Abstract
Specialized metabolites correspond to millions of natural molecules from different chemical families depending on plant taxa that play a key role in ecological interactions during their life cycle. Due to their chemical properties, plants’ specialized metabolites have been exploited for a long time for various industrial applications. However, the limitations in natural population resources as well as the difficulties of their cultivation in terms of production quality or product safety have not always been satisfactory, notably for perennials such as forest trees. Reliable and eco-adapted practices for the production of specialized metabolites such as in vitro cultures provide a useful and powerful alternative to agronomic cultures. Modern omics have allowed the identification of metabolite pathways but have also raised the question of their complex regulation to improve their production. Among the major regulatory players, epigenetics have been shown in recent years to be involved in plant development and the response to environmental variations. Here, the state of the art concerning the epigenetic control of plant specialized metabolite in vitro production as well as the challenges in forest trees are presented.
1. Background and Problem
Specialized metabolites represent over two million natural molecules depending on plant taxa. In opposition to primary metabolites (proteins, lipids, sugars, and nucleic acids) accumulating in substantial amounts and essential for growth and development, secondary metabolites correspond to low-level and tissue-specific molecules playing key roles in ecological interactions with biotic or abiotic factors and in general adaptation to environmental variations [1,2]. Four major groups have been identified in plants: terpenoids, phenolic compounds, alkaloids, and sulfur-containing compounds. These molecules have a wide range of industrial applications in the pharmaceutic, cosmetic, or agri-food sectors. One major limitation for their industrial application is the availability the specialized metabolites. Indeed, their production yield is low and environmentally dependent. The use of natural resources as well as the difficulties in and time for cultivation of some species have led to the development of more reliable and eco-friendly practices through which to produce specialized metabolites using in vitro cultures. The most famous example is the production of paclitaxel by the Taxus tree species [3]. Recently, the current state of knowledge about specialized metabolites in forest trees as well as the opportunities and methods to improve their production was reported [1]. Forest trees are long-living plants, with high natural diversity in terms of species, populations, and individuals. Forest ecosystems are constantly subjected to microclimate changes and biotic interactions, giving rise to a complex diversity of natural compounds that are underestimated for human resources [1]. While omics studies, mostly transcriptomic and metabolomic, have allowed the identification of many metabolite pathways [4,5], in vitro production is still facing difficulties in producing high-yield and low-cost specialized metabolites. One limitation is the complex regulation of these pathways notably by epigenetic mechanisms [6]. Here, the rationale for and evidence of the epigenetic control of specialized metabolites in in vitro production are presented in trees when they exist or in plants before concluding on the corresponding challenges.
2. Epigenetics as a New Omics in the Study of In Vitro Plant Cell Cultures
Epigenetic corresponds to heritable changes through cell division (mitosis or meiosis) affecting genes and/or transposable elements’ activity that cannot be explained by changes in DNA sequence. Epigenetic mechanisms that modulate chromatin dynamics include DNA methylation, histone variants, histone post-translational modifications and to some extent regulation by non-coding RNAs [7]. Epigenetics represents a new layer of information at the interface between genetics and environment, notably in plants that display high phenotypic plasticity and genotype plasticity through environment (GxE) interactions [8]. Accordingly, epigenomics is a new level of omics for revealing phenomes, in direct interaction with genomics, transcriptomics, and downstream processes such as proteomics and metabolomics. It is now widely demonstrated that epigenetics controls plant development and environmental responses to biotic and abiotic stress [9]. Due to the key role of specialized metabolites in the same processes, the interplay between epigenetics and the production of specialized metabolites notably for in vitro cell cultures deserves particular attention [10,11]. While the role of epigenetics, particularly DNA methylation, in plant cell cultures is an over-30-year-old story [12,13,14], recent findings using multi-omics opened up innovative ideas and potential applications. To unravel this complex relationship, it will be necessary to address several points: (1) How does epigenetics affect in vitro developmental processes and vice versa? (2) How does epigenetics affect plant metabolism and vice versa, notably for the specialized metabolites produced? (3) How could epigenetics be used to improve the production of specialized metabolites?
