Abstract
Clonal plantation involves the rooting of cuttings from superior genotypes selected for their hybrid vigor and desired qualities. However, the cuttings of some Eucalyptus species and their hybrid genotypes present difficulties in their rooting capacity. Applying PGPR to cutting growth medium as a root stimulating agent has not been extensively studied for Eucalyptus tree species. We aimed to assess the rooting capacity of cuttings taken from two poor-rooting Eucalyptus hybrid clones of E. grandis × E. nitens through the application of PGPR in nursery trials. Seven rhizospheric bacterial species that demonstrated the ability to produce indole-3-acetic acid and to solubilise phosphate were used to prepare two rhizospheric consortium inoculums in which Pseudomonas-Bacillus strains and non-Pseudomonas-Bacillus were grouped. Inoculums were tested for their rooting stimulating capacity on cuttings of the hybrids GN 018B and GN 010 and compared to the nursery standard indole-3-butyric acid. A total of 320 cuttings were treated. Both hybrid clones demonstrated significant (p < 0.0001) genotype differences for all three growth responses, i.e., total, root, and shoot length. Cuttings of both hybrids demonstrated high survival rates and rooting percentage. Although several rooting architectural configurations were prevalent, the Pseudomonas-Bacillus consortium promoted adventitious root development and fibrosity in GN 018B hybrids.
1. Introduction
Worldwide demand for hardwood from commercial plantations is rising and projected to increase further by 2030, in particular for Eucalyptus species [1]. Traditionally, breeding with Eucalyptus species has involved the methods of intraspecific and interspecific hybridisation and the selection of superior genotypes presenting tolerance to several environmental factors [2,3]. Intraspecific and interspecific Eucalyptus hybrids are multiplied through vegetative propagation, a method widely used for the establishment of clonal plantations [4]. Young Eucalyptus hybrid clonal plants are commercially multiplied by the rooting of cuttings in a nursery [5]. A major drawback that is faced by the commercial forestry industry is the manifestation of poor or variable rooting capacity of certain genotypes, especially Eucalyptus nitens, resulting in considerable losses [6].
The improvement of rooting percentages of clonal cuttings that mitigate losses through poor survival and low rooting percentages will contribute to more efficient and cost-effective clonal forestry practices. Research is thus required to develop improved Eucalyptus rooting regimes that can be applied in commercial forestry nurseries. One of the strategies to improve rooting capacity in tree species is that of using rhizospheric bacteria to stimulate rooting [7,8]. Rhizospheric bacteria that supports plant growth are referred to as plant-growth-promoting rhizobacteria (PGPR). Rhizospheric bacteria can stimulate plant growth and more recently have also shown to increase the rooting of cuttings [7,9,10,11,12].
PGPR are a varied group of rhizospheric bacteria that reside in rhizospheres, and at root surfaces of plants. PGPR are in symbiotic association with their host plants and feed on sloughed-off cells and root exudates while, in return, improving the quality of plant growth either directly and or indirectly [13]. PGPR contribute directly to plant growth either through providing plant-growth-promoting substances or by facilitating the uptake of certain plant nutrients from the soil. PGPR may also promote plant growth indirectly by mitigating or preventing the damaging effect of one or more phytopathogenic microorganisms. A number of non-pathogenic PGPR belonging to the genera Agrobacterium, Alcaligens, Arthrobacter, Azospirillum, Azotobacter, Bacillus, Brassica, Burkholderia, Enterobacter, Klebsiella, Pseudomonas, Serratia and Streptomyces have been reported to enhance plant growth either directly or indirectly [10,14,15].
The exact mechanisms by which PGPR promote plant growth are not fully understood. However, a number of PGPR traits are involved in the promotion of plant growth. One of the more important traits is their ability to produce or change the concentrations of plant growth regulators, such as for auxin, especially indole-3-acetic acid (IAA) [16,17]. PGPR in the rhizosphere synthesises IAA as exogenous auxin [10,18]. PGPR-produced exogenous auxin can also enhance endogenous IAA production in the host plant itself [10]. A high endogenous IAA concentration is normally associated with a high rooting rate at the beginning of the rooting process [19,20]. This complementary production of IAA can result in a peak IAA concentration that initiates the rooting process in a cutting [21,22]. IAA thus stimulates lateral and adventitious root development, thereby increasing the number of nutrient-absorbing surfaces, which results in better assimilation of water and nutrients from the soil [23].
Another important trait is the ability of PCPR to solubilise mineral phosphate and other nutrients [24,25]. The ability of some PGPR to solubilise phosphate allows IAA to promote plant growth. Phosphate-solubilising rhizospheric bacteria increase the available phosphorus for a plant, especially in soils with large amounts of precipitated phosphate [14,25,26]. These rhizospheric bacteria release bound phosphate by secreting a number of organic acids [14,25].
In South Africa, E. grandis and its hybrids are by far the most widely cultivated Eucalyptus genotypes [27,28,29,30]. E. grandis is relatively fast growing and most commonly used as a source of pulpwood, fuel, and timber. The cuttings of this species root relatively well, making it suitable for cloning. However, the cuttings of some of the most sought-after hybrids of E. grandis, particularly E. nitens, have demonstrated relatively poor rooting (personal communication, CSIR, South Africa). The root-promoting effect of PGPR has been demonstrated in a number of forest tree species [10] but has not been extensively studied for Eucalyptus [7,10]. However, not all rhizospheric bacteria act as PGPR. To exploit the root-growth-promoting attributes of rhizospheric bacteria, specific strains need to be isolated and screened for their potential root-promoting abilities. Therefore, the aim of this investigation was to improve the rooting capacity of cuttings of Eucalyptus hybrid clones of E. grandis × E. nitens, which is known for its relatively low rooting ability, through the application of isolated and characterised plant-growth-promoting rhizobacteria.
