Genome-Wide Identification and Expression Analysis of the AlkB Homolog Gene Family in Tamarix chinensis
Abstract
1. Introduction
2. Materials and Methods
2.1. Identificaiton of ALKBH Genes in the T. chinensis Genome
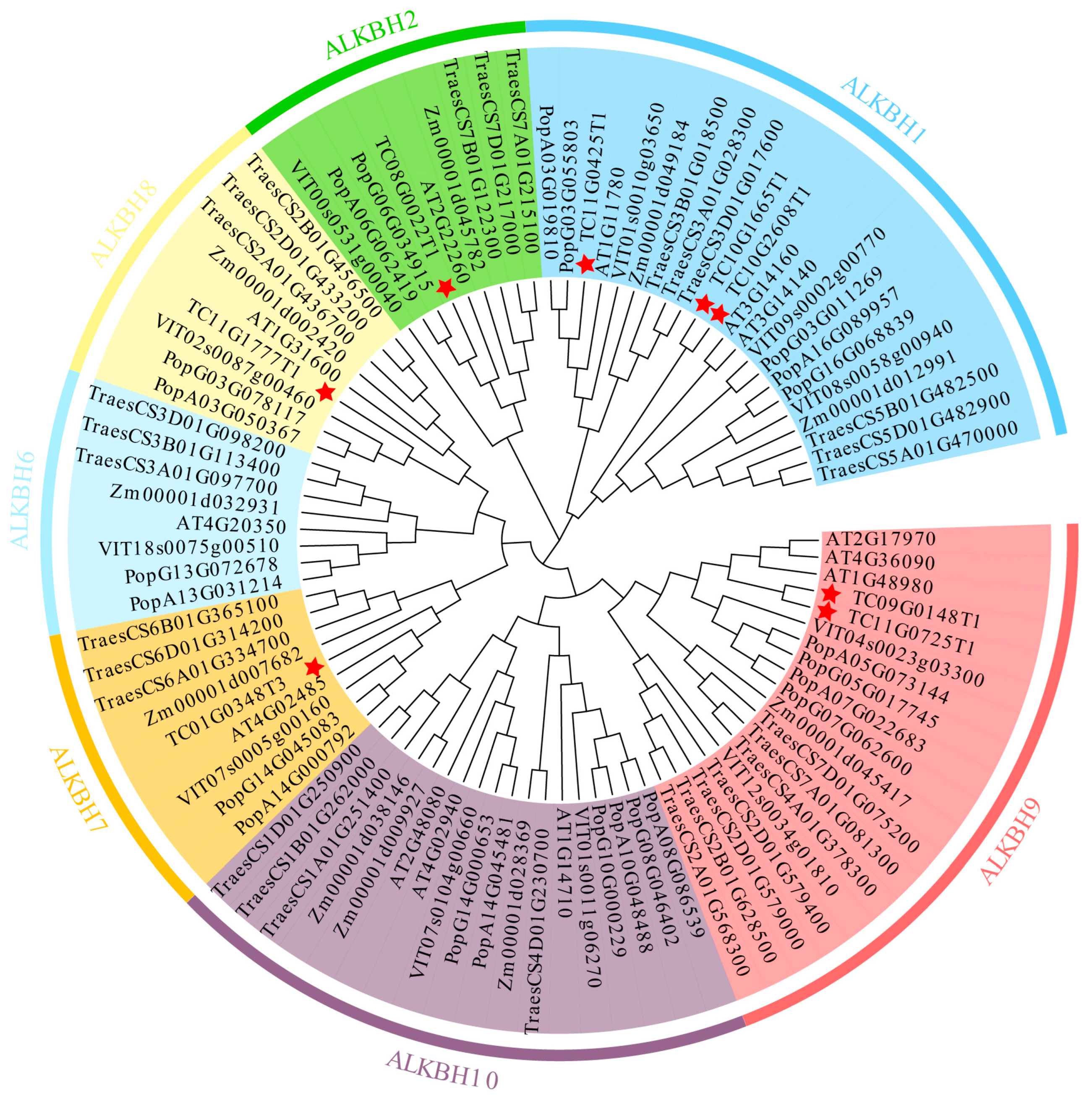
2.2. Chromosomal Mapping and Phylogenetic Analysis of TcALKBH Genes
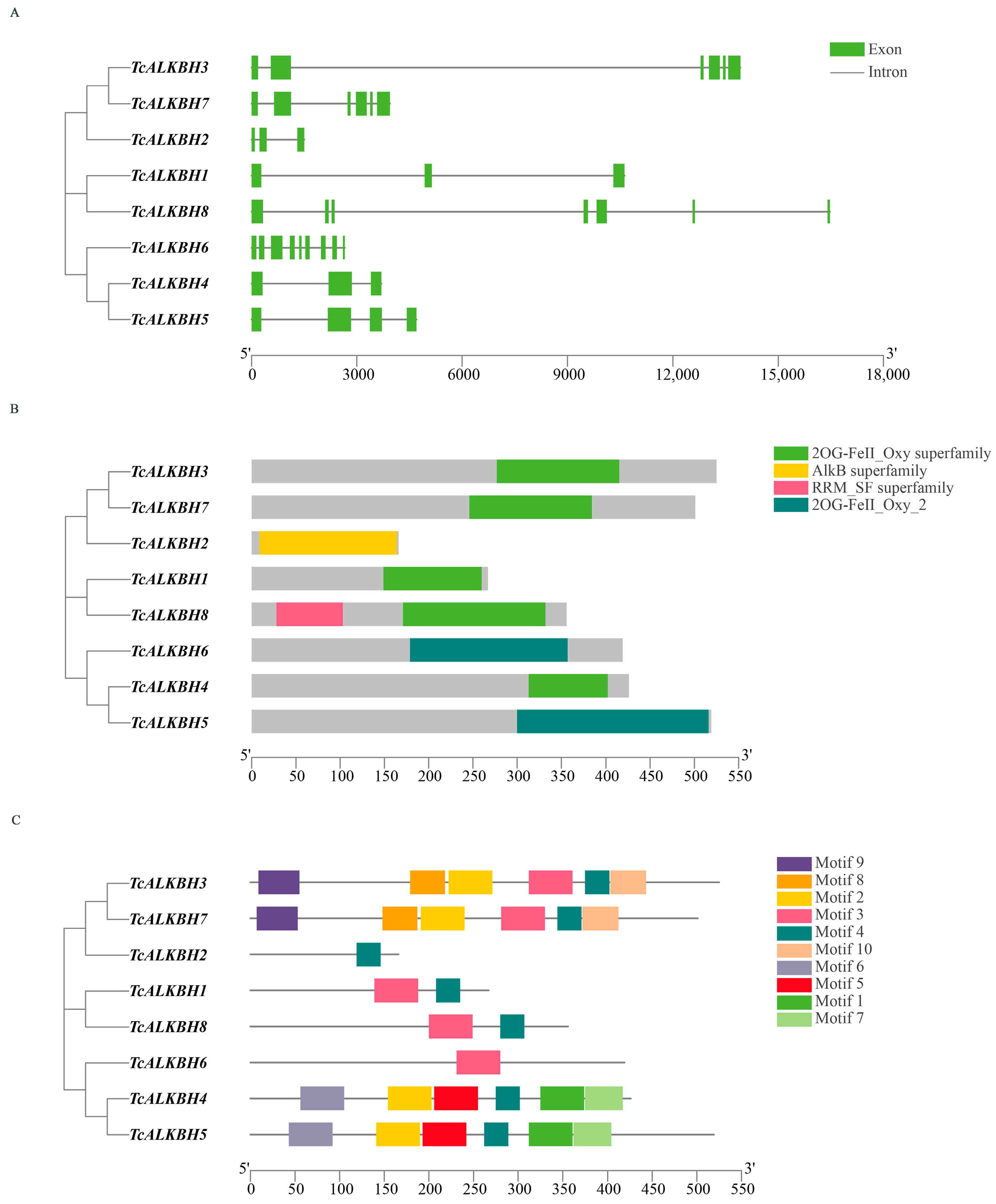
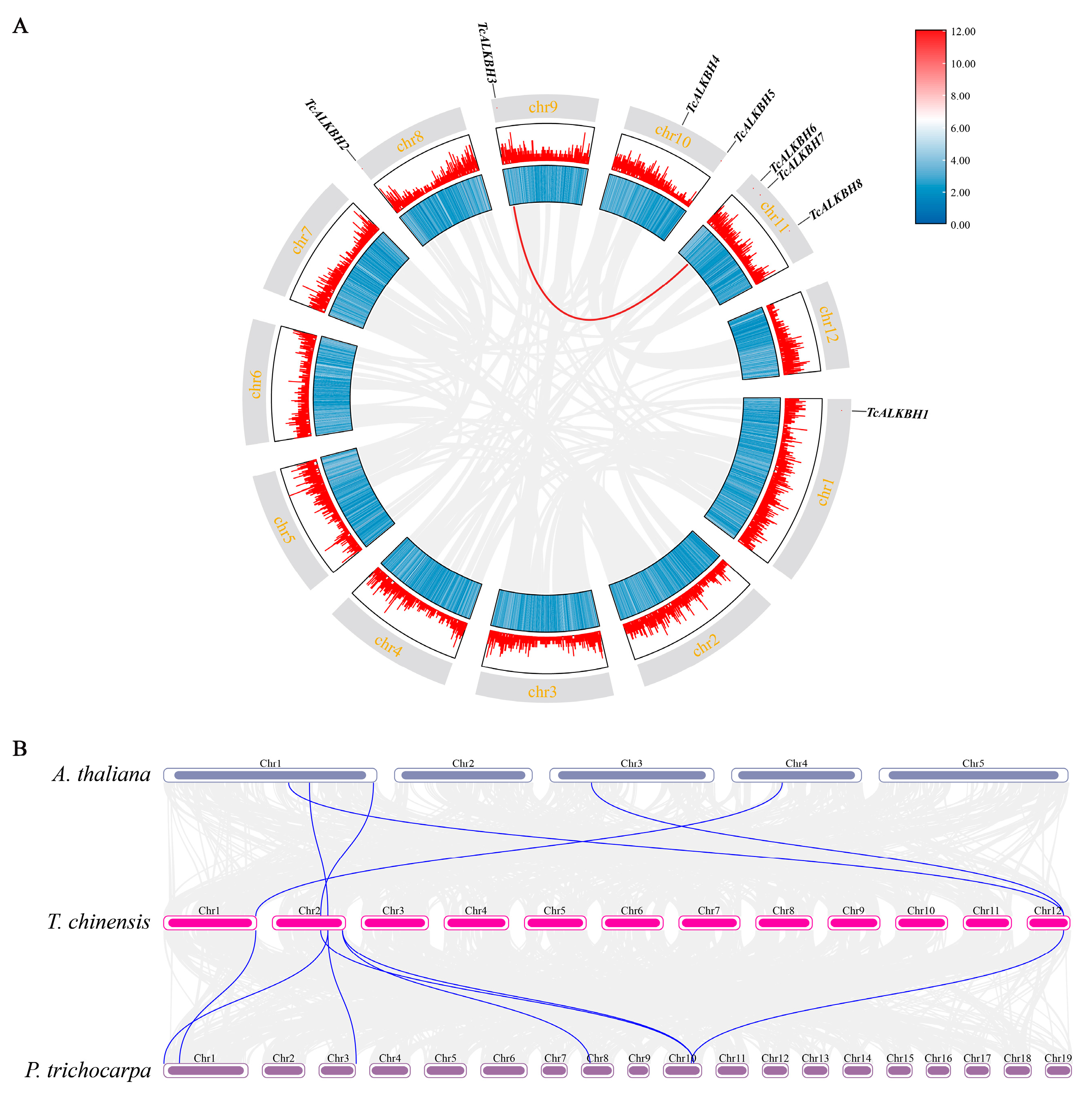
2.3. Gene Structure, Amino Acid Sequence and Gene Collinearity Analysis
2.4. Cis-Acting Elements Analysis of the TcALKBH Genes Promoter
2.5. Plant Materials and Stress Treatment
2.6. qRT-PCR Was Used to Analyze the Expression of TcALKBH Genes
3. Results
3.1. Genome-Wide Identification of TcALKBH Gene in T. chinensis
3.2. Phylogenetic Analysis of TcALKBH Genes
3.3. Gene Structure, Conserved Domain, and Motif Analysis of TcALKBH Genes
3.4. Gene Replication Analysis and Synteny Analysis of the TcALKBH Genes
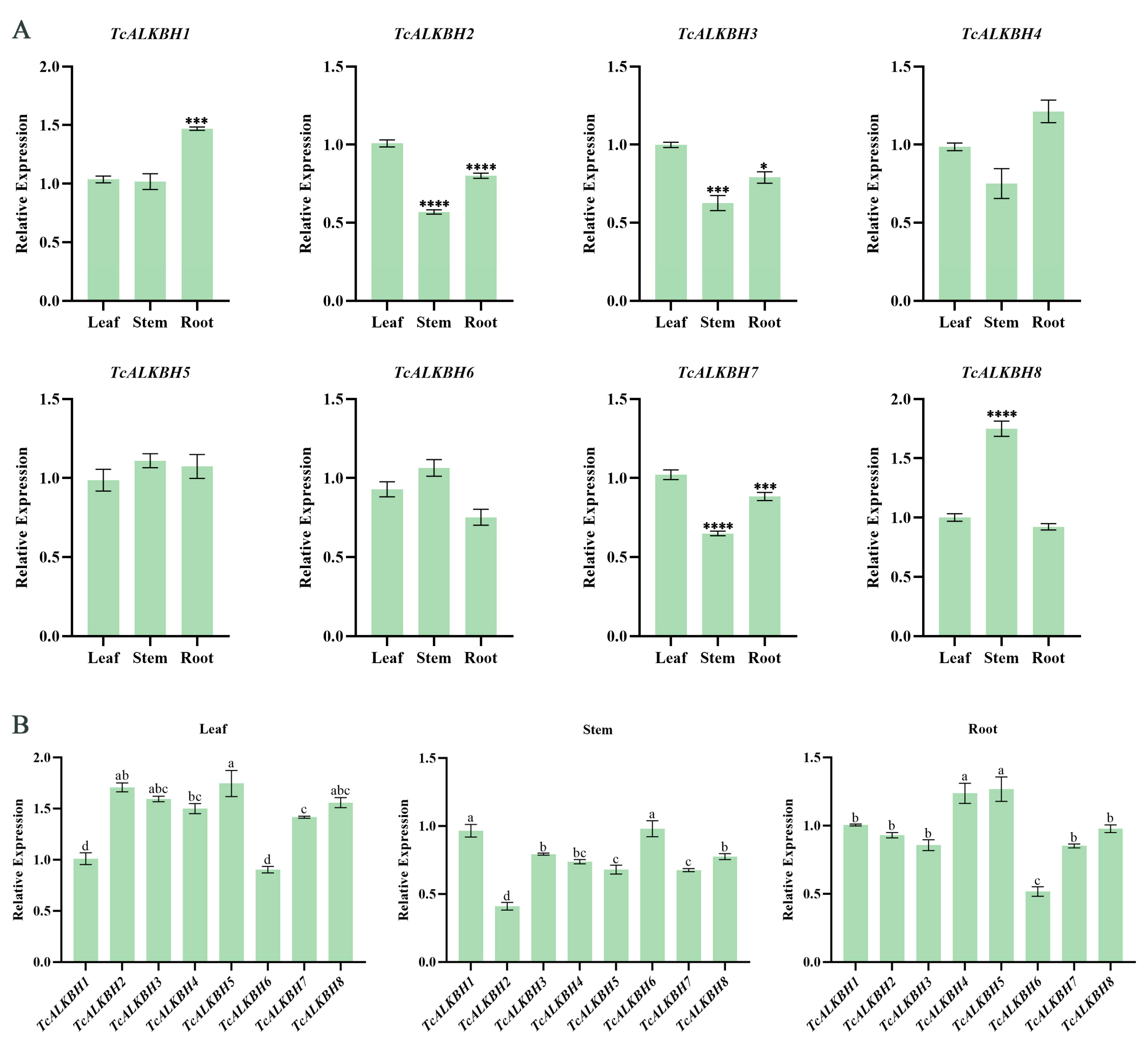
3.5. Tissue-Specific Expression Pattern of TcALKBH Genes
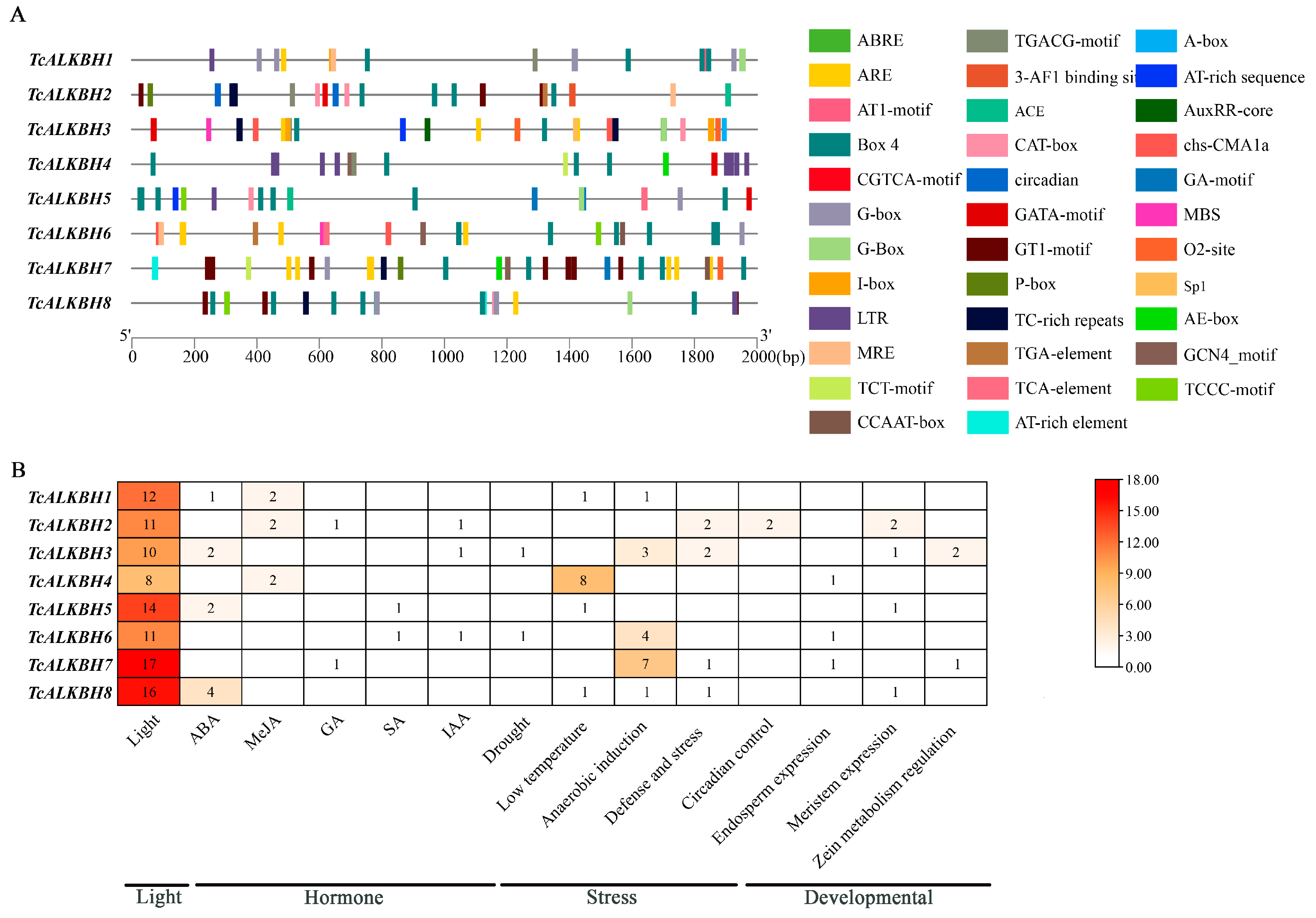
3.6. Analysis of Promoter Cis-Acting Elements of TcALKBH Genes
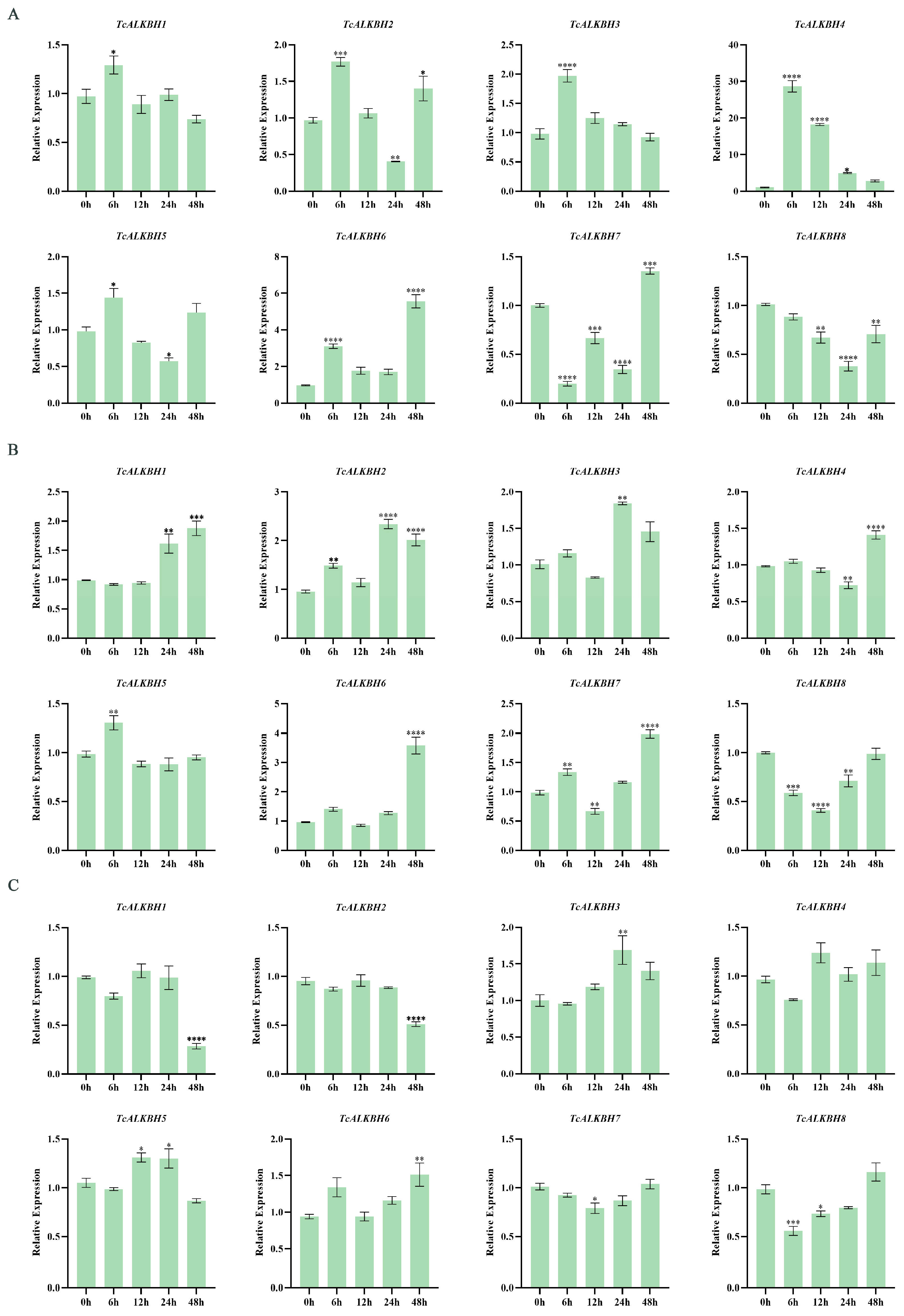