3. Evidence for Epigenetic Control of the Specialized Metabolites In Vitro Production
Plant-cultured cells are usually obtained by inducing dedifferentiation from specialized tissues inheriting some epigenetic modifications [15]. This could be associated to the heterogeneity in their biosynthetic and growth properties [11]. Epigenetic regulation also plays a role in the dynamic reprogramming of cell fate to ensure the correct spatiotemporal expression pattern of the key regulators [16]. For example, the regeneration of distinct pairs of organogenic (shoot or root regeneration) and non-organogenic in vitro sugar beet (Beta vulgaris) cell lines derived from three parental lines showed that DNA methylation is reprogrammed in an organ-specific way during regeneration and that epialleles could be conserved between parental lines [17]. In addition, epigenetic changes arise during in vitro maintenance in relation to their yield variations and viability [11]. The most famous example is derived from studies on tree species; paclitaxel production in Taxus long-term cell cultures showed increased methylation with the prolongation of the culture time in relation to the gradual loss of the taxol biosynthesis capacity [3,18]. Recently, the promoters of three genes involved in taxane biosynthesis, GERANYLGERANYL DIPHOSPHATE SYNTHASE (GGPPS), the first gene of the diterpene pathway, the TAXADIENE SYNTHASE (TXS) gene, involved in the first step of taxane biosynthesis, and the 3′-N-DEBENZOYL-2′-DEOXYTAXOL-N-BEZOYLTRANSFERASE (DBTNBT) gene, the last gene of the pathway, were analyzed to compare the methylation patterns between a new line and a 14-year-old one [19]. While the central promoter of the GGPPS gene is protected from cytosine methylation accumulation, TXS and DBTNBT promoters accumulate methylation at various levels particularly in the old line and may affect the binding of transcription factors. These in vitro-induced epimutations (somaclonal variation) [13] have been well reported and have been shown to induce phenotypic variations in regenerated plants [9,11,14]. Indeed, they have been observed through somatic embryogenesis in the ‘mantle’ epimutant in oil palm (Elaies guineensis) [20] or in norway spruce (Picea abies) epitypes obtained after embryo exposure at different temperatures [21,22]. Finally, these in vitro-induced epimutations can also activate transposable elements (TEs) that generate somatic mutations [12,23,24]. Altogether, developmental processes (in vitro or in planta) are under epigenetic control and in vitro (long-lasting) conditions can induce epi/mutations affecting cell fate and notably their ability to produce specialized metabolites. A better knowledge of epigenetic regulation could help in designing novel strategies to enhance the biotechnological production of specialized metabolites in various species and over time.
While extensive connections between metabolism and epigenetics have been established mostly in animal models, recent reports are also now available on plants [7,25]. Several metabolites are cofactors or targets of chromatin-modifying enzymes. For example, acetyl CoA participates in the biosynthesis of amino acids, lipids and secondary metabolites, and is necessary for histone acetylation. S-Adenosyl-L-methionine (SAM) is also a general methyl donor group involved in both primary and secondary metabolisms and is also required for both DNA and histone methylation. Then, acetylome and methylome are complex networks coordinating nuclear activities (including epigenetic control) with the metabolic status of the cell, allowing its adaption to stress [7,25]. Beyond this complex interaction, recent insights into the hierarchical and combinatorial control of plant specialized metabolism including all regulatory mechanisms known to date have been reported, particularly at the epigenetic level [26]. Recent in planta studies have characterized epigenetic control of the production of various specialized metabolites such as aroma indole compounds [27], anthocyanins [5,28,29], phenylpropanoids [30], glucosinolates [4] or volatile organic compounds (VOCs) [31]. Altogether, genes related to the specialized metabolite pathway are under epigenetic control in the interplay between transcription factors (FTs) and their promoter regions in relation to the phytohormonal and redox state signaling pathways.