2. Materials and Methods
2.1. Isolation of Rhizospheric Bacteria
Rhizospheric bacteria were isolated from rhizospheric soil of ten (10) clones of Eucalyptus hybrid genotype, E. grandis × E. urophylla (GU), which is known for its relatively high rooting capacity. Soil samples were collected in nursery tunnels at Sunshine Seedlings near Pietermaritzburg, South Africa from a 2 mm radius around the roots using a sterile teaspoon. Approximately 60 mL (4 scoops) of soil were collected from each plant, placed in a sterile plastic bag, and kept on ice while in transport to the laboratory. Soil samples (3.5 g) were mixed with 100 mL of sterile distilled water in a conical flask. Flasks were incubated at 25 °C with shaking (120 rpm) for 20 min. Thereafter samples were diluted and plated on potato dextrose agar (PDA) and R2A agar using an easySpiral® Pro automatic plater (interscience, Saint Nom, France). Plates were incubated at 25 °C for 48 h. After incubation, representatives of bacterial colonies presenting different morphological variations (colour, shape, and size) were isolated and purified by sequentially streaking single colonies onto fresh agar plates [31,32]. Purified isolates were cryo-preserved at −80 °C in a Microbank™ (Pro-Lab Diagnostics, Bromborough, UK).
2.2. DNA Extraction, Amplification and Sequencing
Genomic DNA (gDNA) was extracted from bacterial cells using a harsh lysis manual extraction method [33]. gDNA concentration and purity were determined using a NanoDrop™ 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and integrity analysed by 0.8% agarose gel electrophoresis. Targeted amplification of the V1–V9 regions of the 16S rRNA gene was performed using primer set 63-F (5′-CAG GCC TAA CAC ATG CAA GTC-3′) and 1387-R (5′-GGG CGG WGT GTA CAA GGC-3′) [34,35]. Amplification was performed in a C1000™Thermal Cycler (Bio-Rad, Hercules, CA, USA) in 25 μL reactions containing 1 µL of gDNA, 1× ThermoPol® Reaction buffer (20 mM Tris-HCl; 10 mM (NH4)2SO4; 10 mM KCl; 2 mM MgSO4; 0.1% Triton X-100, pH 8.8), 0.2 mM dNTPs, 0.5 μM of each primer, 0.1 μg mL−1 of BSA, and 1 U of Taq DNA Polymerase (New England BioLabs, Ipswich, MA, USA). The following PCR conditions were applied: initial denaturation at 94 °C for 3 min, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, and elongation at 68 °C for 1 min and 30 s. A final elongation step was incorporated at 68 °C for 5 min. Successful amplification was confirmed by electrophoresis on 1% agarose gel. Bands of the appropriate lengths (≈1300 bp) were excised from the gel, purified using an illustra™ GFX™ PCR DNA and Gel Band Purification Kit (GE Healthcare Life Sciences, Chicago, IL, USA), and used as template for sequencing.
Sequencing was performed in single reactions using primers 63-F and 1387-R, as well as internal primers 533-F (5′-GTG CCA GCM GCC GCG GTA A-3′) and 805-R (5′-GAC TAC CAG GGT ATC TAA TC-3′) on an ABI Prism 3130 XL genetic analyser using a BigDye® Terminator V3.1 Cycle Sequencing Kit (Life Technologies, Carlsbad, CA, USA). DNA was prepared for sequencing by precipitation using EDTA and ethanol. Sequences were assembled using DNA Baser Assembler v5 15.0 software, and the contig construct was used to determine sequence homology. Sequence homology searches were undertaken by comparing each of the contig constructs against nucleotide sequence databases (nucleotide collection/16S ribosomal RNA sequences). The BLAST server of the National Centre for Biotechnology Information (NCBI) was used for the respective sequence homology searches using the BLAST algorithm (megablast). Only similarities with a BLAST index of 98% and above were considered for identification (Table 1).

Table 1.
BLAST homology results compared to 16S ribosomal RNA gene partial sequences.
2.3. Screening Bacterial Isolates for IAA Production and Phosphate Solubilisation
A qualitative approach was applied to determine if a bacterial isolate possessed the ability to produce indole-3-acetic acid (IAA) and solubilise phosphate. The bacterial strains were revived from the cryo-preservation by plating on potato dextrose agar (PDA) and incubated for 24 h at 25 °C. Thereafter, single colonies were spot plated onto fresh agar plates and incubated for 48 h at 25 °C. After incubation, 50 mL of Salkowaski reagent was prepared containing 0.5 M FeCl3 in 35% HClO4 in a ratio of 1:5 (v v−1). One millilitre of the Salkowaski reagent was poured in a thin layer over the bacterial growth on the agar plates. Bacterial strains with the ability to produce IAA showed a characteristic pink colouration [17,36]. Phosphate solubilisation was determined by growing isolates on PDA supplemented with 0.5% tribasic phosphate (Ca5HO13P3), 0.05% (NH4)2SO4, 0.02% KCl2, 0.01% MgSO4·7H2O and 0.05% yeast extract. A clear halo was visible around colonies able to solubilise phosphates after incubation at 25 °C for 7 days [37].
2.4. Preparation of Bacterial Inoculum for Field Trials
Growth parameters of the 7 bacterial species selected for field application were determined in suspension culture by monitoring optical density (OD620nm) and colony proliferation (CFU mL−1). OD was measured using a SpectraMax® M3 Multi-Mode Microplate Reader (Molecular Devices, San Jose, CA, USA) and spread plating and enumeration with an easySpiral®Pro and high-resolution Scan® 1200 automatic colony counter, respectively. Growth was followed for a period of 10 h with incubation performed in nutrient broth at 25 °C with shaking (150 rpm).
Separate suspensions were prepared for each bacterial species, which were then mixed prior to the application of the treatments in the field trials (Table 2). Four 300 mL cultures were prepared for each species with cell densities of ≈107 CFU mL−1. Cell suspensions of each species were harvested in mid exponential growth phase at selected time intervals based on growth data. The content of each flask was aseptically transferred to a 500 mL sterile Schott bottle, which contained 45 g glycerol (15% final concentration), then mixed gently and stored at −80 °C. Viability, indole-3-acetic acid production, and phosphate solubilisation of the bacterial colonies were determined before freezing and after thawing (before use in field trials). Schott bottles containing the bacterial inoculums were transported to the field in a mobile freezer at −25 °C and allowed to thaw overnight before application. Two mixtures containing different species were prepared in sterile 2 L Schott bottles immediately prior to application and dispensed using sterile VITLAB® genius2 5 mL bottle-top dispensers (VITLAB, Grossostheim, Germany).

Table 2.
Treatment description and composition used for field trail application.