3.7. Expression Patterns of TcALKBH Genes Under ABA, NaCl, and NaHCO3 Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yang, Y.; Hsu, P.J.; Chen, Y.S.; Yang, Y.G. Dynamic transcriptomic m6A decoration: Writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018, 28, 616–624. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Chen, S.Y.; Jia, G.F. N6-Detection, regulation, and functions of RNA methyladenosine modification in plants. Plant Commun. 2023, 4, 100546. [Google Scholar] [CrossRef]
- Duan, H.C.; Wei, L.H.; Zhang, C.; Wang, Y.; Chen, L.; Lu, Z.K.; Chen, P.R.; He, C.; Jia, G.F. ALKBH10B Is an RNA N6-Methyladenosine Demethylase Affecting Arabidopsis Floral Transition. Plant Cell 2017, 29, 2995–3011. [Google Scholar] [CrossRef] [PubMed]
- Huong, T.T.; Yang, Z.M.; Ngoc, L.T.; Kang, H. ALKBH8B, a Putative RNA Demethylase, Plays a Role in the Response of Arabidopsis to Salt Stress and Abscisic Acid. J. Plant Biol. 2022, 65, 319–330. [Google Scholar] [CrossRef]
- Amara, U.; Shoaib, Y.; Kang, H.S. ALKBH9C, a potential RNA m6A demethylase, regulates the response of Arabidopsis to abiotic stresses and abscisic acid. Plant Cell Environ. 2022, 45, 3566–3581. [Google Scholar] [CrossRef]
- Tang, J.; Yang, J.B.; Lu, Q.; Tang, Q.; Chen, S.Y.; Jia, G.F. The RNA N6-methyladenosine demethylase ALKBH9B modulates ABA responses in Arabidopsis. J. Integr. Plant Biol. 2022, 64, 2361–2373. [Google Scholar] [CrossRef] [PubMed]
