained per sample after quality control procedures, while 10,652,856 raw reads and 352,914 effective reads were obtained by sequencing analysis of the ITS1. These qualified sequences were clustered into bacterial 2383 and fungal 1373 OTUs, varying from 470 to 1060 OTUs per sample (Supplementary Table S2). Furthermore, a total of 103 and 74 distinct bacterial OTUs were identified in the straight-trunk and twisted-trunk groups, respectively, and 2206 bacterial OTUs were common in both groups; 22 and 12 unique fungal OTUs and 1339 fungal OTUs were common in both groups (Figure 1A,B). The analysis of the estimated phloem bacterial (Shannon, 5.31 ± 0.09 vs. 5.30 ± 0.10, p > 0.05; Chao1, 1158.02 ± 33.07 vs. 1186.20 ± 23.86, p > 0.05) and fungal alpha-diversity indices (Shannon, 4.38 ± 0.12 vs. 4.23 ± 0.17, p > 0.05; Chao1, 696.35 ± 14.35 vs. 732.67 ± 15.26, p > 0.05) revealed that straight and twisted-trunk groups harbored similar diversity and richness (Figure 1C,D). Moreover, beta-diversity analysis was used to reveal the phloem microbiota structure. The results of PCoA based on binary_jaccard indicated that dots from both groups were not separated (Figure 1E,F), demonstrating that phloem microbial structure was not strongly influenced by trunk type.
3.2. Community Composition of Phloem Bacterial and Fungal Communities
The total bacterial OTUs were annotated for subsequent analyses, including 31 phyla, 68 classes, 167 orders, 298 families, and 715 genera (Supplementary Table S3) of phloem bacteria. We evaluated the taxonomic distributions across various classification levels. (Figure 2A–C). The findings revealed that Proteobacteria, Firmicutes, and Bacteroidetes were the most prevalent bacterial phyla in the phloem of P. yunnanensis. We also used statistical analysis to distinguish the differential communities (Supplementary Figure S1A). The straight-trunk group was compared with the twisted-trunk group. The relative abundance of the phylum Tenericutes (0.09% and 0.20%, respectively; p < 0.05) is considerably greater in the twisted-trunk group than in the straight-trunk group. In addition, the phylum Elusimicrobia was only detected in the twisted-trunk group, accounting for less than 0.1% of the bacterial community. At the family level, Pseudomonadaceae (0.77% and 2.25%, respectively; p < 0.05), one of the top 10 families of bacteria, was significantly increased in the twisted trunk of P. yunnanensis. Rhodocyclaceae (0.51% and 0.22%, respectively; p < 0.05) was significantly more increased in the straight trunk than the twisted trunk of P. yunnanensis. At the genus level, the abundance rate of Craurococcus (0.01% and 0.23%, respectively; p < 0.05) was much higher in the twisted-trunk group than in the straight-trunk group, while Dechloromonas (0.15% and 0.02%, respectively; p < 0.05) was significantly higher in the straight-trunk group. Pseudomonas (0.77% and 2.25%, respectively; p < 0.05) significantly differed between the straight and twisted trunks of P. yunnanensis. Compositional analysis by LEfSe revealed that twisted-trunk types were enriched with the genera Craurococcus and Pseudomonas, whereas there was no predominant or mentionable microbiota in the straight-trunk group at the genus level (Figure 3A,C).
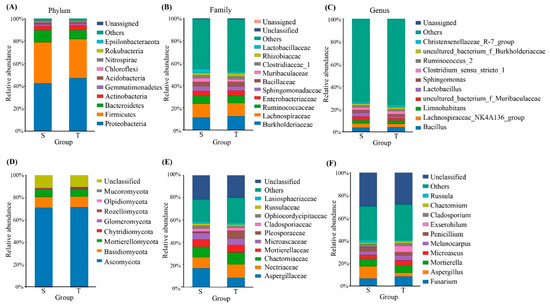
Figure 2.
The taxonomic structure and composition of bacterial and fungal communities at various taxonomic ranks, including phylum, family, and genus. (A), (B), and (C) represent the phylum, family, and genus levels of bacteria, respectively. (D), (E), and (F) represent the phylum, family, and genus levels of fungi, respectively. S: Straight-trunk type of P. yunnanensis; T: Twisted-trunk type of P. yunnanensis.
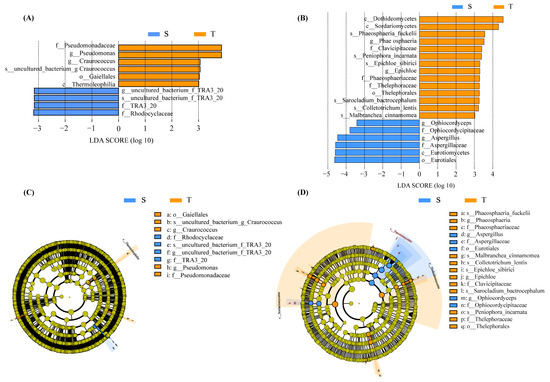
Figure 3.
Differential biomarkers associated with trunk type. The microbial biomarkers in different trunk types of P. yunnanensis for bacteria (A) and fungi (B,C) The varying bacterial taxa associated with different trunk types of P. yunnanensis. (D) The different taxon between different trunk types of P. yunnanensis in fungi. The phylogenetic distribution of differential taxa was visualized by cladogram. The lowercase letters p, c, o, f, g, and s preceding the symbol “-” denote the phylum, class, order, family, genus, and species, respectively. The circles inside and outside of the cladogram indicate the phylum, class, order, family, and genus levels. Coloring: the taxa that do not show significant differences are highlighted in yellow, with orange representing the twisted-trunk group and blue indicating the straight-trunk group. The yellow circles represent the taxa with no significant differences. S: Straight-trunk type of P. yunnanensis; T: Twisted-trunk type of P. yunnanensis.
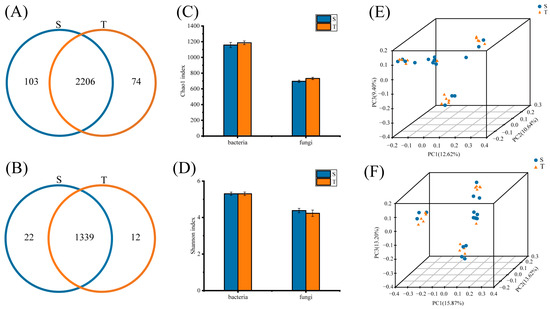
Figure 1.
The preliminary comparison of the diversity of phloem microbiota in P. yunnanensis with straight-trunk and twisted-trunk types. (A) The Venn diagram illustrates the distribution of bacterial OTUs, highlighting both shared and unique ones between the two groups. (B) Venn diagram shows the number of shared and unique fungal OTUs in both groups. Microbial alpha diversity was estimated by Chao1 (C) and Shannon (D). Principal coordinate analysis (PCoA) according to the binary_jaccard bacterial (E) and fungal (F) community distances between straight-trunk and twisted-trunk types.
We also explored the fungal community by ITS sequencing with the same analysis method used for bacterial 16S rRNA sequencing. Three dominant fungal phyla with a relative abundance of more than 1% were identified in both groups (Figure 2D). These three fungi phyla, including the phylum Ascomycota (70.80% and 71.29%, respectively), Basidiomycota (9.51% and 9.42%, respectively) and Mortierellomycota (6.61% and 7.01%, respectively), accounting for 86.92% and 87.72%, respectively, in the straight-trunk and twisted-trunk groups. At the family level (Figure 2E), Aspergillaceae (16.11% and 8.11%, respectively, p < 0.05) and Ophiocordycipitaceae (2.52% and 1.49%, respectively, p < 0.05) were much higher in the straight-trunk group than in the twisted-trunk group (Supplementary Figure S1B). At the genus level (Figure 2F), in addition, the most abundant genera in the straight-trunk group were Aspergillus (9.84% and 3.10%, respectively), Fusarium (6.67% and 8.44%, respectively), and Mortierella (6.53% and 6.81%, respectively). The relative abundance of Aspergillus and Ophiocordyceps was higher in straight-trunked P. yunnanensis than in twisted-trunked P. yunnanensis (6.53% and 6.81%, respectively) (p < 0.05). However, based on LEfSe analysis, a total of four and six differentially fungal abundant taxonomic clades were identified in straight-trunk and twisted-trunk groups respectively (Figure 3B,D). The straight-trunk types were enriched with the Phaeosphaeria and Epichloë, whereas the predominant microbiota in the twisted-trunk group consisted of Ophiocordyceps and Aspergillus, which represented differential species across all treatments.