2.5. Preparation of Treatments
Four rooting treatments were prepared and tested in the nursery setting. The nursery standard (T1) was applied as the control treatment. The nursery standard consisted of the application of commercially available Seradix 2 to cuttings prior to their setting in media. Seradix 2 contains one active ingredient, indole-3-butyric acid (IBA) and is applied by dipping a cutting’s cut edge into Seradix 2 powder before the cutting is set. A commercial biological rooting agent, Eco-T, was also included as a treatment (T2). This biological rooting agent contains live fungus (Trichoderma harzianum) as active ingredient. Two treatments were prepared from bacterial species that were isolated and screened. Treatment 3 (T3) mainly comprised species that were known for their rooting enhancement abilities, including Bacillus aryabhattai, Pseudomonas fluorescens, P. koreensis, and P. putida. Treatment 4 (T4) consisted of the remaining three species Brevibacterium frigoritolerans, Burkholderia phytofirmans, and Chryseobacterium rhizosphaerae. Table 2 shows the compositions of the different treatments that were applied in this study. Apart from the control treatment (T1) that was applied at the setting of the cuttings, the other treatments were applied two days after the cuttings were set in media.
2.6. Field Trials: Set up and Procedure
Cuttings of E. grandis × E. nitens (GN) hybrid genotypes GN 018B and GN 010 were collected from mini-hedge ramets grown in nursery tunnels at Sunshine Seedlings near Pietermaritzburg, South Africa. These hybrids are well suited for cold regions and poor nutrient soil environments. They are relatively fast growing and most commonly used as a source of pulpwood, fuel, and timber in South Africa. Shoots with a length of approximately 7 cm were carefully removed from the mini-hedges with sharp secateurs and placed in a 20 L bucket half filled with water. Each cutting consisted of two nodes, an internode and two pairs of leaves. The buckets containing the cuttings were taken to the cutting room which was constantly exposed to misting irrigation for 30 s every 5 min. The cuttings were prepared by trimming the shoots and leaves with a sharp scissor. The leaves were cut to a size of approximately one third of the original leaf surface area to reduce transpiration and to conserve energy. Finally, cuttings were stored in a water bath until setting in rooting medium.
Prior to the setting of the cuttings, the cuttings were washed in dilute Vapour Gard with Di-1-p-Menthene to reduce moisture stress in the cuttings. The cutting bases were then dipped into the Seradix 2 according to the manufacturer’s specifications. Thereafter, the cuttings were placed in the centre of Unigrow cells filled with rooting media at a depth of approximately 2 cm prior to the application of treatments. The four treatments (T1, T2, T3, and T4) were applied to the respective Eucalyptus hybrid clone cuttings in a Unigrow tray that comprised 128 cells arranged in 16 columns and 8 rows. Each of the four treatments was applied along two rows of 16 cells (32 cuttings) delineated with plastic markers (Figure 1). The procedure was repeated in 10 different trays, with the row allocations for each treatment per tray determined by a random number generator. After all the treatments were applied, the trays were arranged across the nursery floor to be exposed to similar climatic variation in the nursery. The treated cuttings were left in the nursery for eight weeks, at which time the growth measurements were recorded.

Figure 1.
Trays with Unigrow cells filled with rooting media and cuttings (a). Tray consisting of 128 cells arranged in 16 columns and 8 rows. Each of the four treatments (T1, T2, T3, and T4) was applied along two rows of 16 cells (32 cuttings in total) delineated with plastic markers. The four treatments were applied to GN 018B and GN 010 Eucalyptus hybrid clone cuttings in 10 separate trays as independent repeats (b).
2.7. Field Trials: Growth Measurements
Firstly, it was noted whether the cutting had survived. If a cutting had survived, growth measurements were made, and the rooting architecture configuration described. Three growth measurements were made for each surviving cutting, namely, total length of cutting, root length, and shoot length. The rooting architecture of each rooted cutting was described by using six different descriptive configurations. The six different rooting configurations comprised of the number of primary adventitious roots with secondary and tertiary roots (R+), or without secondary and tertiary roots (R−). Separate categories were allocated to the number of primary adventitious roots up to five roots (R1–R5). Cuttings with more than five adventitious roots were placed into a category called “roots-many (RM)”. Rooting architecture data were further categorised into three levels of fibrosity [7]. Level 1, which referred to the lowest level of fibrosity, included the rooting architectural types R1 and R2, including both plus and minus types. Level 2, referred to a medium level of fibrosity, and included the rooting architectural types R3 and R4, also inclusive of both plus and minus types. Level 3, referred to the highest level of fibrosity, and included the rooting architectural types R5 and RM, also inclusive of both R+ and R− types.
2.8. Statistical Analyses of Treatment Outcomes
Summary statistics were calculated for the survival of the cuttings, growth properties of the cuttings, as well as for the rooting architectural configurations of the cuttings. Several analyses of variance (ANOVA) tests and Turkey HSD were performed on the data to ascertain if significant differences existed between the treatments for growth responses. Chi-square tests of independence were performed to test if the different rooting architectural configurations were independent of treatment.
3. Results
3.1. Isolation, Identification and Characterisation of Potential PGPR
A total of 31 bacterial isolates were purified and identified. Eight different bacterial genera were identified with Pseudomonas being the most prevalent, containing four different species. Pseudomonas was followed by Bacillus with two different species. The remaining six genera, Aeromicrobium, Arthrobacter, Brevibacterium, Burkholderia, Chryseobacterium, and Curtobacterium, were each represented by a single species. These 12 unique species were screened for their ability to produce indole-3-acetic acid (IAA) and solubilise phosphate. All the bacteria isolates, except for Aeromicrobium ginsengisoli and Curtobacterium oceanosedimentum, were able to produce IAA. Only seven species also demonstrated an ability to solubilize phosphate (Table 1). The isolates with caption isolate identity numbers indicated in bold were selected for use in field trials.
3.2. Preparation and Monitoring of Bacterial Inoculums for Field Trials
The 7 isolates in Table 2 were propagated in suspension cultures in nutrient broth and cryo-preserved for application in field trials. Harvest times were determined by the target cell density value of ≈107 CFU mL−1. Viability was confirmed after the cultures were thawed for use in field trials (Table 3). IAA production and phosphate solubilisation were also confirmed after thawing; no loss of either capability was noticed in any of the isolates.

Table 3.
