Geographical Differences Weaken the Convergence Effect of the Rhizosphere Bacteria of Rubber Trees
Abstract
1. Introduction
2. Methods
2.1. Study Site
2.2. Soil Sampling
2.3. DNA Extraction and Illumina Sequencing
2.4. Soil Physical and Chemical Property Determination
2.5. Data Analysis
3. Results
3.1. Community Composition
3.2. Community Diversity
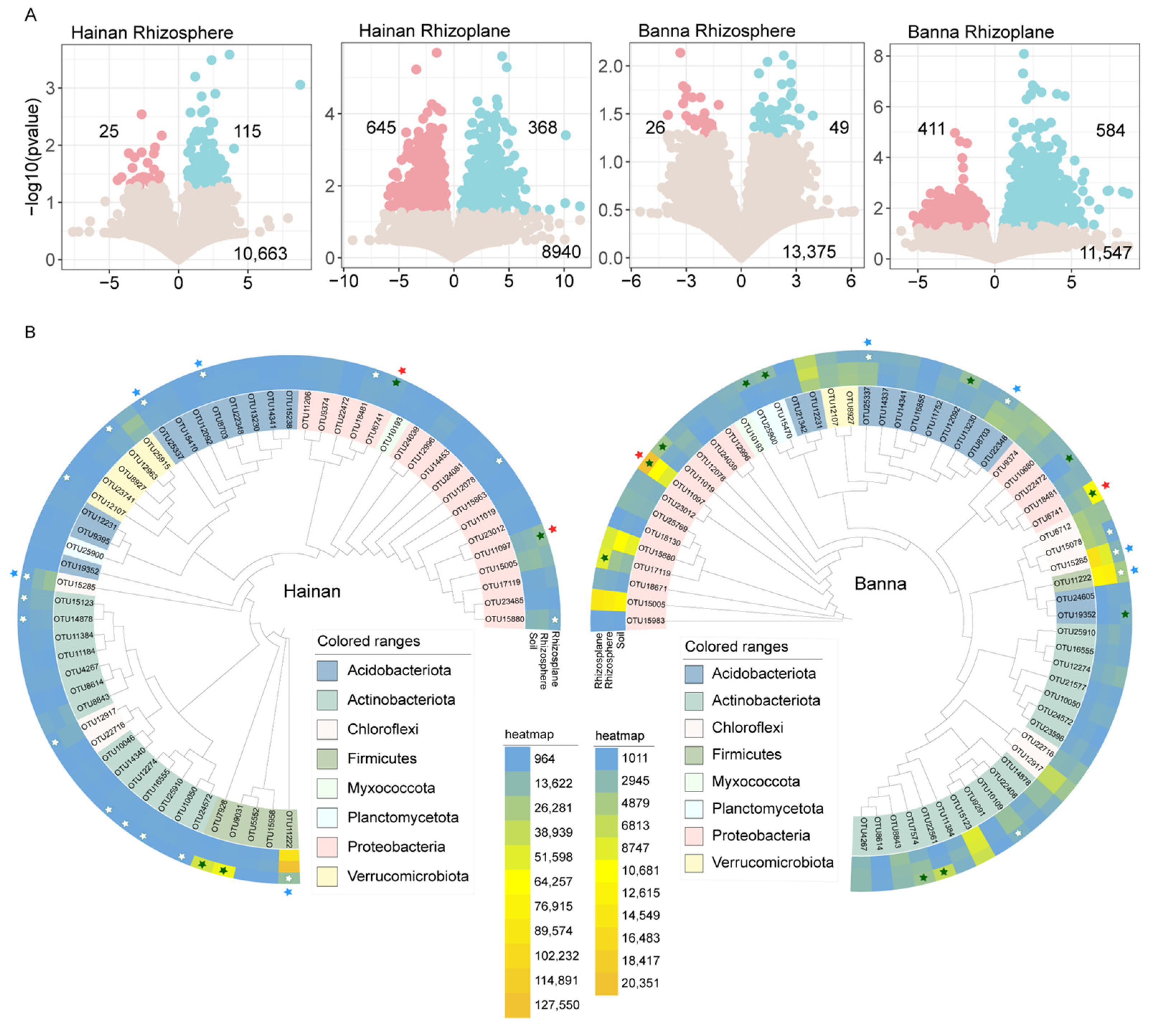
3.3. Enriched and Depleted OTUs
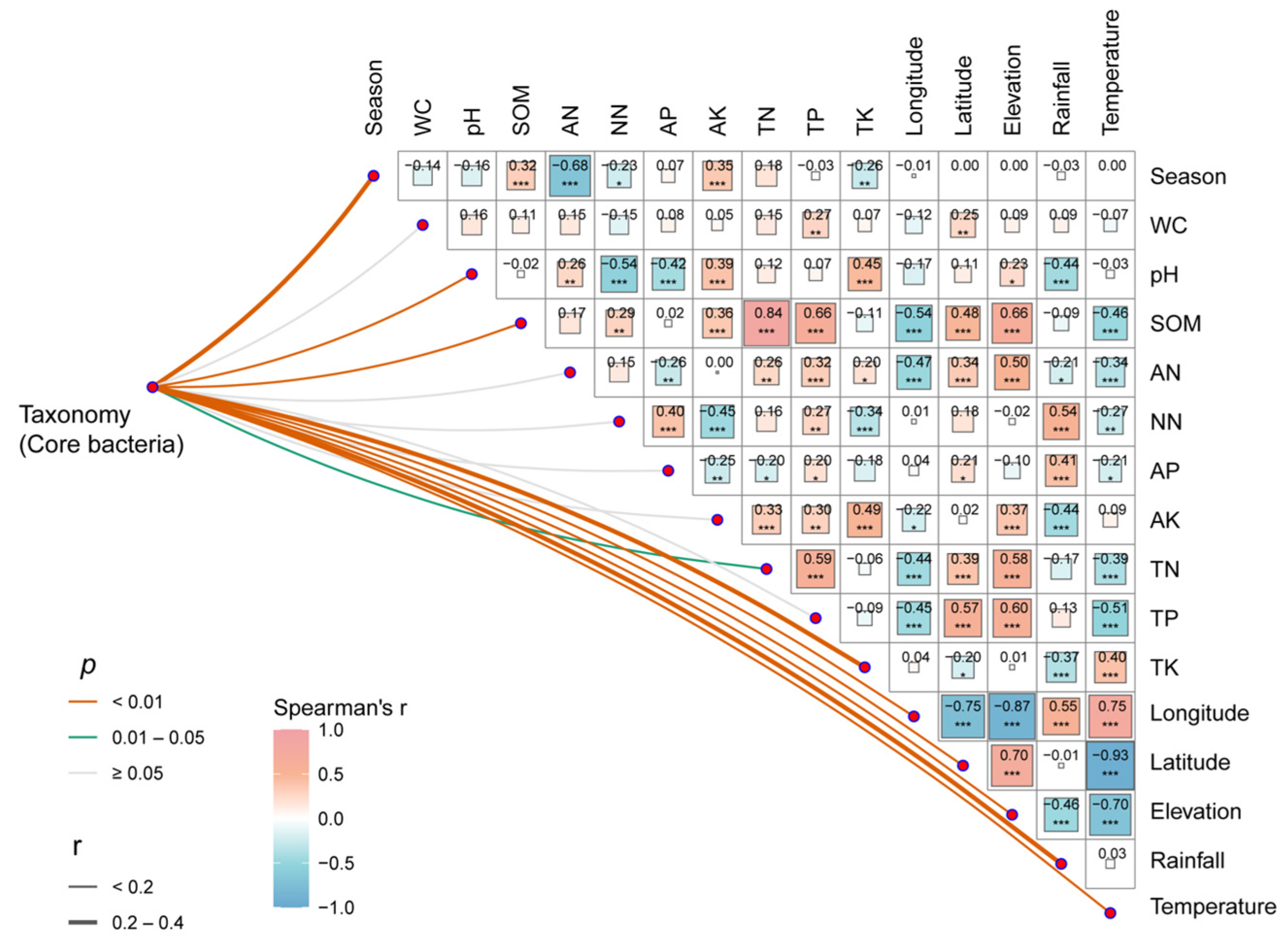
3.4. Correlation of Bacterial Communities with Environmental Factors
4. Discussion
4.1. Convergent Effect of Rhizoplane Bacterial Community
4.2. Geographical Effect of Rhizoplane Bacterial Community
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kuzyakov, Y.; Razavi, B.S. Rhizosphere size and shape: Temporal dynamics and spatial stationarity. Soil Biol. Biochem. 2019, 135, 343–360. [Google Scholar] [CrossRef]
- Fierer, N.; Jackson, R.B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; He, J.Z.; Zhu, Y.G.; Zhang, J.B.; Xu, Z.; Zhang, L.M.; Zheng, Y.M. Differences in soil bacterial diversity: Driven by contemporary disturbances or historical contingencies? ISME J. 2008, 2, 254–264. [Google Scholar] [CrossRef] [PubMed]
- Lan, G.Y.; Wu, Z.X.; Li, Y.W.; Chen, B.Q. The drivers of soil bacterial communities in rubber plantation at local and geographic scales. Arch. Agron. Soil Sci. 2019, 66, 358–369. [Google Scholar] [CrossRef]
- Bahram, M.; Hildebrand, F.; Forslund, S.K.; Anderson, J.L.; Soudzilovskaia, N.A.; Bodegom, P.M.; Bengtsson-Palme, J.; Anslan, S.; Coelho, L.P.; Harend, H.; et al. Structure and function of the global topsoil microbiome. Nature 2018, 560, 233–237. [Google Scholar] [CrossRef]
- Karimi, B.; Terrat, S.; Dequiedt, S.; Saby, N.P.A.; Horriguel, W.; Lelievre, M.; Nowak, V.; Jolivet, C.; Arrouays, D.; Wincker, P.; et al. Biogeography of soil bacteria and archaea across France. Sci. Adv. 2018, 4, eaat1808. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, T.; Li, L.; Li, J.; Feng, Y.; Lu, Q. The Patterns and Drivers of Bacterial and Fungal β-Diversity in a Typical Dryland Ecosystem of Northwest China. Front. Microbiol. 2017, 8, 2126. [Google Scholar] [CrossRef]
- Zhang, Y.; Cong, J.; Lu, H.; Li, G.; Xue, Y.; Deng, Y.; Li, H.; Zhou, J.; Li, D. Soil bacterial diversity patterns and drivers along an elevational gradient on Shennongjia Mountain, China. Microb. Biotechnol. 2015, 8, 739–746. [Google Scholar] [CrossRef]