- Mielecki, D.; Zugaj, D.L.; Muszewska, A.; Piwowarski, J.; Chojnacka, A.; Mielecki, M.; Nieminuszczy, J.; Grynberg, M.; Grzesiuk, E. Novel AlkB Dioxygenases-Alternative Models for In Silico and In Vivo Studies. PLoS ONE 2012, 7, e30588. [Google Scholar] [CrossRef]
- Xu, B.F.; Liu, D.Y.; Wang, Z.R.; Tian, R.X.; Zuo, Y.C. Multi-substrate selectivity based on key loops and non-homologous domains: New insight into ALKBH family. Cell. Mol. Life Sci. 2021, 78, 129–141. [Google Scholar] [CrossRef]
- Lindahl, T.; Sedgwick, B.; Sekiguchi, M.; Nakabeppu, Y. Regulation and expression of the adaptive response to alkylating agents. Annu. Rev. Biochem. 1988, 57, 133–157. [Google Scholar] [CrossRef]
- Sedgwick, B.; Lindahl, T. Recent progress on the Ada response for inducible repair of DNA alkylation damage. Oncogene 2002, 21, 8886–8894. [Google Scholar] [CrossRef]
- Marcinkowski, M.; Pilzys, T.; Garbicz, D.; Steciuk, J.; Zugaj, D.; Mielecki, D.; Sarnowski, T.J.; Grzesiuk, E. Human and Arabidopsis alpha-ketoglutarate-dependent dioxygenase homolog proteins-New players in important regulatory processes. Iubmb Life 2020, 72, 1126–1144. [Google Scholar] [CrossRef]