Harvesting time intervals, OD values, and viability before and after freezing of inoculums for field trial.
3.3. Survival and Rooting of Cuttings
The four treatments were applied to cuttings in the 10 Unigrow trays of each experimental trial. The trays were arranged across the nursery floor from one side to the other. The treated cuttings were left in the nursery for eight weeks, at which time growth measurements were taken. Figure 1 shows the arrangement of the two rows of 10 trays each GN 018B and GN 010 hybrids across the nursery floor.
The number of cuttings that survived after eight weeks of growth was counted. Some of the survived cuttings did not produce roots, and the number of rooted cuttings was also calculated. For GN 018B treatments, more than 90% of the cuttings survived, although treatments 1 and 2 performed better than treatments 3 and 4. The total mean percentage of survived rooted cuttings was greater than 75%. Similarly, more than 90% of the GN 010 cuttings survived. The total mean percentage of survived rooted cuttings was greater than 80% (Supplementary Tables S1 and S2). The control treatment 1 outperformed the other treatments on both survival and rooting for both hybrids (Figure 2).
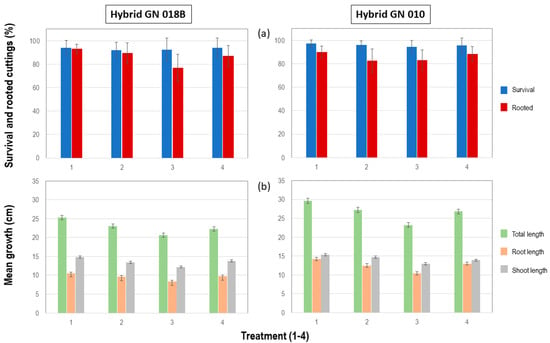
Figure 2.
Graphical representation of (a) survival, rooting and (b) growth parameters for cuttings of both GN 018B and GN 010 hybrid clones after 8 weeks.
3.4. Growth Response of Cuttings
Three growth measurements were made on each rooted cutting. Mean total growth, which included the shoot and root length, showed that the control treatment 1 outperformed all the other treatments by producing the longest rooted cuttings. A similar pattern could be discerned for both mean root length and means shoot length, where the control treatment 1 also produced the longest roots and longest shoots in both GN 018B and GN 010 hybrids (Figure 2).
Two factor ANOVA tests were performed on the respective growth responses to ascertain if significant difference existed between treatments as well as between trays. All growth responses showed significant differences between the different treatments as well as between trays. The tray versus treatment interaction also proved to be significantly different with p values < 0.0001 (Supplementary Tables S3 and S4). Two-factor ANOVA tests were also performed on the three growth responses (total, root, and shoot length) of the two hybrids GN 018B and GN 010. The performances of the two hybrids were significantly different (p < 0.0001 and 0.0147), as was the outcomes of the different treatments (p < 0.0001). However, the relation between hybrid genotypes and treatment was not significant (Table 4). Tukey HSD tests indicated that most combinations of treatments differed significantly. In the case of GN 018B, treatment 4, however, did not differ significantly from T1 and T2 for total length or shoot length nor from T3 for root length. For GN 010, treatment 4 showed no significant differences from T2 for total length, from T1 and T2 for root length, and from T2 and T3 for shoot length (Figure 3).

Table 4.
Two-way ANOVA test performed to compare growth responses on the hybrids GN 018B and GN 010.
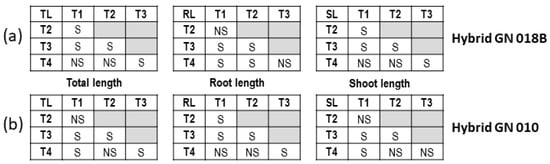
Figure 3.
Tukey HSD tests for hybrids (a) GN 018B and (b) GN 010 comparing treatments 1–4 for growth parameters; total, root, and shoot length.
3.5. Rooting Architecture of Cuttings
In response to the different treatments, the rooting architecture of the cuttings showed different presentations of the number of primary adventitious roots (R) with (R+) or without (R−) secondary and tertiary roots (Supplementary Tables S5 and S6). Similar results were obtained for both hybrids. When the R+ and R− were grouped together, the rooting architectural category with a single primary adventitious root demonstrated the highest percentage of cuttings. The percentage of cuttings showed a descending pattern with an increase in the number of primary adventitious roots up to a total of five roots. The category of many primary adventitious roots demonstrated a higher percentage of cuttings than the two categories with four and five primary adventitious roots (Figure 4).
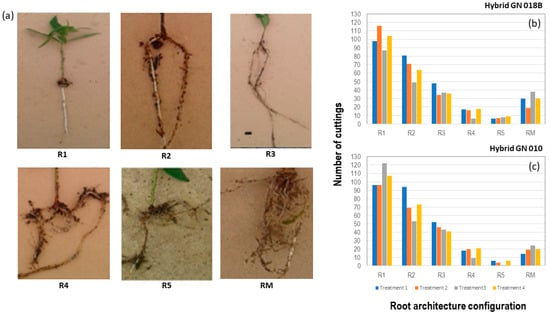
Figure 4.
(a) Key for the identification of rooting architectural configurations. Six different rooting configurations were specified; primary adventitious roots with secondary and tertiary roots (R+) and the number of primary adventitious roots up to five roots (R1–R5). Cuttings with more than five adventitious roots were considered to have “many roots” (RM). Graphical representation of (b) GN 018B and (c) GN 010 cuttings in each treatment with different rooting architectural configuration types.
A chi-square test of independence was performed to test if the different rooting architectural types were independent of the treatment. The test showed that treatment did not affect rooting architecture significantly of GN1018B cuttings (χ2 = 23.672, df = 15, p = 0.0709) or that of GN 010 cuttings (χ2 = 22.996, df = 15, p = 0.0842). The percentages of cuttings that demonstrated different levels of fibrosity were calculated (Table 5). When considering Level 3 (the highest level of fibrosity), T3 and T4 showed increased fibrosity when applied to hybrid GN 018B cuttings, while for GN 010 cuttings, all treatments promoted fibrosity when compared to the control (T1). Goodness of fit chi-square tests were conducted to test to what extent treatments T2, T3, and T4 promoted fibrosity in comparison to the control (T1). Most tests were not significant at α = 0.05. However, the chi-square test for T3 of GN 018B, proved to be highly significant (χ2 = 12.14, df = 2, p = 0.0023).