- Chi, F.; Shen, S.H.; Cheng, H.P.; Jing, Y.X.; Yanni, Y.G.; Dazzo, F.B. Ascending migration of endophytic Rhizobia from roots to leaves inside rice plants and assessment of their benefits to the growth physiology of rice. Appl. Environ. Microbiol. 2005, 71, 7271–7278. [Google Scholar] [CrossRef]
- Lan, G.Y.; Wei, Y.Q.; Li, Y.W.; Wu, Z.X. Diversity and assembly of root-associated microbiomes of rubber trees. Front. Plant Sci. 2023, 14, 1136418. [Google Scholar] [CrossRef]
- Ofek-Lalzar, M.; Sela, N.; Goldman-Voronov, M.; Green, S.J.; Hadar, Y.; Minz, D. Niche and host-associated functional signatures of the root surface microbiome. Nat. Commun. 2014, 5, 4950. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; van Themaat, E.V.L.; Schulze-Lefert, P. Structure and Functions of the Bacterial Microbiota of Plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef]
- Guttman, D.S.; McHardy, A.C.; Schulze-Lefert, P. Microbial genome-enabled insights into plant–microorganism interactions. Nat. Rev. Genet. 2014, 15, 797–813. [Google Scholar] [CrossRef]
- Korenblum, E.; Dong, Y.; Szymanski, J.; Panda, S.; Jozwiak, A.; Massalha, H.; Meir, S.; Rogachev, I.; Aharoni, A. Rhizosphere microbiome mediates systemic root metabolite exudation by root-to-root signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 3874–3883. [Google Scholar] [CrossRef]
- Bakker, P.A.H.M.; Berendsen, R.L.; Doornbos, R.F.; Wintermans, P.C.A.; Pieterse, C.M.J. The rhizosphere revisited: Root microbiomics. Front. Plant Sci. 2013, 4, 165. [Google Scholar] [CrossRef]
- Lareen, A.; Burton, F.; Schafer, P. Plant root-microbe communication in shaping root microbiomes. Plant Mol. Biol. 2016, 90, 575–587. [Google Scholar] [CrossRef]
- Reinhold-Hurek, B.; Bünger, W.; Burbano, C.S.; Sabale, M.; Hurek, T. Roots Shaping Their Microbiome: Global Hotspots for Microbial Activity. Annu. Rev. Phytopathol. 2015, 53, 403–424. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.; Johnson, C.; Santos-Medellin, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef] [PubMed]
- López, J.L.; Fourie, A.; Poppeliers, S.W.M.; Pappas, N.; Sánchez-Gil, J.J.; de Jonge, R.; Dutilh, B.E. Growth rate is a dominant factor predicting the rhizosphere effect. ISME J. 2023, 17, 1396–1405. [Google Scholar] [CrossRef] [PubMed]