- Huong, T.T.; Ngoc, L.N.T.; Kang, H. Functional Characterization of a Putative RNA Demethylase ALKBH6 in Arabidopsis Growth and Abiotic Stress Responses. Int. J. Mol. Sci. 2020, 21, 6707. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.L.; Tian, S.P.; Qin, G.Z. RNA methylomes reveal the m6A-mediated regulation of DNA demethylase gene SlDML2 in tomato fruit ripening. Genome Biol. 2019, 20, 23. [Google Scholar] [CrossRef]
- Tang, J.; Yang, J.B.; Duan, H.C.; Jia, G.F. ALKBH10B, an mRNA m6A Demethylase, Modulates ABA Response During Seed Germination in Arabidopsis. Front. Plant Sci. 2021, 12, 712713. [Google Scholar] [CrossRef]
- Han, R.P.; Shoaib, Y.; Cai, J.; Kang, H.S. ALKBH10B-mediated m6A demethylation is crucial for drought tolerance by affecting mRNA stability in Arabidopsis. Environ. Exp. Bot. 2023, 209, 105306. [Google Scholar] [CrossRef]
- Martínez-Pérez, M.; Gómez-Mena, C.; Alvarado-Marchena, L.; Nadi, R.; Micol, J.L.; Pallas, V.; Aparicio, F. The m6A RNA Demethylase ALKBH9B Plays a Critical Role for Vascular Movement of Alfalfa Mosaic Virus in Arabidopsis. Front. Microbiol. 2021, 12, 745576. [Google Scholar] [CrossRef]