Table 5.
Percentages of cuttings demonstrating different levels of fibrosity for both hybrids.
4. Discussion
The many benefits that Eucalyptus clonal forestry offers have brought about commercial nurseries to take advantage of this avenue of forest tree production. The cloning of trees allows for the preservation of superior genotypes [4]. At the heart of cloning operations lies vegetative reproduction, which allows the commercial forestry industry to mass-produce such superior genotypes of species and hybrids [38]. The vegetative reproduction of Eucalyptus involves the rooting of cuttings. In South African forestry nurseries, cuttings are taken from indoor mini-hedges. For cuttings to develop successfully, adventitious root formation must occur. Adventitious roots increase the number of nutrient-absorbing surfaces and results in a better assimilation of water and nutrients from the soil [23]. Most commercial nursery enterprises apply plant growth promoting regulators, such as auxins, to cuttings to stimulate the formation of adventitious root primordia [39]. However, some valued Eucalyptus genotypes, especially of the subtropical species, and those deployed in low productivity areas consistently demonstrate relatively poor adventitious rooting abilities [6,40].
The exploitation of rhizospheric microbes for their properties in biotechnological applications in clonal forestry, such as for the enhancement of the rooting of cuttings; requires some understanding of the diversity of rhizospheric microorganism communities, conditions of the rhizosphere, and change over time [41,42]. The diversity is often very specific to certain environmental conditions and tree species. Understanding diversity is only the first step, and utilising this information requires the subsequent culturing, purification, identification, and characterisation of microbes for further study or application. We took a targeted approach to isolate, identify, and partially characterise rhizospheric microbes present in the rhizospheres of a Eucalyptus hybrid genotype, E. grandis × E. urophylla (GU), known for its relatively high rooting capacity. Of the 31 bacterial strains that were isolated, 12 were unique species belonging to eight different genera. Seven demonstrated the ability to produce indole-3-acetic acid (IAA) and to solubilise phosphates and were deemed suitable for further study in nursery trials. Three of these species were from the genus Pseudomonas, and one was from the genus Bacillus. The abundance of both genera in the plant rhizosphere is well known, as is their ability to stimulate plant growth and adventitious root development in Eucalyptus [7,10,43]. Therefore, the Pseudomonas and Bacillus isolates were combined as a consortium in T3. Besides the ability to produce IAA and to solubilise phosphate, species of the genus Burkholderia are also able to solubilise zinc into a form so that plants can absorb this mineral [13,44,45,46]. Chryseobacterium spp. are credited for the production of siderophores, which can supply iron to plant roots [47,48]. It is worth mentioning that the specific species investigated in our study have not been applied as a consortium of rooting-enhancing agents of Eucalyptus cuttings and were therefore included in T4.
The preparation of bacterial inoculums for field application required the up-scaling of suspension cultures. Several authors suggest 108 CFU mL−1 as the preferred concentration at which a bacterial inoculum should be applied to the rooting medium of Eucalyptus cuttings [7,8,10]. Not all seven bacterial species in the current study were able to reach a growth concentration of ≈108 CFU mL−1, and to ensure consistency, ≈107 CFU mL−1 was used for field applications, which was still considered suitable [49,50]. Although growth medium optimisation was not an objective of our study, it would be worthwhile to consider other nutrient media to increase inoculum concentration. Nevertheless, all seven bacterial isolates retained viability, IAA production, and phosphate solubilisation functionality after cryo-preservation at −80 °C.
The species of the Bacillus and Pseudomonas were combined into one rooting treatment (T3), while the other isolated bacterial species were combined into a separate rooting treatment (T4). Bacterial consortia can provide different physiological activities simultaneously, which makes it more effective in promoting plant growth compared to the use of single bacterial strains [32]. Rhizospheric rooting treatments T3 and T4 were compared to the standard of general practice in the nursery, which was the application of indole-3-butyric acid (control), and to a commercial product (Eco-T), which contains live Trichoderma harzianum.
All four treatments demonstrated high survival rates and high rooting percentages. The nursery standard outperformed all treatments, although mostly marginally so. The rhizospheric rooting treatment comprising the non-Pseudomonas-Bacillus bacteria (T4), as well as fungus treatment (T2), were closest in performance to the nursery standard, while the Pseudomonas-Bacillus treatment (T3) often showed lower values when compared to the other treatments. The poorer performance of treatment 3 was unexpected since countless studies have demonstrated that species belonging to these genera are proficient PGPR. However, the specific species used in our study have not been used as a consortium before, and it might have been necessary to test compatibility before using them in combination, as is suggested by Zul et al. [32], who prudently verified strain compatibility mainly before using unidentified strains in combination.
A noteworthy outcome of this study was that PGPR treatments influenced the growth parameters of the two Eucalyptus grandis × Eucalyptus nitens hybrid clones differently. These genotypic differences were noted particularly for growth responses of the cuttings and adventitious root development. Both hybrid clones demonstrated significant genotypic differences for all three growth responses, namely, total, root, and shoot length. These outcomes support the findings of [39], who identified genotypic differences in adventitious root development in tree clones of Eucalyptus benthamii × Eucalyptus dunnii. Although genotype-linked growth parameter variation was not an unexpected outcome [39,51], it was peculiar that two clones would exhibit significant differences considering that they were derived through hybridisation between the same two species. GN 010 treated with the non-Pseudomonas-Bacillus consortium (T4) yielded growth responses resembling that of treatment with the commercial product Eco-T, which contains fungus. The Pseudomonas-Bacillus bacterial treatment improved fibrosity of the rooting architecture of cuttings of the GN 018B hybrid. This outcome could be of value when rooted cuttings are planted out in plantations and require further investigation to ascertain any other rooting enhancement traits these bacterial species possess [23,39].