- Turner, T.R.; Ramakrishnan, K.; Walshaw, J.; Heavens, D.; Alston, M.; Swarbreck, D.; Osbourn, A.; Grant, A.; Poole, P.S. Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. ISME J 2013, 7, 2248–2258. [Google Scholar] [CrossRef] [PubMed]
- Walters, W.A.; Jin, Z.; Youngblut, N.; Wallace, J.G.; Sutter, J.; Zhang, W.; Gonzalez-Pena, A.; Peiffer, J.; Koren, O.; Shi, Q.J.; et al. Large-scale replicated field study of maize rhizosphere identifies heritable microbes. Proc. Natl. Acad. Sci. USA 2018, 115, 7368–7373. [Google Scholar] [CrossRef]
- Oh, Y.M.; Kim, M.; Lee-Cruz, L.; Lai-Hoe, A.; Go, R.; Ainuddin, N.; Rahim, R.A.; Shukor, N.; Adams, J.M. Distinctive Bacterial Communities in the Rhizoplane of Four Tropical Tree Species. Microb. Ecol. 2012, 64, 1018–1027. [Google Scholar] [CrossRef]
- Fan, M.; Li, J.; Luan, X.; Yang, L.; Chen, W.; Ma, X.; Yang, Z.; Shangguan, Z. Biogeographical patterns of rhizosphere microbial communities in Robinia pseudoacacia forests along a north–south transect in the Loess Plateau. Geoderma 2023, 435, 116516. [Google Scholar] [CrossRef]
- Li, Y.M.; Yang, Y.; Wu, T.E.; Zhang, H.; Wei, G.H.; Li, Z.F. Rhizosphere bacterial and fungal spatial distribution and network pattern of Astragalus mongholicus in representative planting sites differ the bulk soil. Appl. Soil Ecol. 2021, 168, 104114. [Google Scholar] [CrossRef]
- Xiao, C.; Li, P.; Feng, Z. Monitoring annual dynamics of mature rubber plantations in Banna during 1987–2018 using Landsat time series data: A multiple normalization approach. Int. J. Appl. Earth Obs. Geoinf. 2019, 77, 30–41. [Google Scholar] [CrossRef]
- Xu, X.; Fei, Q.; Wei, Y.; Yang, C.; Zhang, H.; Lan, G. Diversity analysis of rhizosphere fungal communities of rubber trees in rubber plantation area of Hainan and Banna. J. South Agric. 2022, 53, 2392–2402. [Google Scholar] [CrossRef]
- Zhai, D.L.; Cannon, C.H.; Slik, J.W.F.; Zhang, C.P.; Dai, Z.C. Rubber and pulp plantations represent a double threat to Hainan’s natural tropical forests. J. Environ. Manag. 2012, 96, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Walters, W.; Hyde, E.R.; Berg-Lyons, D.; Ackermann, G.; Humphrey, G.; Parada, A.; Gilbert, J.A.; Jansson, J.K.; Caporaso, J.G.; Fuhrman, J.A.; et al. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 2016, 1, e00009-15. [Google Scholar] [CrossRef]