- Yang, H.J.; Xia, J.B.; Cui, Q.; Liu, J.T.; Wei, S.C.; Feng, L.; Dong, K.K. Effects of different Tamarix chinensis-grass patterns on the soil quality coastal saline soil in the Yellow River Delta, China. Sci. Total Environ. 2021, 772, 145501. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.N.; Fang, H.C.; Liang, Q.; Dong, Y.H.; Wang, C.X.; Yan, L.P.; Ma, X.M.; Zhou, R.; Lang, X.Y.; Gai, S.S.; et al. Genomic analyses provide insights into the evolution and salinity adaptation of halophyte Tamarix chinensis. Gigascience 2023, 12, giad053. [Google Scholar] [CrossRef]
- Shen, H.; Zhou, Y.; Liao, C.G.; Xie, Q.L.; Chen, G.P.; Hu, Z.L.; Wu, T. The AlkB Homolog SlALKBH10B Negatively Affects Drought and Salt Tolerance in Solanum lycopersicum. Int. J. Mol. Sci. 2024, 25, 173. [Google Scholar] [CrossRef]
- Zhao, Y.; Guo, Q.; Cao, S.; Tian, Y.T.; Han, K.J.; Sun, Y.H.; Li, J.; Yang, Q.S.; Ji, Q.J.; Sederoff, R.; et al. Genome-wide identification of the AlkB homologs gene family, PagALKBH9B and PagALKBH10B regulated salt stress response in Populus. Front. Plant Sci. 2022, 13, 994154. [Google Scholar] [CrossRef]
- Berardini, T.Z.; Reiser, L.; Li, D.H.; Mezheritsky, Y.; Muller, R.; Strait, E.; Huala, E. The arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis 2015, 53, 474–485. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, 16. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Wu, Y.; Li, J.W.; Wang, X.; Zeng, Z.H.; Xu, J.; Liu, Y.L.; Feng, J.T.; Chen, H.; He, Y.H.; et al. TBtools-II: A “one for all, all for one”bioinformatics platform for biological big-data mining. Mol. Plant. 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11 Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Subramanian, B.; Gao, S.H.; Lercher, M.J.; Hu, S.N.; Chen, W.H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.Y.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Liu, J.H.; Xia, J.B.; Fang, Y.M.; Li, T.; Liu, J.T. Effects of Salt-Drought Stress on Growth and Physiobiochemical Characteristics of Tamarix chinensis Seedlings. Sci. World J. 2014, 2014, 765840. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhou, L.L.; Gao, G.T.; Tang, R.K.; Liu, J.Y.; Wang, Y.Y.; Liang, Z.C.; Tian, S.P.; Qin, G.Z. Redox modification of m6A demethylase SlALKBH2 in tomato regulates fruit ripening. Nat. Plants 2025, 11, 218–233. [Google Scholar] [CrossRef]
- Zhao, J.; Yang, T.F.; Liu, P.; Liu, H.J.; Zhang, H.; Guo, S.C.; Liu, X.Y.; Chen, X.G.; Chen, M.J. Genome-Wide Identification of the Soybean AlkB Homologue Gene Family and Functional Characterization of GmALKBH10Bs as RNA m6A Demethylases and Expression Patterns under Abiotic Stress. Plants 2024, 13, 2491. [Google Scholar] [CrossRef]