5. Conclusions
Improving the rooting capacity of Eucalyptus hybrid clone cuttings, which is known for its low rooting ability, remains an interesting prospect. The plant-growth-promoting rhizobacteria (PGPR) that were successfully isolated, identified, and characterised in this study were only comparable to a nursery standard and a commercially available fungal rooting enhancer. It is important to note that the consortium of three non-Pseudomonas-Bacillus species able to elicit the same growth response as the commercial products have not been previously applied to Eucalyptus cuttings in a commercial nursery setting. Furthermore, the consortium of four Pseudomonas-Bacillus isolates were able to increase the root fibrosity of GN 018B cuttings. Unexpected growth responses and rooting architecture differences were observed between clones from hybridisations between the same two tree species and should be taken into account for future experimental designs. Further investigations based on the same approach used in this study should expand the plant-growth-promoting selection criteria used for potential rhizosphere bacterial isolates to include siderophore production capabilities and species compatibility testing for consortium applications. Extending the treatment time for nursery trials could also be necessary to ensure the proper establishment of the PGPR consortia in the rhizosphere. The continued attempt to improve rooting percentages of Eucalyptus cuttings will not only mitigate financial losses, which Eucalyptus commercial nursery growers are currently experiencing, but will also allow for the maximum use of the limited space in nursery environments.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/f14091848/s1, Table S1: Survival and rooted number (%) of cuttings per tray per treatment for GN 018B, Table S2: Survival and rooted number (%) of cuttings per tray per treatment for GN 010, Table S3: ANOVA tests for the growth responses, total length, root length, and shoot length of GN 018B cuttings, Table S4: ANOVA tests for the growth responses, total length, root length, and shoot length of GN 010 cuttings, Table S5: Rooting architecture, showing number (%) of cuttings with primary adventitious roots with (+) or without (−) secondary and tertiary roots per treatment of hybrid GN 018B, Table S6: Rooting architecture, showing number of cuttings (%) with primary adventitious roots with (+) or without (−) secondary and tertiary roots per treatment of hybrid GN 010.
Author Contributions
Conceptualisation, C.N., A.F. and O.d.S.; methodology, C.N., A.F. and O.d.S.; validation, C.N. and A.F.; formal analysis, C.N. and A.F.; resources, A.F. and O.d.S.; data curation, C.N., A.F. and O.d.S.; writing—original draft preparation C.N.; writing—review and editing, C.N., A.F. and O.d.S.; visualisation, O.d.S.; supervision, A.F. and O.d.S.; project administration, A.F.; funding acquisition, A.F. and O.d.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the South African National Research Foundation (NRF) under grant number UID: 107624, Central University of Technology, Free State, and the Free State and Department of Education.
Data Availability Statement
This research is an extract from Chimdi Nwigwe’s Ph.D. Thesis. The full thesis with all the data can be viewed on the Central University of Technology, Free Sate Institutional DSpace repository http://hdl.handle.net/11462/1927.
Acknowledgments
The authors thank Cay van der Merwe for her assistance with the statistical analyses.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lock, P.; Legg, P.; Whittle, L.; Black, S. Global Outlook for Wood Markets to 2030 Projections of Future Production, Consumption and Trade Balance; Australian Bureau of Agricultural and Resource Economics and Sciences: Canberra, Australia, 2021.
- van Wyk, G.; Verryn, S. The Basic Principles of Tree Breeding in South Africa. In South African Forestry Handbook; Owen, D., Ed.; The South African Institute of Forestry: Pretoria, South Africa, 2000; Volume 1, pp. 61–68. [Google Scholar]
- Pita-Barbosa, A.; Oliveira, L.A.; de Barros, N.F.; Hodecker, B.E.R.; Oliveira, F.S.; Araújo, W.L.; Martins, S.C. V Developing a Roadmap to Define a Potential Ideotype for Drought Tolerance in Eucalyptus. For. Sci. 2023, 69, 101–114. [Google Scholar] [CrossRef]
- de Assis, T.; Fett-Neto, A.; Couto Alfenas, A. Current Techniques and Prospects for the Clonal Propagation of Hardwoods with Emphasis on Eucalyptus. In Plantation Forest Biotechnology for the 21st Century, 2004; Walter, C., Carson, M., Eds.; Research Signpost: Kerala, India, 2004; pp. 303–333. [Google Scholar]
- Ruaud, J.; Lawrence, N.; Pepper, S.; Potts, B.; Borralho, N. Genetic Variation of in Vitro Rooting Ability with Time in Eucalyptus Globulus. Silvae. Genet. 1999, 48, 4–7. [Google Scholar]
- Fogaça, C.M.; Fett-Neto, A.G. Role of Auxin and Its Modulators in the Adventitious Rooting of Eucalyptus Species Differing in Recalcitrance. Plant Growth Regul. 2005, 45, 1–10. [Google Scholar] [CrossRef]
- Díaz, K.; Valiente, C.; Martínez, M.; Castillo, M.; Sanfuentes, E. Root-Promoting Rhizobacteria in Eucalyptus Globulus Cuttings. World J. Microbiol. Biotechnol. 2009, 25, 867–873. [Google Scholar] [CrossRef]