- Wei, Y.Q.; Lan, G.Y.; Wu, Z.X.; Chen, B.Q.; Quan, F.; Li, M.M.; Sun, S.Q.; Du, H.N. Phyllosphere fungal communities of rubber trees exhibited biogeographical patterns, but not bacteria. Environ. Microbiol. 2022, 24, 3777–3790. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.K. Methods for Soil and Agriculture Chemistry Analysis; Chinese Agricultural Science and Technology Press: Beijing, China, 1999; pp. 20–35. [Google Scholar]
- Ji, N.; Liang, D.; Clark, L.V.; Sacks, E.J.; Kent, A.D. Host genetic variation drives the differentiation in the ecological role of the native Miscanthus root-associated microbiome. Microbiome 2023, 11, 216. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Abdelfattah, A.; Wisniewski, M.; Schena, L.; Tack, A.J.M. Experimental evidence of microbial inheritance in plants and transmission routes from seed to phyllosphere and root. Environ. Microbiol. 2021, 23, 2199–2214. [Google Scholar] [CrossRef] [PubMed]
- He, R.; Zeng, J.; Zhao, D.; Wang, S.; Wu, Q.L. Decreased spatial variation and deterministic processes of bacterial community assembly in the rhizosphere of Phragmites australis across the Middle–Lower Yangtze plain. Mol. Ecol. 2021, 31, 1180–1195. [Google Scholar] [CrossRef] [PubMed]
- Xiong, C.; Zhu, Y.G.; Wang, J.T.; Singh, B.; Han, L.L.; Shen, J.P.; Li, P.P.; Wang, G.B.; Wu, C.F.; Ge, A.H.; et al. Host selection shapes crop microbiome assembly and network complexity. New Phytol. 2020, 229, 1091–1104. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Shenhav, L.; Thompson, M.; Joseph, T.A.; Briscoe, L.; Furman, O.; Bogumil, D.; Mizrahi, I.; Pe’er, I.; Halperin, E. FEAST: Fast expectation-maximization for microbial source tracking. Nat. Methods 2019, 16, 627–632. [Google Scholar] [CrossRef]
- Jiao, S.; Yang, Y.; Xu, Y.; Zhang, J.; Lu, Y. Balance between community assembly processes mediates species coexistence in agricultural soil microbiomes across eastern China. ISME J. 2020, 14, 202–216. [Google Scholar] [CrossRef]
- Attia, S.; Russel, J.; Mortensen, M.S.; Madsen, J.S.; Sørensen, S.J. Unexpected diversity among small-scale sample replicates of defined plant root compartments. ISME J. 2021, 16, 997–1003. [Google Scholar] [CrossRef]