- Li, Y.; Dong, X.M.; Ma, J.Y.; Sui, C.X.; Jian, H.J.; Lv, D.Q. Genome-Wide Identification and Expression Analysis of the ALKB Homolog Gene Family in Potato (Solanum tuberosum L.). Int. J. Mol. Sci. 2024, 25, 10984. [Google Scholar] [CrossRef]
- Huang, A.J.; Wang, Y.; Gu, P.P.; Yang, Z.X.; Han, J.N.; Yi, L. Genome-Wide Identification and Characterization of the AlkB Gene Family in Sweet Orange (Citrus sinensis). Curr. Issues Mol. Biol. 2023, 45, 122–133. [Google Scholar] [CrossRef]
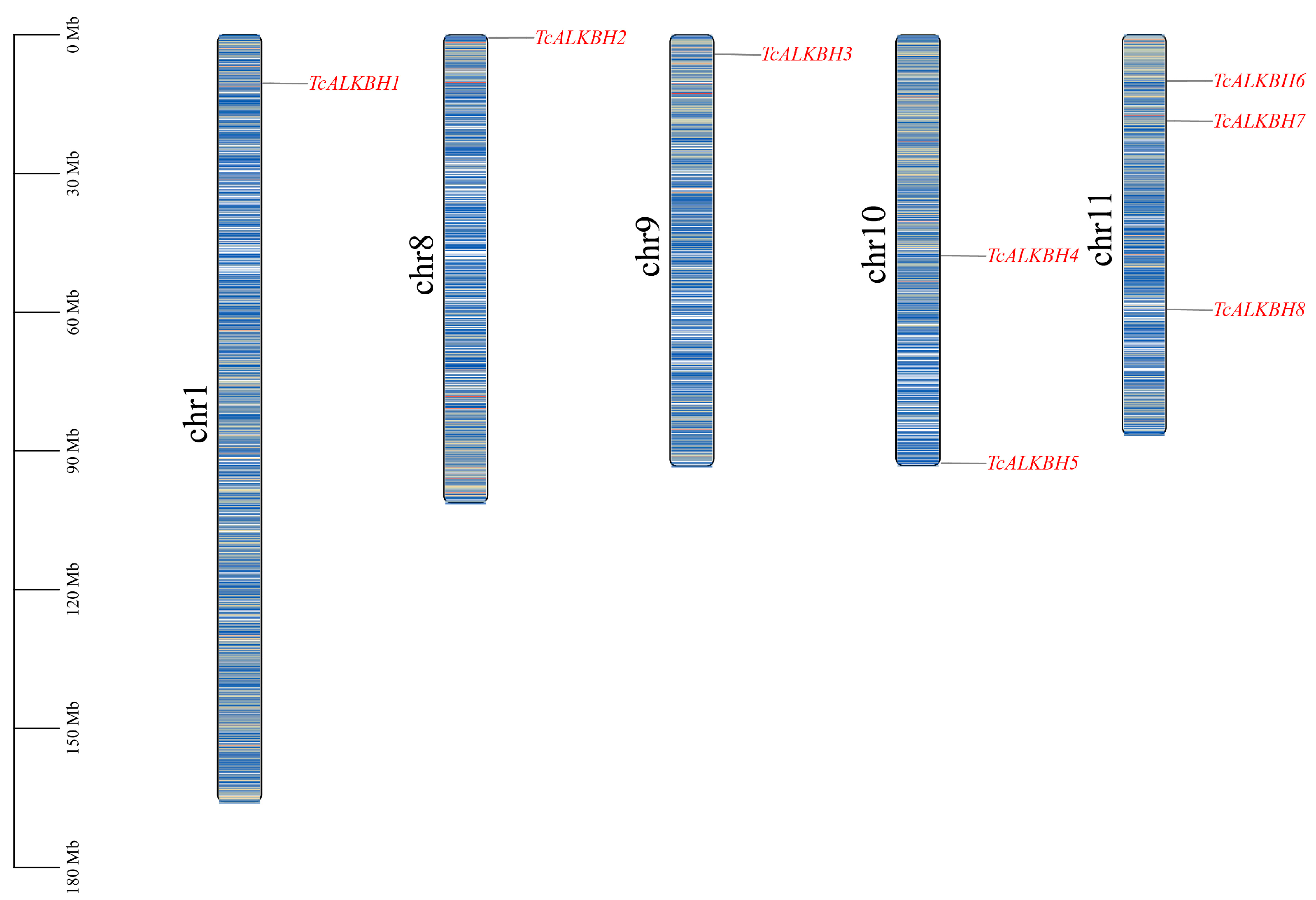
| Gene ID | Gene Name | Locus | CDS (bp) | Protein Length (aa) | MW (kDa) | Aliphatic Index | Gravy | pI | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|
| TC01G0348T3 | TcALKBH1 | Chr1 | 801 | 266 | 30.25 | 80.56 | −0.366 | 4.75 | nucleus |
| TC08G0022T1 | TcALKBH2 | Chr8 | 498 | 165 | 18.67 | 75.03 | −0.593 | 9.16 | nucleus |
| TC09G0148T1 | TcALKBH3 | Chr9 | 1575 | 524 | 59.26 | 72.02 | −0.633 | 6.49 | nucleus |
| TC10G1665T1 | TcALKBH4 | Chr10 | 1278 | 425 | 46.68 | 64.49 | −0.536 | 9.24 | nucleus |
| TC10G2608T1 | TcALKBH5 | Chr10 | 1557 | 518 | 57.46 | 70.21 | −0.55 | 9.26 | chloroplast |