Recent studies using an integrative multi-omics approach have brought new insights: Shirai et al. (2021) showed that only the methylation of CG sites located in promoter regions of genes associated with specialized metabolites have a selective sweep signature in A. thaliana populations [32]. Dugé de Bernonville et al. (2020), investigating the developmental and environmental methylomes of Catharanthus roseus, have unraveled the epigenetic regulation of specialized metabolisms (indolic alkaloids). Epigenetic control was shown to target transcription factors that in turn may regulate the expression of enzyme-encoding genes [33]. Finally, Zhou et al. (2021), studying artemisinin production in Artemisia annua glandular trichomes (GT) versus leaves (with GT removed via gentle brushing), using the assay for transposase-accessible chromatin with sequencing (ATAC-seq), found that several genes involved in artemisinin biosynthesis have more accessible regions in GT compared to that in the leaf. This implies that these genes might be regulated by chromatin accessibility [34], which could also have been evaluated using other technical approaches with DNase I (DNase-Seq), formaldehyde-assisted isolation of regulatory elements (FAIRE-Seq), or micrococcal nuclease digestion (MNase-seq). Altogether, epigenetics may play a key role in controlling plant specialized metabolite pathways but the machinery behind this process remains unclear, awaiting further research, notably using in vitro systems.
Recently, Sanchez-Muñoz et al. (2019) and Ghosh et al. (2021) summarized studies on epigenetic changes during in vitro long-term maintenance, the mechanisms involved and their potential applications [10,11]. They proposed that de novo methylation process(es) involving domain-rearranged methylase (DRM) enzymes may be responsible for the decline in secondary metabolism during in vitro culture maintenance. Accordingly, a promising strategy will be to reverse this decline, allowing the recovery of the original production ratios or more generally allowing the control of cell fate and the ability of cells to produce specialized metabolites. The use of epigenetic drugs on Beta vulgaris cell cultures was shown to control their differentiation state as well as the production of phenolic compounds in relation to their redox state [35,36]. In Vitis amurensis cell cultures, the use of the demethylating agent azacytosine (AzaC) restored the transgene transcription rate and the initial concentrations of resveratrol. The treatment did not cause a loss of biomass, indicating its suitability for use in long-term biofactories to avoid methylation accumulation [37]. Recently, grapevine cell suspensions (Gamay Teinturier) treated with the hypomethylating drug zebularine, were shown to display increased anthocyanin accumulation in the light, and induced production in the dark. Interestingly, the expression of the UDP GLUCOSE: FLAVONOID-3-O-GLUCOSYLTRANSFERASE (UFGT) gene, known to be critical for anthocyanin biosynthesis, was shown to be regulated by DNA methylation [38]. Finally, using histone deacetylase (HDAC) inhibitors in bamboo cultured plant cells (Bambusa multiplex), Nomura et al. (2021) succeeded in activating cryptic secondary metabolite biosynthesis [39].
4. Challenges and Opportunities
Potential applications of epigenetics for crop improvement and particularly using in vitro systems have been extensively reviewed [8,9,10,11]. The focus here is on forest trees, which represent an important and scarcely exploited source of natural specialized metabolites with a wide range of applications for human [1] (Figure 1). The generation of epi/genetically modified cell lines using various technical approaches (genetic, pharmacologic, and based on genomic edition) specifically blocking the DRM pathway or affecting phytohormonal or redox signaling pathways could provide resistance to silencing and ensure stable or even increased (cryptic) highly productive bioreactor cultures (Figure 1). For example, epigenetic marks such as DNA methylation can induce gene silencing [7,9] and epi/genetically modified cell lines could prevent gene silencing, leading to a decrease in secondary metabolite production during cell culture. This is of the highest importance since the loss of yields during cell culture maintenance is a major barrier in the optimization and scale-up of plant cell biofactories for their commercial application [10]. Another strategy could be to induce epigenetic modifications activating transposition [40] (Figure 1). Thieme et al. (2017) induced in Arabidopsis thaliana the inhibition of DNA methylation and/or RNA polymerase II activity and observed the strong stress-dependent mobilization of the heat-responsive copia-like retrotransposon ONSEN. These transpositions have introduced novel genetic (stable) variations. This strategy could be tested on cell lines and be evaluated for the potential impact on their production of specialized metabolites. Another potential contribution of epigenetics could be the use of epimarkers to identify the clonal fidelity of the regenerants or their origin, or to select or predict highly producing in vitro material [9] (Figure 1). For example, in sugar beet cell lines, Maury et al. (2012) reported epimarkers distinguishing three pairs of organogenic and non-organogenic cell lines conserved between parental lines [17].