3.3. Microbial Functional Group Analysis
To obtain more complete information on the functions of the phloem microorganisms of P. yunnanensis, the prediction tool PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) was used to determine the functional characteristics of the bacterial communities in the straight-trunk and twisted-trunk groups. At the second level, putative functional determinations revealed little difference. The processes involved in amino acid metabolism, membrane transport, energy production, and the handling of cofactors and vitamins were the most abundant predicted KEGG pathways in all types of P. yunnanensis (Supplementary Figure S2A).
The fungal functional group (guild) of the OTUs was inferred using FUNGuild. Regarding the guild mode of the fungal community (Supplementary Figure S2B), the twisted-trunk types showed a higher abundance of the plant pathogen and lower abundance of ectomycorrhizal than the straight-trunk types; however, these differences were not significant. In addition, many uncharacterized saprotrophs were enriched by KEGG in both tissue types.
3.4. Metabolomics Analyses Between Two Different Trunk Types of P. yunnanensis
To determine the features and differences in phloem metabolite of straight-trunk and twisted-trunk types, a total of 30 samples were analyzed using metabolomics on the UPLC-MS/MS platform in both positive and negative ion modes. The PCA in the negative mode showed a relatively obvious separation of the types among the groups with straight trunks and those with twisted trunks (Figure 4A,B). In the positive ionization mode, 1668 metabolites were detected, while in the negative ionization mode, 779 metabolites were detected. (Figure 4C,D, Supplementary Table S4). Fractional metabolites were further annotated to specific categories via search against the human metabolome database (HMDB) (Supplementary Table S5), Kyoto Encyclopedia of Genes and Genomes (KEGG) (Supplementary Table S6), and Lipidmaps database (Supplementary Table S7).
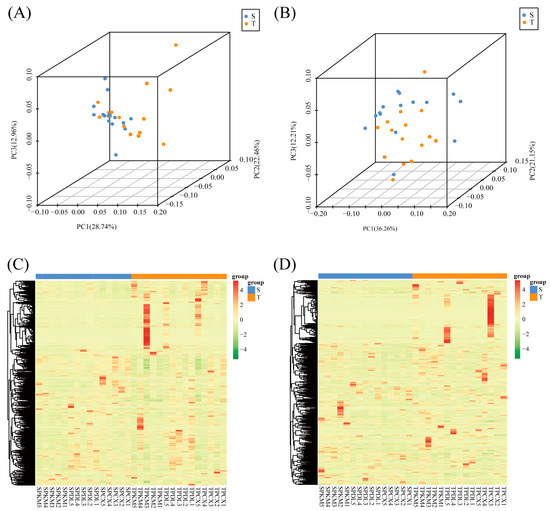
Figure 4.
Multivariate statistical analysis of identified metabolites between the straight-trunk and twisted-trunk types of P. yunnanensis. (A,B) Principal component analysis (PCA) score plots in positive ion and negative ion modes. (C,D) Heat map depicting metabolites in both positive and negative ion modes. The horizontal axis represents the samples, while the vertical axis indicates the metabolites. The color blocks reflect the normalized relative abundances of the metabolites. Straight-trunk type of P. yunnanensis; T: Twisted-trunk type of P. yunnanensis.
To identify the key metabolites responsible for the difference, p value < 0.05 combined with the |log2 (FC)| ≥ 1.5 results were considered to be differential metabolites in the straight-trunk and twisted-trunk types of P. yunnanensis (Supplementary Table S8). In total, we identified 162 and 65 differential metabolites in positive and negative ion modes, respectively. Compared to the straight-trunk group, 84 metabolites were significantly increased in the twisted-trunk group, and 78 metabolites were significantly decreased in positive ion mode (Figure 5A). 52 metabolites were significantly increased and 13 metabolites were significantly decreased in negative ion mode (Figure 5B). In particular, the content ofL-histidine, L-tryptophan, baccatin III, carnitine, geldanamycin and echinacoside were significantly increased in the twisted-trunk group, whereas 5(S)-HETE, trans-Zeatin, gluconolactone, D-mannose, phytosphingosine, and sodium gluconate were evidently reduced. Following this, the metabolites showing significant differences were analyzed for their associated pathways using MetaboAnalyst v4.0. The most prominent pathways enriched in both positive and negative modes are shown in Figure 5C,D, and Supplementary Table S9. Among enriched pathways, four pathways with significant differences were the beta-alanine metabolism, metabolism of Jasmonic acid and trans-Zeatin metabolism, linoleic acid metabolism, and pentose phosphate pathway, which involved 12 potential biomarkers: uracil, pantothenate, dehydromyodesmone, adenosine, trans-Zeatin, Jasmonic acid, dihomo-gamma-linolenic acid, 13-OxoODE, sodium gluconate, 2-dehydro-3-deoxy-D-gluconate and 1-Stearoyl-2-hydroxy-sn-glycero-3-phosphocholine.
3.5. Co-Occurrence Network and Metabolites Correlated with Keystone Genus
To further explore the underlying connection between microbiota and metabolites, we chose to analyze differential microorganisms (genus level) and metabolites (Supplementary Table S10) to generate a heatmap depicting the interactions between metabolites and microbes based on Spearman’s correlation coefficient. As shown in Supplementary Figure S3 and Supplementary Table S11, there were different degrees of correlation between microorganisms and metabolites. Among them, Ile Asp Lys Asn, Carnitine and Craurococcus; L-Tryptophan and Pseudomonas; and 4′,5-Dihydroxy-7-methoxyflavanone and Chryseobacterium showed strong positive correlations (correlation coefficient > 0.7).

Figure 5.
Differential metabolites and metabolic pathways associated with trunk type of P. yunnanensis. (A,B) Volcano plots depicting the differential metabolites in both positive and negative ion modes were generated. The red dots in the graph represent upward adjustments, the blue dots downward adjustments, and the size of the origin represents the difference multiplier. (C,D) Enrichment analysis of metabolic pathways in positive ion and negative ion modes.
4. Discussion
The characteristics of wood hold considerable importance for the economic value of wood-based products, as well as for sustainable development of forest resources [43]. Trunk twisting occurs in many tree species and is particularly prominent and pronounced in coniferous species, including but not limited to P. sylvestris [44], P. radiata [45] and P. abies [46]. Endophytes that engage with host plants are recognized for promoting the synthesis of a wide range of metabolites, contributing to their diversity and abundance in the plants [47]. However, because the mechanism of action of metabolites and microorganisms on different trunk morphogenesis of P. yunnanensis is still unclear, it is essential to systematically study the effects of microorganisms and metabolism on trunk morphology and their correlation to analyze the occurrence and development of trunk types. In this study, high-throughput sequencing and UHPLC-QTOF-MS were used to evaluate differences in bacterial and fungal communities as well as metabolites in P. yunnanensis phloem as a result of the straight-trunk and twisted-trunk types.