- Liu, Y.M.; Zheng, F.; Liu, Z.H.; Lan, H.B.; Cui, Y.H.; Gao, T.G.; Roitto, M.; Wang, A.F. Enhanced Root and Stem Growth and Physiological Changes in Pinus Bungeana Zucc. Seedlings by Microbial Inoculant Application. Forests 2022, 13, 1836. [Google Scholar] [CrossRef]
- Zafar, M.; Abbasi, M.K.; Khan, M.A.; Khaliq, A.; Sultan, T.; Aslam, M.; Jammu, A. Effect of Plant Growth-Promoting Rhizobacteria on Growth, Nodulation and Nutrient Accumulation of Lentil Under Controlled Conditions. Pedosphere 2012, 22, 848–859. [Google Scholar] [CrossRef]
- Teixeira, D.A.; Couto Alfenas, A.; Gonçalves Mafia, R.; Ferreira, E.M.; de Siqueira, L.; Maffia, L.A.; Mounteer, A.H. Rhizobacterial promotion of eucalypt rooting and growth. Braz. J. Microbiol. 2007, 38, 118–123. [Google Scholar] [CrossRef]
- Erturk, Y.; Ercisli, S.; Haznedar, A.; Cakmakci, R. BR Effects of Plant Growth Promoting Rhizobacteria (PGPR) on Rooting and Root Growth of Kiwifruit (Actinidia Deliciosa) Stem Cuttings. Biol. Res. 2010, 43, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Santoyo, G.; Alberto Urtis-Flores, C.; Damián Loeza-Lara, P.; del Carmen Orozco-Mosqueda, M.; Glick, B.R. Biology Rhizosphere Colonization Determinants by Plant Growth-Promoting Rhizobacteria (PGPR). Biology 2021, 10, 475. [Google Scholar] [CrossRef]
- Ahmad, F.; Ahmad, I.; Khan, M.S. Screening of Free-Living Rhizospheric Bacteria for Their Multiple Plant Growth Promoting Activities. Microbiol. Res. 2008, 163, 173–181. [Google Scholar] [CrossRef]
- Ji, S.H.; Gururani, M.A.; Chun, S.C. Isolation and Characterization of Plant Growth Promoting Endophytic Diazotrophic Bacteria from Korean Rice Cultivars. Microbiol. Res. 2014, 169, 83–98. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Rajkumar, M.; Zhang, C.; Freitas, H. Inoculation of Brassica oxyrrhina with Plant Growth Promoting Bacteria for the Improvement of Heavy Metal Phytoremediation under Drought Conditions. J. Hazard. Mater. 2016, 320, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Arshad, M.; Frankenberger, W. Soil Microbial Ecology. In Microbial Production of Plant Growth Regulators; Blaine, F., Metting, J., Eds.; Mercel and Dekker, Inc.: New York, NY, USA, 1993; pp. 307–347. [Google Scholar]
- Glickmann, E.; Dessaux, Y. A Critical Examination of the Specificity of the Salkowski Reagent for Indolic Compounds Produced by Phytopathogenic Bacteria. Appl. Env. Microbiol. 1995, 61, 793–796. [Google Scholar] [CrossRef]
- Ljung, K.; Bhalerao, R.P.; Sandberg, G. Sites and Homeostatic Control of Auxin Biosynthesis in Arabidopsis during Vegetative Growth. Plant J. 2001, 28, 465–474. [Google Scholar] [CrossRef]
- Caboni, E.; Lauri, P.; Tonelli, M.; Falasca, G.; Damiano, C. Root Induction by Agrobacterium rhizogenes in Walnut. Plant Sci. 1996, 8, 203–208. [Google Scholar] [CrossRef]
- Blazkova, A.; Sotta, B.; Tranvan, H.; Maldiney, R.; Bonnet, M.; Einhorn, J.; Kerhoas, L.; Miginiac, E. Auxin Metabolism and Rooting in Young and Mature Clones of Sequoia sempervirens. Physiol. Plant 2006, 99, 73–80. [Google Scholar] [CrossRef]
- Gaspar, T.; Kevers, C.; Penel, C.; Greppin, H.; Reid, D.M.; Thorpe, T.A. Plant hormones and plant growth regulators in plant tissue culture. Vitr. Cell Dev. Biol. Plant 1996, 32, 272–289. [Google Scholar] [CrossRef]
- Gatineau, F.; Fouche, J.; Kevers, J.; Hausman, J.; Gaspar, T. Quantitative Variations of Indolyl Compounds Including IAA, IAA-Aspartate and Serotonin in Walnut Microcuttings during Root Induction. Biol. Plant. 1997, 39, 131–137. [Google Scholar] [CrossRef]
- Dilfuza, E. Indole-Acetic Acid Production by Root Associated Bacteria and Its Role in Plant Growth and Development. In Auxins: Structures, Biosynthesis and Functions; Keller, A., Fallon, M.D., Eds.; Nova Science Publishers Inc.: Hauppauge, NY, USA, 2011; pp. 1–13. [Google Scholar]
- Gaur, A. Physiological Functions of Phosphate Solubilizing Micro-Organisms. In Phosphate Solubilizing Micro-Organisms as Biofertilizers; Gaur, A., Ed.; Omega Scientific Publishers: New Delhi, India, 1990; pp. 16–72. [Google Scholar]
- De Freitas, J.R.; Banerjee, M.R.; Germida, J.J. Phosphate-Solubilizing Rhizobacteria Enhance the Growth and Yield but Not Phosphorus Uptake of Canola (Brassica Napus L.). Biol. Fertil. Soils 1997, 4, 358–364. [Google Scholar] [CrossRef]
- Pikovskaya, R. Mobilization of Phosphorus in Soil Connection with the Vital Activity of Some Microbial Species. Microbiology 1948, 17, 362–370. [Google Scholar]
- Poyton, R. Tree Planting in Southern Africa: The Eucalypts; Department of Forestry, Southern African Regional Commission for the Conservation and Utilisation of the Soil: Pretoria, South Africa, 1979. [Google Scholar]
- Chetty, S. The Development of Clone-Unspecific Micropropagation Protocols for Three Commercially Important Eucalyptus Hybrids. Master’s Thesis, University of Natal, Durban, South Africa, 2001. [Google Scholar]
- Komakech, C.; Swain, T.L.; Fossey, A. Growth Potential of Eucalyptus cypellocarpa as an Alternative Species for the Mid-Altitude Summer Rainfall Region of South Africa. South For. 2013, 75, 149–154. [Google Scholar] [CrossRef]
- McMahon, L.; George, B.; Hean, R. Eucalyptus camaldulensis Species Summary. Primefacts 2010, 1054, 1–6. [Google Scholar]
- González, P.; Sossa, K.; Rodríguez, F.; Sanfuentes, E. Rhizobacteria Strains as Promoters of Rooting in Hybrids of Eucalyptus nitens × Eucalyptus globulus. Chil. J. Agric. Res. 2018, 78, 3–12. [Google Scholar] [CrossRef]