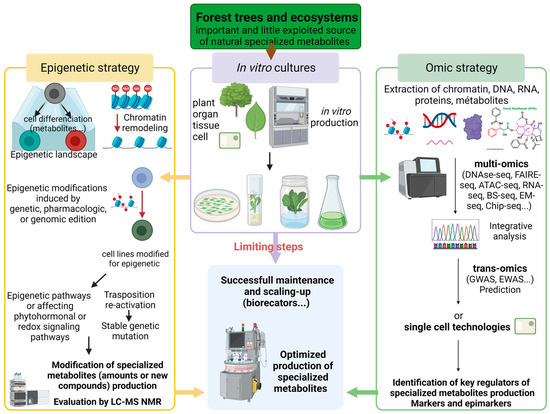
Figure 1.
Global strategy using epigenetics and omics for the optimization of specialized metabolite production from tree in vitro cultures. (Created with BioRender.com; agreement number: XA26BESHD9).
5. Conclusions
The optimization of forest tree in vitro producing systems could also be inspired by recent publications on epigenetics as an additional tool for fungal biotechnology [41,42] (Figure 1). For example, coupling the high potential of epigenetic manipulation with the discovery of new molecules, i.e., using the LC-MS-NMR strategy for unlocking the chemical diversity encoded by fungal genomes [43], could now be applied to plants in in vitro systems. The next method to overcome the bottlenecks of in vitro systems and go beyond fragmentary and correlative studies will be to use genomic methodologies at the whole-genome level [11,44]. Indeed, it is important beyond genes to consider their regulatory sequences as well as transposable elements, and to develop multi-omics approaches with RNA-seq, proteomics, chromatin immuno-precipitation (Chip)-seq, bisulfite (BS) or enzymatic (EM)-seq, or trans-omics strategies with genome-wide association studies (GWAS) or epigenetic €EGWAS, but also to develop single-cell (sc) technologies such as scATAC-seq on one dedicated tree in an in vitro producing system from initiation to maintenance and scale-up (Figure 1).
Funding
This research was supported by ‘Le Studium Loire Valley Institute for Advanced Studies’ in the frame of the research network ‘CosmeNovIC’ (plant natural product production via novel in vitro culture approaches for improved cosmeceutical efficiency and security; https://www.lestudium-ias.com/research-networks/consortia/cosmenovic-plant-natural-products-production-novel-vitro-culture, accessed on 15 July 2019).
Data Availability Statement
No new data were created.
Acknowledgments
The author thanks the European COST actions EPICATCH (CA19125) and COPYTREE (CA21157) for their help in building research networks and discussions on epigenetics and in vitro techniques for trees, respectively.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Nawrot-Chorabik, K.; Sułkowska, M.; Gumulak, N. Secondary Metabolites Produced by Trees and Fungi: Achievements So Far and Challenges Remaining. Forests 2022, 13, 1338. [Google Scholar] [CrossRef]
- Erb, M.; Kliebenstein, D.J. Plant Secondary Metabolites as Defenses, Regulators, and Primary Metabolites: The Blurred Functional Trichotomy. Plant Physiol. 2020, 184, 39–52. [Google Scholar] [CrossRef]
- Liu, Y.; Zhao, F.; Wang, Q.; Zhao, Q.; Hou, G.; Meng, Q. Current Perspectives on Paclitaxel: Focus on Its Production, Delivery and Combination Therapy. Mini Rev. Med. Chem. 2023, 23, 1780–1796. [Google Scholar] [CrossRef] [PubMed]
- Mitreiter, S.; Gigolashvili, T. Regulation of Glucosinolate Biosynthesis. J. Exp. Bot. 2021, 72, 70–91. [Google Scholar] [CrossRef]
- Cappellini, F.; Marinelli, A.; Toccaceli, M.; Tonelli, C.; Petroni, K. Anthocyanins: From Mechanisms of Regulation in Plants to Health Benefits in Foods. Front. Plant Sci. 2021, 12, 748049. [Google Scholar] [CrossRef] [PubMed]