Microorganisms form complex microbial communities, regulate nutrient cycling, and affect plant growth and ecosystem sustainability [12,48]. The microbial communities linked to plant hosts are likely influenced by numerous environmental and host-specific factors, such as plant phenotype and ecological niche [49]. This study demonstrates that the phloem microbiome of P. yunnanensis does not differ significantly between the types with straight trunks and those with twisted trunks, and the compositions of bacterial and fungal communities were also similar in the straight-trunk type compared to the twisted-trunk type. This corroborates with previous findings that there is a strong diversity of rhizosphere bacterial and fungal communities in both trunk types [50]. Despite this, we found some differences in microorganisms between the straight-trunk and twisted-trunk phloem of P. yunnanensis. Our findings indicate that the bacterial microbiome associated with twisted trunks exhibited a marginally greater number of OTUs and higher alpha diversity compared to the straight-trunk group, although this difference was not significant, as observed in a previous study in rhizosphere microbiome [50]. Additionally, this study found that the microbial communities linked to P. yunnanensis were affected by the host’s niche, with notable differences observed between the rhizosphere soil and phloem. We found that bacterial OTU numbers, Chao 1, and Shannon index in the phloem were significantly higher than those in the rhizosphere soil (Supplementary Figure S4A). Conversely, fungal OTU numbers and Chao 1 index were significantly lower than those in the rhizosphere soil. However, no significant difference was shown in different niches for the Shannon index (Supplementary Figure S4B). In addition, the Venn diagram shows the quantity of common and distinct bacterial and fungal microbiome groups in both niches (Supplementary Figure S4C). At the genus level, two common dominant bacterial groups and two common dominant fungal groups (relative abundance ≥ 1.0%) were detected in both niches. Among them, Sphingomonas and Penicillium were more abundant in phloem than in rhizosphere soil, whereas Russula was significantly depleted in phloem than in rhizosphere soil (Supplementary Figure S4D,E). Moreover, beta-diversity analysis indicated that the dots from both groups were separated, revealing significant differences in the community composition of both bacteria and fungi (Supplementary Figure S4F,G). The results of the PERMANOVA analysis indicated that niches accounted for 18.76% of the variation in bacterial communities (p < 0.01) and 11.91% in fungal communities (p < 0.01) (Supplementary Table S12). It also demonstrated that microbial structure in P. yunnanensis was strongly influenced by niches. We speculated that this was probably due to the differences in various microbial habitats (i.e., phloem contacts nutrients, while soil contacts air).
In addition, we also observed significant variations in some bacteria and fungi, which may play important roles in the development of the trunk. Pseudomonas is a well-characterized pathogen [51]. However, the importance of the genus Pseudomonas in plants has been reported in many metagenomic studies. Due to its potential pro-growth effects and possible antagonistic effects on plant pathogens, it has been studied as a model for plant–microbe interactions [52,53,54,55]. For example, the endophytic bacteria Pseudomonas isolated from the stem of the Sedum alfredii, which produces phytohormone, promoted the growth of the host [56]. The study additionally discovered numerous fungal pathogens that were previously known as the pathogenic microorganism responsible for white spot disease in corn, such as Phaeosphaeria [57], which is also the main composition of fungal communities in living needles of P. heldreichi [58]. Epichloë live partly or wholly in plants and have the effects of improving drought resistance, cold resistance, disease and insect resistance and promoting growth of host plants [59,60]. At present, it has a wide range of application value in planting and animal husbandry and attracted much attention as a new microbial resource [61]. Ophiocordyceps is a classic entomopathogenic fungus [62]. However, according to the latest report, this fungus can also live inside plant tissues and exhibits a complete set of genes that encode plant cell wall-degrading carbohydrate-active enzymes (CAZymes) [63]. The cell wall was previously shown to be intimately related to the formation of a spiral grain [64]. Endophytic Aspergillus has demonstrated a rich diversity of secondary metabolites, including alkaloids, sterols, phenylenones, xanthones, cytochalasin, and butenolactones, exhibiting a large variety of biological activities, including cytotoxic activities, antifungal, anti-inflammatory, anti-cancer and antibacterial, which ameliorate the growth of their hosts [65]. It has a great interest in exploitation in medicine, agriculture, and industry [66]. Lastly, and most notably, the presence of the Dechloromonas species, which have the ability to remove nitrogen and phosphorus, may be an important factor contributing to this difference [67]. The straight-trunk type is significantly higher than the twisted-trunk type. In the straight-trunk type, the phosphorus degradation efficiency is higher, while in the twisted-trunk type, phosphorus is not degraded in a timely manner, leading to growth differences between the two types. The growth pattern differences in the twisted-trunk type, along with environmental factors, contribute to the significant differences between the straight-trunk and twisted-trunk types. According to our previous study, the effective phosphorus content in the rhizosphere soil of different trunk types of P. yunnanensis differed significantly, and the rhizosphere was also enriched with a multitude of microorganisms related to phosphorus metabolism and catabolism [50]. Perhaps in the next step, we could pay more attention to the role of phosphorus in the growth and development of P. yunnanensis. The microbes mentioned above play a crucial role in maintaining the health of their host. We speculated that the altering of these microbes may further induce the trunk type of P. yunnanensis.
Endophytes can secrete or induce plants to produce a diverse array of metabolites, each with distinct biological functions, crucial for mediating communication between microbiota and host cells [68]. In addition, these metabolites can also act on other tissues through systemic circulations, thereby more systematically regulating host health and phenotype [69]. Thus, we conducted untargeted metabolomics to explore the differences in phloem metabolites in P. yunnanensis with different trunk types. The results showed that the differential metabolites are mainly enriched in 4 metabolic pathways, including beta-alanine metabolism, metabolism of Jasmonic acid and trans-Zeatin metabolism, linoleic acid metabolism, and pentose phosphate pathway. These metabolic pathways, along with the 12 metabolites they involve, could be crucial in influencing the development of the trunk type in P. yunnanensis. Beta-alanine is a nonproteinogenic beta-amino acid that occurs in all living organisms [70]. Jasmonic acid, we detected a marked alteration in the beta-alanine metabolism pathway, along with a noticeable reduction in uracil and a significant increase in pantothenate levels. It has been reported that uracil, one of the four bases that make up RNA in cells, is often added to the culture medium as a microbial growth factor [71]. Additionally, uracil could promote the growth of bacteria and increase the viable count of the strain [72]. Pantothenate, commonly referred to as vitamin B5, plays an essential role as a key precursor in the biosynthetic pathway of coenzyme A (CoA) as well as carrier proteins. Additionally, it plays a role in the release of CoA from cells [73,74].
Plant hormones are essential regulators of various processes in plant growth, development, reproduction, and survival of forest trees [75]. In our work, plant hormone signal transduction was found to be a metabolic pathway for differential metabolite enrichment. Abnormal plant morphogenesis can result from disruptions in auxin signaling within the plant system [76]. Therefore, we proposed that auxin signaling influences the formation of the twisted-trunk type during the P. yunnanensis development, consistent with the idea that the presence of auxin promotes the development of spiral grain in trees such as Picea abies and Abies balsamea [77]. Additionally, several phytohormones and their derivatives are also known to be implicated in plant defense signaling pathways that induce resistance in response to environmental stress and pathogens, e.g., Jasmonic acid, as a signal transmitter, promoted the accumulation of lignin in rice in interaction with Rhizoctonia solani [78]. Over the past years, cytokinins have been shown to be crucial in the defense mechanisms of plant immunity, such as trans-Zeatin [79,80]. Trans-Zeatin is an active cytokinin in all plant species [81] and functions mainly as a cytokinin. In the last few years, trans-Zeatin has played an important role in plant immunity [79,80]. Stress treatments can induce changes in phytohormone levels that enhance plant resistance, such as heat stress this is usually accompanied by an increase in Jasmonic acid content and a decrease in trans-Zeatin content, as in our findings [82]. Jasmonic acid is a key player in plant defense, but its function shows variability in the face of different types of pathogens. Instead, it was utilized by pathogens in response to living and semi-living nutritive pathogens [83]. In the intercropping of dead-body nutrient-type pathogens, increased JA content activates the expression of JA signaling pathway and defense-related genes, thus increasing plant resistance to dead-body nutrient-type pathogens inhibiting the catabolism of JA, thus accumulating high levels of JA in the plant and enhancing resistance [84,85]. In the mutualism of living and semi-living trophic pathogens, living trophic pathogens can activate the JA pathway and promote their infestation in a number of ways, and pathogens can activate the JA pathway and interfere with plant immunity by synthesizing Jasmonic acid analogs [86]. In addition, Pseudomonas syringae is able to synthesize coronin (COR), a Jasmonic acid analog that binds to the Jasmonic acid receptor COI1, leading to the degradation of JAZ proteins and activation of the JA pathway [87]. Jasmonic acid has a high potential for exploitation in interactions with microorganisms.