- Walker, T.S.; Bais, H.P.; Grotewold, E.; Vivanco, J.M. Root Exudation and Rhizosphere Biology. Plant Physiol. 2003, 132, 44–51. [Google Scholar] [CrossRef]
- Bai, Y.; Muller, D.B.; Srinivas, G.; Garrido-Oter, R.; Potthoff, E.; Rott, M.; Dombrowski, N.; Munch, P.C.; Spaepen, S.; Remus-Emsermann, M.; et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nature 2015, 528, 364–369. [Google Scholar] [CrossRef]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; del Rio, T.G.; et al. Defining the core Arabidopsis thaliana root microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef]
- Thiergart, T.; Durán, P.; Ellis, T.; Vannier, N.; Garrido-Oter, R.; Kemen, E.; Roux, F.; Alonso-Blanco, C.; Ågren, J.; Schulze-Lefert, P.; et al. Root microbiota assembly and adaptive differentiation among European Arabidopsis populations. Nat. Ecol. Evol. 2019, 4, 122–131. [Google Scholar] [CrossRef]
- Perez-Jaramillo, J.E.; Carrion, V.J.; Bosse, M.; Ferrao, L.F.V.; de Hollander, M.; Garcia, A.A.F.; Ramirez, C.A.; Mendes, R.; Raaijmakers, J.M. Linking rhizosphere microbiome composition of wild and domesticated Phaseolus vulgaris to genotypic and root phenotypic traits. ISME J. 2017, 11, 2244–2257. [Google Scholar] [CrossRef] [PubMed]
- Agler, M.T.; Ruhe, J.; Kroll, S.; Morhenn, C.; Kim, S.T.; Weigel, D.; Kemen, E.M. Microbial Hub Taxa Link Host and Abiotic Factors to Plant Microbiome Variation. PLoS Biol. 2016, 14, e1002352. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Sui, Y.; Yu, Z.; Shi, Y.; Chu, H.; Jin, J.; Liu, X.; Wang, G. High throughput sequencing analysis of biogeographical distribution of bacterial communities in the black soils of northeast China. Soil Biol. Biochem. 2014, 70, 113–122. [Google Scholar] [CrossRef]
- Mueller, R.C.; Belnap, J.; Kuske, C.R. Soil bacterial and fungal community responses to nitrogen addition across soil depth and microhabitat in an arid shrubland. Front. Microbiol. 2015, 6, 891. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Smalla, K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 2009, 68, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Gottel, N.R.; Castro, H.F.; Kerley, M.; Yang, Z.; Pelletier, D.A.; Podar, M.; Karpinets, T.; Uberbacher, E.; Tuskan, G.A.; Vilgalys, R.; et al. Distinct microbial communities within the endosphere and rhizosphere of Populus deltoides roots across contrasting soil types. Appl. Environ. Microbiol. 