- Kliebenstein, D.J. Is Specialized Metabolite Regulation Specialized? J. Exp. Bot. 2023, 74, 4942–4948. [Google Scholar] [CrossRef]
- Mladenov, V.; Fotopoulos, V.; Kaiserli, E.; Karalija, E.; Maury, S.; Baranek, M.; Segal, N.; Testillano, P.S.; Vassileva, V.; Pinto, G.; et al. Deciphering the Epigenetic Alphabet Involved in Transgenerational Stress Memory in Crops. Int. J. Mol. Sci. 2021, 22, 7118. [Google Scholar] [CrossRef]
- Maury, S.; Sow, M.D.; Le Gac, A.-L.; Genitoni, J.; Lafon-Placette, C.; Mozgova, I. Phytohormone and Chromatin Crosstalk: The Missing Link for Developmental Plasticity? Front. Plant Sci. 2019, 10, 395. [Google Scholar] [CrossRef]
- Kakoulidou, I.; Avramidou, E.V.; Baránek, M.; Brunel-Muguet, S.; Farrona, S.; Johannes, F.; Kaiserli, E.; Lieberman-Lazarovich, M.; Martinelli, F.; Mladenov, V.; et al. Epigenetics for Crop Improvement in Times of Global Change. Biology 2021, 10, 766. [Google Scholar] [CrossRef]
- Sanchez-Muñoz, R.; Moyano, E.; Khojasteh, A.; Bonfill, M.; Cusido, R.M.; Palazon, J. Genomic Methylation in Plant Cell Cultures: A Barrier to the Development of Commercial Long-Term Biofactories. Eng. Life Sci. 2019, 19, 872–879. [Google Scholar] [CrossRef]
- Ghosh, A.; Igamberdiev, A.U.; Debnath, S.C. Tissue Culture-Induced DNA Methylation in Crop Plants: A Review. Mol. Biol. Rep. 2021, 48, 823–841. [Google Scholar] [CrossRef] [PubMed]
- Peschke, V.M.; Phillips, R.L.; Gengenbach, B.G. Discovery of Transposable Element Activity Among Progeny of Tissue Culture—Derived Maize Plants. Science 1987, 238, 804–807. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.T.H. DNA Methylation in Plants and Its Role in Tissue Culture. Genome 1989, 31, 717–729. [Google Scholar] [CrossRef]
- Kaeppler, S.M.; Phillips, R.L. Tissue Culture-Induced DNA Methylation Variation in Maize. Proc. Natl. Acad. Sci. USA 1993, 90, 8773–8776. [Google Scholar] [CrossRef] [PubMed]
- Wibowo, A.; Becker, C.; Durr, J.; Price, J.; Spaepen, S.; Hilton, S.; Putra, H.; Papareddy, R.; Saintain, Q.; Harvey, S.; et al. Partial Maintenance of Organ-Specific Epigenetic Marks during Plant Asexual Reproduction Leads to Heritable Phenotypic Variation. Proc. Natl. Acad. Sci. USA 2018, 115, E9145–E9152. [Google Scholar] [CrossRef]
- Jing, T.; Ardiansyah, R.; Xu, Q.; Xing, Q.; Müller-Xing, R. Reprogramming of Cell Fate During Root Regeneration by Transcriptional and Epigenetic Networks. Front. Plant Sci. 2020, 11, 317. [Google Scholar] [CrossRef] [PubMed]
- Maury, S.; Trap-Gentil, M.-V.; Hébrard, C.; Weyens, G.; Delaunay, A.; Barnes, S.; Lefebvre, M.; Joseph, C. Genic DNA Methylation Changes during in Vitro Organogenesis: Organ Specificity and Conservation between Parental Lines of Epialleles. Physiol. Plant 2012, 146, 321–335. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Li, L.; Wu, W.; Li, M.; Yu, X.; Yu, L. Assessment of Genetic and Epigenetic Variation during Long-Term Taxus Cell Culture. Plant Cell Rep. 2012, 31, 1321–1331. [Google Scholar] [CrossRef]
- Escrich, A.; Cusido, R.M.; Bonfill, M.; Palazon, J.; Sanchez-Muñoz, R.; Moyano, E. The Epigenetic Regulation in Plant Specialized Metabolism: DNA Methylation Limits Paclitaxel In Vitro Biotechnological Production. Front. Plant Sci. 2022, 13, 899444. [Google Scholar] [CrossRef]