Therefore, we speculate that the twisted-trunk type P. yunnanensis may have been subjected to more stress in comparison with the straight-trunk type. Lipids, one predominant category of pine, are essential components of the plasma membrane, which could enhance the biotic and abiotic stress adaptation of plants [88]. Significant changes were also shown in linoleic acid metabolism pathways and reductions in several associated metabolites such as dihomo-gamma-linolenic acid and 13-OxoODE. According to current reports, linoleic acid plays an important role in plant immunity to pathogens; for example, it can be deposited as cuticle monomers and oligomers to strengthen the cuticle and act as a barrier to entry of pathogens [89]. Additionally, we also found significant differences in the pentose phosphate pathway between the two different trunk types, with sodium gluconate and phenolic compound 2-methoxyhydroquinonebeing enriched in the straight-trunk type and 2-dehydro-3-deoxy-D-gluconate being enriched in the twisted-trunk type. Sodium gluconate is a small-molecule organic compound known for its excellent ability to penetrate, retain moisture, and adhere effectively [90]. Research has shown that sodium gluconate supports seedling development, boosts resistance to drought, and elevates chlorophyll levels across various plant studies [90,91,92]. The change in phenolic compound content is usually associated with the disease resistance of trees, which may be the activation of defense-related genes, thus increasing the disease resistance of plants [93,94].
In this study, we also discovered a relationship or interaction between the phloem microbiota and metabolites, which enhanced our comprehension of the functions of endophytic microbiota and metabolites, such as Collimonads and pantothenate, Aspergillus and phytosphingosine, Chryseobacterium and berberine, Craurococcus and carnitine, Jasmonic acid as well as Pseudomonas and L-Tryptophan. Among them, Collimonads are part of the Oxalobacteraceae family, which is widely recognized for the antifungal properties of its members [95]. Up to now, six strains (C. fungivorans, C. pratensis, C. arenae, C. anthrihumi, C. silvisoli, and C. humicola) have been described [96,97]. Endophytic bacterial Chryseobacterium is a kind of bacteria that not only produces bioactive compounds with antimicrobial activity against bacterial pathogens but also promotes growth in plants [98,99]. Studies have reported that phytosphingosine and berberine have an effective inhibition of bacteria and fungi [100,101]. Carnitine, a kind of quaternary ammonium compound (QAC), can be used by Pseudomonas as osmoprotectants and nutrient source [102]. Studies suggest that a solitary QAC released during the germination process may play a key role in promoting bacterial proliferation as seeds undergo colonization [103]. Additionally, carnitine is produced during the germination of bean seeds, particularly at the stages of radicle emergence and elongation, where it plays a significant role in promoting the growth of the sprouting seeds [103]. Ile Asp Lys Asn may play an important role as a basic amino acid in anabolic life metabolism, substance synthesis. Carnitine is present in plants and some fungi, and it is worth mentioning that it may be more abundant in mushrooms compared to plants, and L-carnitine regulates the levels of acyl-CoA and CoA in mitochondria and cytoplasm [11]. Accordingly, we speculate that the activities of Craurococcus are more relevant to Ile Asp Lys Asn and Carnitine content or that they play some possible role in the morphogenesis of twisted-trunk types. The growth of Pseudomonas acidophilus in the presence of tryptophan leads to the formation of a tryptophan transport system, which affects enzyme activity and product production [104]. Interestingly, we also found between 4′,5-Dihydroxy-7-methoxyflavanone, and Chryseobacterium. 4′,5-Dihydroxy-7-methoxyflavanone is found in plants such as motherwort and belongs to the class of flavonoids, which may play some important roles in antimicrobial activity [105]. Chryseobacterium has the potential to help plants dissolve insoluble, soluble phosphorus to increase its availability to plants, and its genome also carries important genes related to photosynthesis, growth, and ethylene degradation in plants, which may have the function of promoting plant growth and resisting adverse stresses [106]. Based on the above results, we speculated that the interaction between microbiota and metabolites may be one of the effects in the development of the trunk type of P. yunnanensis.
In fact, this study is a further step in our research on the twisted-trunk type of P. yunnanensis. Previously, we have analyzed the rhizosphere microbial diversity and composition between the straight-trunk and twisted-trunk types in detail [7]. In this study, we focused on the differences in phloem endophytic microorganisms and metabolites between the two different trunk types of P. yunnanensis. We consider that the interrelationships of microorganisms and metabolites are extremely complex and variable because the interrelationships involve so many kinds of microorganisms and metabolites and the relationships may change at any time due to a wide range of biotic and abiotic influences. What is certain, however, is that the study has also clarified some research directions for us. (1) Hormones have an important role to play in trunk form, but how does this work? Additionally, what role do microorganisms play in this process? (2) The effective phosphorus content in the rhizosphere soils of P. yunnanensis with different trunk shapes varies significantly, and there are numerous microorganisms related to phosphorus metabolism in the rhizosphere microorganisms. In this study, we have also found differential microorganisms related to phosphorus metabolism as well as differential metabolites. Thus, what is the special role of phosphorus in the growth and development of P. yunnanensis, and what influences do microorganisms have in phosphorus metabolism and transport? Undoubtedly, these are the key contents that we will focus on and research in the future.
5. Conclusions
In summary, our results suggested that both the abnormalities of microbiota and metabolism between the straight-trunk and twisted-trunk types were closely linked with the twisted-trunk occurrence and development of P. yunnanensis. We found some possible relationships between trunk types and bacterial genera, such as Pseudomonas, Dechloromonas, and Craurococcus bacteria, as well as the fungus taxa Phaeosphaeria, Aspergillus and Epichloë, providing the basis for future studies on the improvement and prevention of the twisted-trunk type of P. yunnanensis. Metabolomics analysis uncovered 162 and 65 differentially metabolites in positive and negative ion modes, respectively, between the straight-trunk and twisted-trunk types. Four metabolic pathways showed significant differences, including beta-alanine metabolism, metabolism of Jasmonic acid and trans-Zeatin metabolism in plant hormone signal transduction, linoleic acid metabolism, and pentose phosphate pathway. Plant hormones, phenolic compound, and lipids and their derivatives were the major different metabolites between the two different trunk types, which were found to form dense interactions with other metabolites and microorganisms, suggesting their crucial roles in the formation of twisted-trunk P. yunnanensis. However, the mechanism of the interaction between these endophytes, metabolites, and their hosts still needs a lot of research and repeated verification, due to the lack of research on the interaction between endophytes and differential metabolites, as well as the research on the metabolic pathways of different metabolites of endophytes in their hosts.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/f16040574/s1, Supplementary Table S1. Sample information and geographic location; Supplementary Table S2. Number of processed sequencing reads and estimates for the diversity of each microbial community; Supplementary Table S3. Relative abundance of microorganisms (bacteria and fungi) from 30 samples; Supplementary Table S4. All metabolites from 30 samples were detected; Supplementary Table S5. Human metabolome database (HMDB) assignment of identified metabolites; Supplementary Table S6. Kyoto Encyclopedia of Genes and Genomes database (KEGG) assignments of identified metabolites; Supplementary Table S7. Lipidmaps assignments of identified metabolites; Supplementary Table S8. All differential metabolites from 30 samples were detected; Supplementary Table S9. Kyoto Encyclopedia of Genes and Genomes database (KEGG) assignments of different metabolites; Supplementary Table S10. All differential metabolites from 30 samples were detected for correlation analysis; Supplementary Table S11. Correlations coefficients between phlome microbiome and metabolites; Supplementary Table S12. PERMANOVAs of the influence of niches on the microbial communities (Supplementary_data). Supplementary Figure S1. Statistical analysis of differential bacteria (A) and fungi (B) between straight and twisted trunk types of P. yunnanensis at the phylum, families, and genus levels. S: Straight trunk type of P. yunnanensis; Supplementary Figure S2. Relative abundance of predicted phloem bacterial (A) and fungal (B) functions in straight and twisted trunk by PICRUSt2 using KEGG Orthologs (hierarchy level 2) and FUNGuild based on the OTUs obtained from ITS rRNA sequencing respectivel; Supplementary Figure S3. Correlation analysis of phloem microorganisms and metabolites of P. yunnanensis; Supplementary Figure S4. Differences of bacteria and fungi between phlome and rhizosphere soil of P. yunnanensi (Supplementary_Material).