- Zul, D.; Elviana, M.; Ismi, K.R.N.; Tassyah, K.R.; Siregar, B.A.; Gafur, A.; Tjahjono, B. Potential of PGPR Isolated from Rhizosphere of Pulpwood Trees in Stimulating the Growth of Eucalyptus pellita F. Muell. Int. J. Agric. Technol. 2022, 18, 401–420. [Google Scholar]
- Labuschagne, M.; Albertyn, J. Cloning of an Epoxide Hydrolase-Encoding Gene from Rhodotorula mucilaginosa and Functional Expression in Yarrowia Lipolytica. Yeast 2007, 24, 69–78. [Google Scholar] [CrossRef]
- Marchesi, J.R.; Sato, T.; Weightman, A.J.; Martin, T.A.; Fry, J.C.; Hiom, S.J.; Wade, W.G. Design and Evaluation of Useful Bacterium-Specific PCR Primers That Amplify Genes Coding for Bacterial 16S RRNA. Appl. Environ. Microbiol. 1998, 64, 795–799. [Google Scholar] [CrossRef]
- Abellan-Schneyder, I.; Matchado, M.S.; Reitmeier, S.; Sommer, A.; Sewald, Z.; Baumbach, J.; List, M.; Neuhaus, K. Primer, Pipelines, Parameters: Issues in 16S RRNA Gene Sequencing. mSphere 2021, 6, e01202-20. [Google Scholar] [CrossRef]
- Bric, J.M.; Bostock, R.M.; Silverstonet, S.E. Rapid In Situ Assay for Indoleacetic Acid Production by Bacteria Immobilized on a Nitrocellulose Membrane. Appl. Environ. Microbiol. 1991, 57, 535–538. [Google Scholar] [CrossRef]
- Whitelaw, M.A.; Harden, T.J.; Helyar, K.R. Phosphate Solubilisation in Solution Culture by the Soil Fungus Penicillium radicum. Soil Biol. Biochem. 1999, 31, 655–665. [Google Scholar] [CrossRef]
- Ferreira, E.; Alfenas Couto, A.; Mafia Gonzalves, R.; Leite Garcia, H.; Sartorio Cardoso, R.; Filho Penchel, R.; Helio, L. Determination of the optimum time for rooting of mini-cuttings of Eucalyptus spp. CLONES. Reshvista Arvore 2004, 28, 183–187. [Google Scholar] [CrossRef]
- Brondani, G.E.; Wendling, I.; Brondani, A.E.; Araujo, M.A.; da Silva, A.L.L.; Gonçalves, A.N. Dynamics of adventitious rooting in mini-cuttings of Eucalyptus benthamii × Eucalyptus dunnii. Acta Sci. Agron. 2012, 34, 169–178. [Google Scholar] [CrossRef]
- Brondani, G.E.; Ferreira Dutra, L. Eucalyptus benthamii × Eucalyptus dunnii Minicutting Technique: (II) Minicutting Survival and Rooting in Relation to Collection and Seasons. Cienc. Florest. Santa Maria 2010, 20, 453–465. [Google Scholar] [CrossRef]
- Di Cello, F.; Bevivino, A.; Chiarini, L.; Fani, R.; Paffetti, D.; Tabacchioni, S.; Dalmastri, A.C. Biodiversity of a Burkholderia cepacia Population Isolated from the Maize Rhizosphere at Different Plant Growth Stages. Appl. Environ. Microbiol. 1997, 63, 4485–4493. [Google Scholar] [CrossRef]
- Nwigwe, C.; Fossey, A.; de Smidt, O. Characterisation of Eucalyptus Rhizospheric Communities Using Fatty Acid Methyl Ester (FAME) Profile Analysis. S. Afr. J. Plant Soil 2021, 38, 116–125. [Google Scholar] [CrossRef]
- Lan, Y.; Liao, L.; Yao, X.; Ye, S. Synergistic Effects of Nitrogen and Plant Growth-Promoting Rhizobacteria Inoculation on the Growth, Physiological Traits and Nutrient Absorption of Intercropped Eucalyptus urophylla × Eucalyptus grandis and Dalbergia odorifera. Trees Struct. Funct. 2023, 37, 319–330. [Google Scholar] [CrossRef]
- Armada, E.; Probanza, A.; Roldán, A.; Azcón, R. Native Plant Growth Promoting Bacteria Bacillus thuringiensis and Mixed or Individual Mycorrhizal Species Improved Drought Tolerance and Oxidative Metabolism in Lavandula dentata Plants. J. Plant Physiol. 2016, 192, 1–12. [Google Scholar] [CrossRef]
- Gontia-Mishra, I.; Sapre, S.; Tiwari, S. Zinc Solubilizing Bacteria from the Rhizosphere of Rice as Prospective Modulator of Zinc Biofortification in Rice. Rhizosphere 2017, 3, 185–190. [Google Scholar] [CrossRef]
- Saeed, Q.; Xiukang, W.; Haider, F.U.; Kučerik, J.; Mumtaz, M.Z.; Holatko, J.; Naseem, M.; Kintl, A.; Ejaz, M.; Naveed, M.; et al. Rhizosphere Bacteria in Plant Growth Promotion, Biocontrol, and Bioremediation of Contaminated Sites: A Comprehensive Review of Effects and Mechanisms. Int. J. Mol. Sci. 2021, 22, 10529. [Google Scholar] [CrossRef]
- Radzki, W.; Gutierrez Mañero, F.J.; Algar, E.; Lucas García, J.A.; García-Villaraco, A.; Ramos Solano, B. Bacterial Siderophores Efficiently Provide Iron to Iron-Starved Tomato Plants in Hydroponics Culture. Antonie Van Leeuwenhoek 2013, 104, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Chhetri, G.; Kim, I.; Kim, J.; So, Y.; Seo, T. Chryseobacterium tagetis Sp. Nov., a Plant Growth Promoting Bacterium with an Antimicrobial Activity Isolated from the Roots of Medicinal Plant (Tagetes patula). J. Antibiot. 2022, 75, 312–320. [Google Scholar] [CrossRef]
- Zaspel, I.; Ewald, D. Tri-Trophic Interactions in the Rhizosphere and Root-Health Nematode-Fungal-Bacterial Interrelationships. In Promotion of Root Development and Root Growth of Forest Plants by Rhizobacteria; International Organization for Biological and Integrated Control of Noxious Animals and Plants, West Palaearctic Regional Section: Wageningen, The Netherlands, 2001; pp. 161–167. [Google Scholar]
- González-Díaz, A.; Ojeda-Morales, M.E.; Hernández-Rivera, M.A.; Córdova-Bautista, Y.; Díaz-Flores, L.L.; López-Lázaro, J.D.L.S.; Álvarez-Ramírez, J.G. Effect of Biofertilizers Application on the Growth of Eucalyptus grandis Seedlings under Greenhouse Conditions. J. Plant Nutr. 2019, 42, 2560–2576. [Google Scholar] [CrossRef]
- Haldar, S.; Sengupta, S. Plant-Microbe Cross-Talk in the Rhizosphere: Insight and Biotechnological Potential. Open Microbiol. J. 2015, 9, 1–7. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).