- Ong-Abdullah, M.; Ordway, J.M.; Jiang, N.; Ooi, S.-E.; Kok, S.-Y.; Sarpan, N.; Azimi, N.; Hashim, A.T.; Ishak, Z.; Rosli, S.K.; et al. Loss of Karma Transposon Methylation Underlies the Mantled Somaclonal Variant of Oil Palm. Nature 2015, 525, 533–537. [Google Scholar] [CrossRef]
- Kvaalen, H.; Johnsen, Ø. Timing of Bud Set in Picea Abies Is Regulated by a Memory of Temperature during Zygotic and Somatic Embryogenesis. New Phytol. 2008, 177, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Carneros, E.; Yakovlev, I.; Viejo, M.; Olsen, J.E.; Fossdal, C.G. The Epigenetic Memory of Temperature during Embryogenesis Modifies the Expression of Bud Burst-Related Genes in Norway Spruce Epitypes. Planta 2017, 246, 553–566. [Google Scholar] [CrossRef] [PubMed]
- Orłowska, R.; Machczyńska, J.; Oleszczuk, S.; Zimny, J.; Bednarek, P.T. DNA Methylation Changes and TE Activity Induced in Tissue Cultures of Barley (Hordeum vulgare L.). J. Biol. Res.-Thessalon. 2016, 23, 19. [Google Scholar] [CrossRef] [PubMed]
- Quesneville, H. Twenty Years of Transposable Element Analysis in the Arabidopsis thaliana Genome. Mobile DNA 2020, 11, 28. [Google Scholar] [CrossRef] [PubMed]
- Leung, J.; Gaudin, V. Who Rules the Cell? An Epi-Tale of Histone, DNA, RNA, and the Metabolic Deep State. Front. Plant Sci. 2020, 11, 181. [Google Scholar] [CrossRef]
- Lacchini, E.; Goossens, A. Combinatorial Control of Plant Specialized Metabolism: Mechanisms, Functions, and Consequences. Annu. Rev. Cell Dev. Biol. 2020, 36, 291–313. [Google Scholar] [CrossRef]
- Yang, J.; Zhou, X.; Wu, S.; Gu, D.; Zeng, L.; Yang, Z. Involvement of DNA Methylation in Regulating the Accumulation of the Aroma Compound Indole in Tea (Camellia sinensis) Leaves during Postharvest Processing. Food Res. Int. 2021, 142, 110183. [Google Scholar] [CrossRef]
- Li, W.-F.; Ning, G.-X.; Mao, J.; Guo, Z.-G.; Zhou, Q.; Chen, B.-H. Whole-Genome DNA Methylation Patterns and Complex Associations with Gene Expression Associated with Anthocyanin Biosynthesis in Apple Fruit Skin. Planta 2019, 250, 1833–1847. [Google Scholar] [CrossRef]
- Sicilia, A.; Scialò, E.; Puglisi, I.; Lo Piero, A.R. Anthocyanin Biosynthesis and DNA Methylation Dynamics in Sweet Orange Fruit [Citrus sinensis L. (Osbeck)] under Cold Stress. J. Agric. Food Chem. 2020, 68, 7024–7031. [Google Scholar] [CrossRef]
- Dong, N.-Q.; Lin, H.-X. Contribution of Phenylpropanoid Metabolism to Plant Development and Plant–Environment Interactions. J. Integr. Plant Biol. 2021, 63, 180–209. [Google Scholar] [CrossRef]
- Picazo-Aragonés, J.; Terrab, A.; Balao, F. Plant Volatile Organic Compounds Evolution: Transcriptional Regulation, Epigenetics and Polyploidy. Int. J. Mol. Sci. 2020, 21, 8956. [Google Scholar] [CrossRef] [PubMed]
- Shirai, K.; Sato, M.P.; Nishi, R.; Seki, M.; Suzuki, Y.; Hanada, K. Positive Selective Sweeps of Epigenetic Mutations Regulating Specialized Metabolites in Plants. Genome Res. 2021, 31, 1060–1068. [Google Scholar] [CrossRef] [PubMed]