Author Contributions
Conceptualization, C.H. and P.L.; methodology, H.L.; software, P.G.; validation, Y.Q., Y.L. and C.W.; formal analysis, X.Z.; investigation, J.F.; resources, X.Z.; data curation, P.L.; writing—original draft preparation, H.L. and J.F.; writing—review and editing, W.Y. and D.Z.; visualization, J.F.; supervision, P.L.; project administration, D.Z.; funding acquisition, D.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Youth Talents Special Project of Yunnan Province “Xingdian Talents Support Program” (XDYC-QNRC-2022-0232).
Data Availability Statement
The data presented in this study are deposited in the NCBI repository, accession numbers are PRJNA892766 and PRJNA892770.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Liu, D.; Du, K.C.; Wang, A.D.; Meng, D.L.; Li, J.L. Secondary Metabolites from the Fresh Leaves of Pinus yunnanensis Franch. Chem. Biodivers. 2022, 19, e202100707. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zheng, W.; Han, M.; Su, J. Progress in Genetic Improvement of Pinus yunnanensis and Analysis of Its Breeding Strategies. West. J. For. Sci. 2010, 39, 104–110. [Google Scholar] [CrossRef]
- He, C.; Wu, Z.; Shen, D.; Gan, P.; Zhou, A.; Zong, D. Study on the Genetic Basis of Different Stem Forms of Pinus yunnanensis Based on SRAP Markers. J. Southwest For. Univ. Nat. Sci. 2021, 41, 1–10+197. Available online: https://kns.cnki.net/kcms2/article/abstract?v=Zw74qSZOFgjJZlaQOtHk1mM5ynHUuRjSlZLhnAoaU0A14-75uh1ETfH3Y1GjlesUOykrJ4B9snzbdAz9q5jShOuPTHmteQ1qLmCu81AUVQF7RpxKMhNRCpsUimI4XoflgDdfOC62DUkTlsiN0IefKyFS-V2uMSZ4BPhqYRuXtWjCYRzjghGL6ztGFOKqfEIm_g4FqZcvZ48=&uniplatform=NZKPT&language=CHS (accessed on 20 March 2025).
- Hong, Y.; Song, G.; Huang, R.; Jiang, H.-Q. A Preliminary Study on Genetic Variation and Relationships of Pinus yunnanensis and Its Closely Related Species. J. Integr. Plant Biol. 2000, 42, 107–110. [Google Scholar]
- Luo, L.; Xu, L. A study on the effect of oblique grain on the physical and mechanical properties of Pinus yunnanensis wood. For. Sci. 1983, 382–389+448. Available online: https://kns.cnki.net/kcms2/article/abstract?v=Zw74qSZOFgi0459KVPkuXKUSJ-57m0XU86g8C_uy6bzf-jfAxFvhWkq7RkSSqNLU-38DImPTiOZlQHAzfX9MLLY1kZsUDPmcD4F124Tr6K3y-nrZX6URGlMyvTfJE_fxgArPvpy28ZnbaBH6G7jksqTm7eKrVjPZzsufAGHvG3pW_Q8Uokve9mQMRLU_8ZYy&uniplatform=NZKPT&language=CHS (accessed on 20 March 2025).
- Luo, L.; Han, M.; Zheng, W.; Su, J.; Li, W.; Zheng, S.; Gong, J. The Causes and Classification of Low-Quality and Low-Efficiency Forests of Pinus yunnanensis. West. For. Sci. 2009, 38, 94–99. [Google Scholar] [CrossRef]
- Zhou, A.; Zong, D.; Luo, J.; Shen, D.; He, R.; Tian, B.; Xu, Y.; He, C. AFLP Analysis on Genetic Variation of Stem Forms in Pinus yunnanensis. Mol. Plant Breed. 2016, 14, 186–194. [Google Scholar] [CrossRef]
- Wang, C.; Wang, J.; Jiang, H.-Q. Morphological Characteristics of Stem of Pinus yunnanensis and Its Related Species in Different Habitats. J. West China For. Sci. 2009, 38, 23–27+125. [Google Scholar] [CrossRef]
- Kumar, A.; Verma, J.P. Does plant—Microbe interaction confer stress tolerance in plants: A review? Microbiol. Res. 2018, 207, 41–52. [Google Scholar] [CrossRef]
- Sharma, M.; Sudheer, S.; Usmani, Z.; Rani, R.; Gupta, P. Deciphering the Omics of Plant-Microbe Interaction: Perspectives and New Insights. Curr. Genom. 2020, 21, 343–362. [Google Scholar] [CrossRef]
- Rospond, B.; Chłopicka, J. Funkcje biologiczne L-karnityny i jej zawartość w wybranych produktach spozywczych [The biological function of L-carnitine and its content in the particular food examples]. Przegl. Lek. 2013, 70, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Zhu, J.; Zhao, X.; Shi, J.; Jiang, C.; Shao, D. Beneficial effects of endophytic fungi colonization on plants. Appl. Microbiol. Biotechnol. 2019, 103, 3327–3340. [Google Scholar] [CrossRef]
- De-La-Pena, C.; Loyola-Vargas, V.M. Biotic interactions in the rhizosphere: A diverse cooperative enterprise for plant productivity. Plant Physiol. 2014, 166, 701–719. [Google Scholar] [CrossRef]
- Rosier, A.; Bishnoi, B.; Lakshmanan, V.; Sherrier, D.J.; Bais, H.P. A perspective on inter-kingdom signaling in plant-beneficial microbe interactions. Plant Mol. Biol. 2016, 90, 537–548. [Google Scholar] [CrossRef]
- Le Cocq, K.; Gurr, S.J.; Hirsch, P.R.; Mauchline, T.H. Exploitation of endophytes for sustainable agricultural intensification. Mol. Plant Pathol. 2017, 18, 469–473. [Google Scholar] [CrossRef]
- Nataraja, K.N.; Suryanarayanan, T.S.; Shaanker, R.U.; Senthil-Kumar, M.; Oelmüller, R. Plant–microbe interaction: Prospects for crop improvement and management. Plant Physiol. Rep. 2019, 24, 461–462. [Google Scholar] [CrossRef]
- Feussner, I.; Polle, A. What the transcriptome does not tell—Proteomics and metabolomics are closer to the plants’ patho-phenotype. Curr. Opin. Plant Biol. 2015, 26, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Sun, D.; Chen, L.; An, Y. Integrative analysis of the microbiome and metabolome in understanding the causes of sugarcane bitterness. Sci. Rep. 2021, 11, 6024. [Google Scholar] [CrossRef]
- Li, Y.-H.; Yang, Y.-Y.; Wang, Z.-G.; Chen, Z. Emerging Function of Ecotype-Specific Splicing in the Recruitment of Commensal Microbiome. Int. J. Mol. Sci. 2022, 23, 4860. [Google Scholar] [CrossRef]
- Kang, S.H.; Cho, H.S.; Cheong, H.; Choongmin, R.; Kim, J.H.; Park, S.H. Two bacterial entophytes eliciting both plant growth promotion and plant defense on pepper (Capsicum annuum L.). J. Microbiol. Biotechnol. 2007, 17, 96. [Google Scholar] [CrossRef]
- Scherling, C.; Kristina, U.; Ewald, D.; Wolfram, W. A metabolic signature of the beneficial interaction of the endophyte Paenibacillus sp. isolate and in vitro-grown poplar plants revealed by metabolomics. Mol. Plant-Microbe Interact. 2009, 22, 1032–1037. [Google Scholar] [CrossRef] [PubMed]
- Balmer, D.; De Papajewski, D.V.; Planchamp, C.; Glauser, G.; Mauch-Mani, B. Induced resistance in maize is based on organ-specific defence responses. Plant J. Cell Mol. Biol. 2013, 74, 213–225. [Google Scholar] [CrossRef]
- Ważny, R.; Rozpądek, P.; Domka, A.; Jędrzejczyk, R.J.; Nosek, M.; Hubalewska-Mazgaj, M.; Lichtscheidl, I.; Kidd, P.; Turnau, K. The effect of endophytic fungi on growth and nickel accumulation in Noccaea hyperaccumulators. Sci. Total Environ. 2021, 768, 144666. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar]
- Chong, J.; Xia, J. Computational Approaches for Integrative Analysis of the Metabolome and Microbiome. Metabolites 2017, 7, 62. [Google Scholar] [CrossRef]