| TC11G0425T1 | TcALKBH6 | Chr11 | 1257 | 418 | 46.71 | 80.29 | −0.336 | 7.69 | cytosol |
| TC11G0725T1 | TcALKBH7 | Chr11 | 1503 | 500 | 56.13 | 67.62 | −0.673 | 8.19 | chloroplast |
| TC11G1777T1 | TcALKBH8 | Chr12 | 1068 | 355 | 39.45 | 77.15 | −0.33 | 6.47 | nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Guo, W.; Yin, H.; Ji, K.; Yu, Q. Genome-Wide Identification and Expression Analysis of the AlkB Homolog Gene Family in Tamarix chinensis. Forests 2025, 16, 470. https://doi.org/10.3390/f16030470
Zhang J, Guo W, Yin H, Ji K, Yu Q. Genome-Wide Identification and Expression Analysis of the AlkB Homolog Gene Family in Tamarix chinensis. Forests. 2025; 16(3):470. https://doi.org/10.3390/f16030470
Chicago/Turabian StyleZhang, Jingjing, Wenhui Guo, Huijuan Yin, Kongshu Ji, and Qiong Yu. 2025. "Genome-Wide Identification and Expression Analysis of the AlkB Homolog Gene Family in Tamarix chinensis" Forests 16, no. 3: 470. https://doi.org/10.3390/f16030470
APA StyleZhang, J., Guo, W., Yin, H., Ji, K., & Yu, Q. (2025). Genome-Wide Identification and Expression Analysis of the AlkB Homolog Gene Family in Tamarix chinensis. Forests, 16(3), 470. https://doi.org/10.3390/f16030470