- Dugé de Bernonville, T.; Maury, S.; Delaunay, A.; Daviaud, C.; Chaparro, C.; Tost, J.; O’Connor, S.E.; Courdavault, V. Developmental Methylome of the Medicinal Plant Catharanthus roseus Unravels the Tissue-Specific Control of the Monoterpene Indole Alkaloid Pathway by DNA Methylation. Int. J. Mol. Sci. 2020, 21, 6028. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Huang, Y.; Wang, Q.; Guo, D. Chromatin Accessibility Is Associated with Artemisinin Biosynthesis Regulation in Artemisia annua. Molecules 2021, 26, 1194. [Google Scholar] [CrossRef]
- Causevic, A.; Delaunay, A.; Ounnar, S.; Righezza, M.; Delmotte, F.; Brignolas, F.; Hagège, D.; Maury, S. DNA Methylating and Demethylating Treatments Modify Phenotype and Cell Wall Differentiation State in Sugarbeet Cell Lines. Plant Physiol. Biochem. 2005, 43, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Causevic, A.; Gentil, M.-V.; Delaunay, A.; El-Soud, W.A.; Garcia, Z.; Pannetier, C.; Brignolas, F.; Hagège, D.; Maury, S. Relationship between DNA Methylation and Histone Acetylation Levels, Cell Redox and Cell Differentiation States in Sugarbeet Lines. Planta 2006, 224, 812–827. [Google Scholar] [CrossRef]
- Tyunin, A.P.; Kiselev, K.V.; Zhuravlev, Y.N. Effects of 5-Azacytidine Induced DNA Demethylation on Methyltransferase Gene Expression and Resveratrol Production in Cell Cultures of Vitis amurensis. Plant Cell Tiss. Organ. Cult. 2012, 111, 91–100. [Google Scholar] [CrossRef]
- Kong, J.; Garcia, V.; Zehraoui, E.; Stammitti, L.; Hilbert, G.; Renaud, C.; Maury, S.; Delaunay, A.; Cluzet, S.; Lecourieux, F.; et al. Zebularine, a DNA Methylation Inhibitor, Activates Anthocyanin Accumulation in Grapevine Cells. Genes 2022, 13, 1256. [Google Scholar] [CrossRef]
- Nomura, T.; Yoneda, A.; Ogita, S.; Kato, Y. Activation of Cryptic Secondary Metabolite Biosynthesis in Bamboo Suspension Cells by a Histone Deacetylase Inhibitor. Appl. Biochem. Biotechnol. 2021, 193, 3496–3511. [Google Scholar] [CrossRef]
- Thieme, M.; Lanciano, S.; Balzergue, S.; Daccord, N.; Mirouze, M.; Bucher, E. Inhibition of RNA Polymerase II Allows Controlled Mobilisation of Retrotransposons for Plant Breeding. Genome Biol. 2017, 18, 134. [Google Scholar] [CrossRef]
- Poças-Fonseca, M.J.; Cabral, C.G.; Manfrão-Netto, J.H.C. Epigenetic Manipulation of Filamentous Fungi for Biotechnological Applications: A Systematic Review. Biotechnol. Lett. 2020, 42, 885–904. [Google Scholar] [CrossRef] [PubMed]
- Serrano, R.; González-Menéndez, V.; Martínez, G.; Toro, C.; Martín, J.; Genilloud, O.; Tormo, J.R. Metabolomic Analysis of The Chemical Diversity of South Africa Leaf Litter Fungal Species Using an Epigenetic Culture-Based Approach. Molecules 2021, 26, 4262. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Zou, Z.-M. Discovery of New Secondary Metabolites by Epigenetic Regulation and NMR Comparison from the Plant Endophytic Fungus Monosporascus eutypoides. Molecules 2020, 25, 4192. [Google Scholar] [CrossRef] [PubMed]
- Dugé de Bernonville, T.; Daviaud, C.; Chaparro, C.; Tost, J.; Maury, S. From Methylome to Integrative Analysis of Tissue Specificity. Methods Mol. Biol. 2022, 2505, 223–240. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).