- Mishra, S.; Priyanka; Sharma, S. Metabolomic Insights Into Endophyte-Derived Bioactive Compounds. Front. Microbiol. 2022, 13, 835931. [Google Scholar] [CrossRef]
- Heuberger, A.L.; Robison, F.M.; Lyons, S.M.A.; Broeckling, C.D.; Prenni, J.E. Evaluating plant immunity using mass spectrometry-based metabolomics workflows. Front. Plant Sci. 2014, 5, 291. [Google Scholar] [CrossRef]
- Wolf, R.T.S. Phloem Transport: Cellular Pathways and Molecular Trafficking. Annu. Rev. Plant Biol. 2009, 60, 207–221. [Google Scholar] [CrossRef]
- Lucas, W.J.; Groover, A.; Lichtenberger, R.; Furuta, K.; Yadav, S.-R.; Helariutta, Y.; He, X.-Q.; Fukuda, H.; Kang, J.; Brady, M.S.; et al. The plant vascular system: Evolution, development and functions. J. Integr. Plant Biol. 2013, 55, 294–388. [Google Scholar] [CrossRef]
- Yoo, S.C.; Chen, C.; Rojas, M.; Daimon, Y.; Ham, B.K.; Araki, T.; Lucas, W. Phloem long-distance delivery of FLOWERING LOCUS T (FT) to the apex. Plant J. 2013, 75, 456–468. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhang, C.X.; Chen, R.; He, S.Y. Challenging battles of plants with phloem-feeding insects and prokaryotic pathogens. Proc. Natl. Acad. Sci. USA 2019, 116, 23390–23397. [Google Scholar] [CrossRef] [PubMed]
- Miyashima, S.; Roszak, P.; Sevilem, I.; Toyokura, K.; Helariutta, Y. Mobile PEAR transcription factors integrate positional cues to prime cambial growth. Nature 2019, 565, 490–494. [Google Scholar] [CrossRef] [PubMed]
- Smetana, O.; Makila, R.; Lyu, M.; Amiryousefi, A.; Rodriguez, F.S.; Wu, M.F.; Sole-Gil, A.; Gavarron, M.L.; Siligato, R.; Miyashima, S. High levels of auxin signalling define the stem-cell organizer of the vascular cambium. Nature 2019, 565, 485–489. [Google Scholar] [CrossRef]
- Mago, T.; Salzberg, S.L. FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Khaleel, S.S.; Huang, H.; Wu, C.H. Software for pre-processing Illumina next-generation sequencing short read sequences. Source Code Biol. Med. 2014, 9, 8. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Elmar, P.; Christian, Q.; Katrin, K.; Fuchs, B.M.; Wolfgang, L.; Jrg, P.; Oliver, G.F. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef]
- Kõljalg, U.; Larsson, K.H.; Abarenkovenrik, K.; Nilsson, R.H.; Alexander, I.; Eberhardt, U.; Erland, S.; Høiland, K.; Kjøller, R.; Larsson, E.; et al. UNITE: A database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Nhytologist 2005, 166, 1063–4068. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2: An improved and customizable approach for metagenome inference. bioRxiv 2020. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.R.; Schilling, J.; Kennedy, P. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Ulvcrona, A.U.; Ulvcrona, T.; Nilsson, U.; Lundmark, T. Stand density and fertilization effects on aboveground allocation patterns and stem form of Pinus yunnanensis in young stands. Scand. J. For. Res. 2014, 29, 197–209. [Google Scholar] [CrossRef]
- Shavnin, S.A.; Golikov, D.Y.; Montile, A.A. Rotational Activity of Trunk Parts in Scots Pine Trees: Seasonal Dynamics and Patterns. Dokl. Biol. Sci. 2022, 503, 63–67. [Google Scholar] [CrossRef]
- Gapare, W.; Hathorn, A.; Kain, D.; Matheson, C.; Wu, H. Inheritance of spiral grain in the juvenile core of Pinus yunnanensis. Can. J. For. Res. 2006, 37, 116–127. [Google Scholar] [CrossRef]
- Hannrup, B.; Säll, H.; Jansson, G. Genetic parameters for spiral grain in Scots pine and Norway spruce. Silvae Genet. 2003, 52, 215–220. [Google Scholar] [CrossRef]
- Pang, Z.; Xu, J.; Yi, C. Linking Plant Secondary Metabolites and Plant Microbiomes: A Review. Front. Plant Sci. 2021, 11, 621276. [Google Scholar] [CrossRef]
- Hiruma, K.; Gerlach, N.; Sacristán, S.; Nakano, R.T.; Hacquard, S.; Kracher, B.; Neumann, U.; Ramírez, D.; Bucher, M.; O’Connell, R.J.; et al. Root Endophyte Colletotrichum tofieldiae Confers Plant Fitness Benefits that Are Phosphate Status Dependent. Cell 2016, 165, 464–474. [Google Scholar] [CrossRef]
- Cregger, M.A.; Veach, A.M.; Yang, Z.K.; Crouch, M.J.; Vilgalys, R.; Tuskan, G.A.; Schadt, C.W. The Populus holobiont: Dissecting the effects of plant niches and genotype on the microbiome. Microbiome 2018, 6, 31. [Google Scholar] [CrossRef]
- Li, P.; Zong, D.; Gan, P.; Li, H.; Wu, Z.; Li, F.; Zhao, C.; Li, L.; He, C. Comparison of the diversity and structure of the rhizosphere microbial community between the straight and twisted-trunk types of Pinus yunnanensis. Front. Microbiol. 2023, 14, 1066805. [Google Scholar] [CrossRef]
- Xin, X.-F.; Kvitko, B.; He, S.Y. Pseudomonas syringae: What it takes to be a pathogen. Nat. Rev. Microbiol. 2018, 16, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Raaijmakers, J.M.; Irene, D.B.; Ole, N.; Marc, O. Natural functions of lipopeptides from Bacillus and Pseudomonas: More than surfactants and antibiotics. Fems Microbiol. Rev. 2010, 34, 1037–1062. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Grube, M.; Schloter, M.; Smalla, K. Unraveling the plant microbiome: Looking back and future perspectives. Front. Microbiol. 2014, 5, 148. [Google Scholar] [CrossRef]
- Nadana, R.V.G. Isolation of Pseudomonas Species from Rhizospheric Soil and its Antagonistic Effect on Plant Pathogen. Int. J. Res. Appl. Sci. Eng. Technol. 2021, 9, 2405–2408. [Google Scholar] [CrossRef]
- Munakata, Y.; Heuson, E.; Daboudet, T.; Deracinois, B.; Duban, M.; Hehn, A.; Coutte, F.; Slezack-Deschaumes, S. Screening of Antimicrobial Activities and Lipopeptide Production of Endophytic Bacteria Isolated from Vetiver Roots. Microorganisms 2022, 10, 209. [Google Scholar] [CrossRef]
- Bao, C.; Sha, L.; Yingjie, W.; Jiayuan, Y.; Qiong, W.; Xiaomeng, X.; Fengshan, P.; Khan, K.Y.; Ying, F.; Xiaoe, Y. The Effects of the Endophytic Bacterium Pseudomonas fluorescens Sasm05 and IAA on the Plant Growth and Cadmium Uptake of Sedum alfredii Hance. Front. Microbiol. 2017, 8, 2538. [Google Scholar] [CrossRef]
- Ghaderi, F.; Habibi, A.; Sharifnabi, B. Phylogenetic Analysis of Phaeosphaeria Species Using Mating Type Genes and Distribution of Mating Types in Iran. Plant Pathol. J. 2022, 38, 78–79. [Google Scholar] [CrossRef]
- Lazarevi, J.; Menkis, A. Fungal Diversity in the Phyllosphere of Pinus heldreichii H. Christ—An Endemic and High-Altitude Pine of the Mediterranean Region. Diversity 2020, 12, 172. [Google Scholar] [CrossRef]
- Laihonen, M.; Saikkonen, K.; Helander, M.; de Aldana, B.R.V.; Zabalgogeazcoa, I.; Fuchs, B. Epichloë Endophyte-Promoted Seed Pathogen Increases Host Grass Resistance Against Insect Herbivory. Front. Microbiol. 2021, 12, 786619. [Google Scholar] [CrossRef]