2011, 77, 5934–5944. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Nuccio, E.E.; Shi, Z.J.; He, Z.; Zhou, J.; Firestone, M.K. The interconnected rhizosphere: High network complexity dominates rhizosphere assemblages. Ecol. Lett. 2016, 19, 926–936. [Google Scholar] [CrossRef] [PubMed]
- Berhongaray, G.; Janssens, I.A.; King, J.S.; Ceulemans, R. Fine root biomass and turnover of two fast-growing poplar genotypes in a short-rotation coppice culture. Plant Soil 2013, 373, 269–283. [Google Scholar] [CrossRef] [PubMed]


| Environment Variable | Hainan | Banna |
|---|---|---|
| Water content (%) | 19.58 ± 6.07 | 20.53 ± 8.27 |
| pH | 4.08 ± 0.37 | 4.31 ± 0.69 |
| Organic matter (%) | 1.5 ± 0.74 a | 2.62 ± 0.97 b |
| Ammonia nitrogen (mg·kg−1) | 7.82 ± 7.87 a | 14.73 ± 8.5 b |
| Nitrate nitrogen (mg·kg−1) | 9.06 ± 4.91 | 11.13 ± 8.25 |
| Available phosphorus (mg·kg−1) | 4.43 ± 4.35 | 3.7 ± 2.71 |
| Available potassium (mg·kg−1) | 44.85 ± 25.35 | 56.78 ± 50.53 |
| Total nitrogen (g·kg−1) | 0.15 ± 0.06 a | 0.26 ± 0.11 b |
| Total phosphorus (g·kg−1) | 0.06 ± 0.03 a | 0.1 ± 0.04 b |
| Total potassium (g·kg−1) | 1.71 ± 1.56 | 0.94 ± 0.65 |
| Elevation (m) | 155.89 ± 20.16 a | 665.11 ± 149.53 b |
| Rainfall (mm) | 1634.66 ± 324.18 | 1513.75 |
| Air temperature (°C) | 24.56 ± 0.69 a | 21.58 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Wei, Y.; Lan, G. Geographical Differences Weaken the Convergence Effect of the Rhizosphere Bacteria of Rubber Trees. Forests 2024, 15, 415. https://doi.org/10.3390/f15030415
Xu X, Wei Y, Lan G. Geographical Differences Weaken the Convergence Effect of the Rhizosphere Bacteria of Rubber Trees. Forests. 2024; 15(3):415. https://doi.org/10.3390/f15030415
Chicago/Turabian StyleXu, Xinnuo, Yaqing Wei, and Guoyu Lan. 2024. "Geographical Differences Weaken the Convergence Effect of the Rhizosphere Bacteria of Rubber Trees" Forests 15, no. 3: 415. https://doi.org/10.3390/f15030415
APA StyleXu, X., Wei, Y., & Lan, G. (2024). Geographical Differences Weaken the Convergence Effect of the Rhizosphere Bacteria of Rubber Trees. Forests, 15(3), 415. https://doi.org/10.3390/f15030415