- Zhong, R.; Bastías, D.A.; Zhang, X.; Li, C.; Nan, Z. Vertically Transmitted Epichloë Systemic Endophyte Enhances Drought Tolerance of Achnatherum inebrians Host Plants through Promoting Photosynthesis and Biomass Accumulation. Fungi 2022, 8, 512. [Google Scholar] [CrossRef]
- Newman, J.A.; Gillis, S.; Hager, H.A. Costs, Benefits, Parasites and Mutualists: The Use and Abuse of the Mutualism-Parasitism Continuum Concept for Epichloë Fungi. bioRxiv. 2021. [Google Scholar] [CrossRef]
- Wang, Z.; Li, M.; Ju, W.; Ye, W.; Xue, L.; Boufford, D.E.; Gao, X.; Yue, B.; Liu, Y.; Pierce, N.E. The entomophagous caterpillar fungus Ophiocordyceps sinensis is consumed by its lepidopteran host as a plant endophyte. Fungal Ecol. 2020, 47, 100989. [Google Scholar] [CrossRef]
- de Menezes, T.A.; Aburjaile, F.F.; Quintanilha-Peixoto, G.; Tomé, L.M.R.; Fonseca, P.L.C.; Mendes-Pereira, T.; Araújo, D.S.; Melo, T.S.; Kato, R.B.; Delabie, J.H.C.; et al. Unraveling the Secrets of a Double-Life Fungus by Genomics: Ophiocordyceps australis CCMB661 Displays Molecular Machinery for Both Parasitic and Endophytic Lifestyles. Fungi 2023, 9, 110. [Google Scholar] [CrossRef] [PubMed]
- Pyszynski, W. Mechanism of formation of spiral grain in Aesculus stems: Dissymmetry of deformation of stems caused by cyclic torsion. Acta Soc. Bot. Pol. 2015, 46, 501–522. [Google Scholar] [CrossRef]
- Hagag, A.; Abdelwahab, M.F.; El-Kader, A.M.A.; Fouad, M.A. The endophytic Aspergillus strains: A bountiful source of natural products. J. Appl. Microbiol. 2022, 132, 4150–4169. [Google Scholar] [CrossRef] [PubMed]
- Toghueo, R.M.K.; Boyom, F.F. Endophytic Penicillium species and their agricultural, biotechnological, and pharmaceutical applications. 3 Biotech 2020, 10, 107. [Google Scholar] [CrossRef]
- Zhao, W.; Bi, X.; Peng, Y.; Bai, M. Research advances of the phosphorus-accumulating organisms of Candidatus Accumulibacter, Dechloromonas and Tetrasphaera: Metabolic mechanisms, applications and influencing factors. Chemosphere 2022, 307, 135675. [Google Scholar] [CrossRef]
- Hedden, P.; Sponsel, V. A Century of Gibberellin Research. J. Plant Growth Regul. 2015, 34, 740–760. [Google Scholar] [CrossRef]
- Hartmann, M.; Zeier, T.; Bernsdorff, F.; Reichel-Deland, V.; Kim, D.; Hohmann, M.; Scholten, N.; Schuck, S.; Bräutigam, A.; Hölzel, T.; et al. Flavin Monooxygenase-Generated N-Hydroxypipecolic Acid Is a Critical Element of Plant Systemic Immunity. Cell 2018, 173, 456. [Google Scholar] [CrossRef]
- López-Sámano, M.; Beltrán, L.F.L.-A.; Sánchez-Thomas, R.; Dávalos, A.; Villaseñor, T.; García-García, J.D.; García-de los Santos, A. A novel way to synthesize pantothenate in bacteria involves β-lanine synthase present in uracil degradation pathway. MicrobiologyOpen 2020, 9, e1006. [Google Scholar] [CrossRef]
- Wright, L.D.; Miller, C.S. Uracil in growth and pyrimidine nucleotide synthesis of Lactobacillus bulgaricus 09. Proc. Soc. Exp. Biol. Med. 1952, 81, 131–134. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Wang, J.; E, J.; Cheng, Z.; Zhang, J. The effect of uracil on the freeze-dried survival rate of Lactobacillus plantarum LIP-1 and its mechanism of action. Trans. Chin. Soc. Agric. Eng. 2022, 38, 308–316. [Google Scholar]
- Lobley, C.M.C.; Schmitzberger, F.; Kilkenny, M.L.; Whitney, H.; Ottenhof, H.H.; Chakauya, E.; Webb, M.E.; Birch, L.M.; Tuck, K.L.; Abell, C. Structural insights into the evolution of the pantothenate-biosynthesis pathway. Biochem Soc Trans 2003, 31, 563–571. [Google Scholar] [CrossRef]
- Leonardi, R.; Jackowski, S. Biosynthesis of Pantothenic Acid and Coenzyme A. Ecosal Plus 2007, 2. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Lou, K.; Li, C. Promotion of plant growth by phytohormone-producing endophytic microbes of sugar beet. Biol. Fertil. Soils 2009, 45, 645–653. [Google Scholar] [CrossRef]
- Bian, X.; Qu, C.; Zhang, M.; Li, D.; Jiang, J.; Liu, G. Transcriptome analysis provides new insights into leaf shape variation in birch. Trees: Struct. Funct. 2019, 33, 1265–1281. [Google Scholar] [CrossRef]
- Eklund, L.; Säll, H.; Linder, S. Enhanced growth and ethylene increases spiral grain formation in Picea abies and Abies balsamea trees. Trees 2003, 17, 81–86. [Google Scholar] [CrossRef]
- Suharti, W.S.; Nose, A.; Zheng, S.H. Metabolomic study of two rice lines infected by Rhizoctonia solani in negative ion mode by CE/TOF-MS. J. Plant Physiol. 2016, 1, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Hinsch, J.; Galuszka, P.; Tudzynski, P. Functional characterization of the first filamentous fungal tRNA-isopentenyltransferase and its role in the virulence of Claviceps purpurea. New Phytol. 2016, 211, 980–992. [Google Scholar] [CrossRef]
- Mishra, D.C.; Arora, D.; Budhlakoti, N.; Solanke, A.U.; Mithra, S.V.A.C.; Kumar, A.; Pandey, P.S.; Srivastava, S.; Kumar, S.; Farooqi, M.S. Identification of Potential Cytokinin Responsive Key Genes in Rice Treated With trans-Zeatin Through Systems Biology Approach. Front. Genet. 2022, 12, 780599. [Google Scholar] [CrossRef]
- Gajdosová, S.; Spíchal, L.; Kamínek, M.; Hoyerová, K.; Novák, O.; Dobrev, P.I.; Galuszka, P.; Klíma, P.; Gaudinová, A.; Žižková, E.; et al. Distribution, biological activities, metabolism, and the conceivable function of cis-Zeatin-type cytokinins in plants. J. Exp. Bot. 2011, 62, 2827–2840. [Google Scholar] [CrossRef] [PubMed]
- Perostová, S.; Jaroová, J.; Dobrev, P.; Hlusková, L.; Motyka, V.; Filepová, R.; Knirsch, V.; Gaudinova, A.; Kieber, J.; Vaňková, R. Heat Stress Targeting Individual Organs Reveals the Central Role of Roots and Crowns in Rice Stress Responses. Front. Plant Sci. 2022, 12, 799249. [Google Scholar] [CrossRef]
- Mengiste, T. Plant immunity to necrotrophs. Annu. Rev. Phytopathol. 2012, 50, 267–294. [Google Scholar] [CrossRef]
- Yuan, H.M.; Liu, W.C.; Lu, Y.T. CATALASE2 Coordinates SA-Mediated Repression of Both Auxin Accumulation and JA Biosynthesis in Plant Defenses. Cell Host Microbe 2017, 21, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Song, R.F.; Yuan, H.M.; Li, T.T.; Wang, L.F.; Lu, K.K.; Guo, J.X.; Liu, W.C. Overexpressing the N-terminus of CATALASE2 enhances plant jasmonic acid biosynthesis and resistance to necrotrophic pathogen Botrytis cinerea B05.10. Mol Plant Pathol. 2021, 22, 1226–1238. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Yang, C.; Liu, J. Phytopathogenic bacterial effectors. Science Bulletin. 2023, 68, 4895–4917. Available online: https://kns.cnki.net/kcms2/article/abstract?v=jNHD1hIvxn0DsP0kHfLF5LuMZb05LXk0DkkbkQpXL2kkW4cxyYdBISTX-e7db4264z35RJa7UJilRHmIwHjS3hJzOsTB0DuPTRcbR3ge-m6-lyFCojsBB5IOTemcGQGLU1atdskxezuY0HHYNWrQGPWjuJtLqrGBOV1kBaX5_yDoDBr-C4NlGngmEbXjaKXpqMYS79gfnog=&uniplatform=NZKPT&language=CHS (accessed on 20 March 2025).
- Darino, M.; Chia, K.S.; Marques, J.; Aleksza, D.; Soto-Jiménez, L.M.; Saado, I.; Uhse, S.; Borg, M.; Betz, R.; Bindics, J.; et al. Ustilago maydis effector Jsi1 interacts with Topless corepressor, hijacking plant jasmonate/ethylene signaling. New Phytol 2021, 229, 3393–3407. [Google Scholar] [CrossRef]
- Nilsson, A.K.; Fahlberg, P.; Johansson, O.N.; Hamberg, M.; Andersson, M.X.; Ellerström, M. The activity of HYDROPEROXIDE LYASE 1 regulates accumultation of galactolipids containing 12-oxo-phytodienoic acid in Arabidopsis. J. Exp. Bot. 2016, 64, 5133–5144. [Google Scholar] [CrossRef]
- Kumar, A.; Yogendra, K.N.; Karre, S.; Kushalappa, A.C.; Dion, Y.; Choo, T.M. WAX INDUCER1 (HvWIN1) transcription factor regulates free fatty acid biosynthetic genes to reinforce cuticle to resist Fusarium head blight in barley spikelets. J. Exp. Bot. 2016, 67, 4127–4139. [Google Scholar] [CrossRef]
- Zhu, K.; Gu, S.; Liu, J.; Luo, T.; Khan, Z.; Zhang, K.; Hu, L. Wood Vinegar as a Complex Growth Regulator Promotes the Growth, Yield, and Quality of Rapeseed. Agronomy 2021, 11, 510. [Google Scholar] [CrossRef]
- Yu, H.; Lin, Z.; Li, Y.; Yuan, L.; Zhao, B. Effects of spraying low molecular organic compounds on growth and nutrients uptake of rape (Brassica Chinensis L.). J. Plant Nutr. Fertil. 2014, 20, 1560–1568. [Google Scholar] [CrossRef]
- Gourkhede, P.H.; Patil, V.D.; Narale, S. Effect of Foliar Feeding of Gluconate and EDTA Chelated Plant Nutrients on Yield, Quality and Nutrient Concentration of Bt-Cotton. Int. J. Trop. Agric. 2015, 33, 1875–1879. [Google Scholar]
- Marra, R.; Coppola, M.; Pironti, A.; Grasso, F.; Lombardi, N.; D’Errico, G.; Sicari, A.; Censi, S.B.; Woo, S.L.; Rao, R. The Application of Trichoderma Strains or Metabolites Alters the Olive Leaf Metabolome and the Expression of Defense-Related Genes. J. Fungi 2020, 6, 369. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Wang, X.; Yan, X.; Ke, J.; Yang, Y.; Li, Y.; Xu, D.; Zhuo, Z.; Yan, X. Research progress on the application of transcriptomics and metabolomics in tree fungal disease defense responses. World For. Res. 2022, 35, 27–32. [Google Scholar] [CrossRef]
- Mosquera, S.; Leveau, J.H.J.; Stergiopoulos, I. Repeated exposure of Aspergillus niger spores to the antifungal bacterium Collimonas fungivorans Ter331 selects for delayed spore germination. Appl. Environ. Microbiol. 2021, 87, e00233-21. [Google Scholar] [CrossRef]
- Li, J.; Pan, M.; Zhang, X.; Zhou, Y.; Feng, G.-D.; Zhu, H. Collimonas silvisoli sp. nov. and Collimonas humicola sp. nov., two novel species isolated from forest soil. Microbiol. Soc. 2021, 71, 005061. [Google Scholar] [CrossRef]
- Uroz, S.; Geisler, O.; Fauchery, L.; Lami, R.; Rodrigues, A.M.S.; Morin, E.; Leveau, J.H.J.; Oger, P. Genomic and transcriptomic characterization of the Collimonas quorum sensing genes and regulon. FEMS Microbiol. Ecol. 2022, 98, 1–22. [Google Scholar] [CrossRef]
- Akinsanya, M.A.; Goh, J.K.; Lim, S.P.; Adeline Su Yien Ting, A.S.Y. Diversity, antimicrobial and antioxidant activities of culturable bacterial endophyte communities in Aloe vera. FEMS Microbiol. Lett. 2015, 362, fnv184. [Google Scholar] [CrossRef]
- Jeong, J.J.; Sang, M.K.; Wan, L.D.; Choi, I.G.; Kim, K.D. Chryseobacterium phosphatilyticum sp. nov., a phosphate-solubilizing endophyte isolated from cucumber (Cucumis sativus L.) root. Int. J. Syst. Evol. Microbiol. 2019, 69, 610–615. [Google Scholar] [CrossRef]
- Singburaudom, N. The alkaloid Berberine isolated from Coscinium fenestratum is an inhibitor of phytopathogenic fungi. J. Biopestic. 2015, 8, 28–36. [Google Scholar] [CrossRef]
- Glenz, R.; Kaiping, A.; Göpfert, D.; Weber, H.; Lambour, B.; Sylvester, M.; Fröschel, C.; Mueller, M.J.; Osman, M.; Waller, F. The major plant sphingolipid long chain base phytosphingosine inhibits growth of bacterial and fungal plant pathogens. Sci. Rep. 2022, 12, 1081. [Google Scholar] [CrossRef] [PubMed]
- Kleber, H.P. Bacterial carnitine metabolism. FEMS Microbiol. Lett. 1997, 147, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Millican, M. The Role of Plant-Derived Quaternary Ammonium Compounds, Including Carnitine and Choline-O-sulfate, on the Biology of the Plant Pathogen Pseudomonas syringae and Its Interactions with the Host Species Phaseolus vulgaris. Doctoral Thesis, Iowa State University, Ames, IA, USA, 2015. [Google Scholar] [CrossRef]
- Rosenfeld, H.; Feigelson, P. Product induction in Pseudomonas acidovorans of a permease system which transports L-tryptophan. J. Bacteriol. 1969, 97, 705–714. [Google Scholar] [CrossRef] [PubMed]
- Kelly, Y.L.; Yinghao, W.; Tszking, L.; Chuenfai, K.; Wingping, Y.; Zhongzhen, Z. Correlation between quality and geographical origins of Leonuri Herba revealed by the qualitative fngerprint profling and quantitative determination of chemical components. Chin. Med. 2022, 17, 2–14. [Google Scholar] [CrossRef]
- Jung, H.; Lee, D.; Lee, S.; Kong, H.J.; Park, J.; Seo, Y.S. Comparative genomic analysis of Chryseobacterium species: Deep insights into plant-growth-promoting and halotolerant capacities. Microb. Genom. 2023, 9, 001108. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).