Abstract
Castanopsis hystrix, a timber resource from Southeast Asia, is characterized by rapid growth and high yield, but plantation quality and efficiency improvements are required. Twenty-year-old C. hystrix experimental forests in Yulin, Liuzhou, and Pingxiang in Guangxi Province, China, comprising 21 open-pollinated half-sib families, were used in this study. Genetic variations in the growth (tree height, diameter at breast height (DBH), and volume (V)) and morphological (height to live crown base (HCB), crown width (CW), and branch angle) traits were assessed, and the genetic parameters were estimated to clarify the genotype × environment interaction effects. The average values for the tree height, DBH, V, HCB, CW, and branch angle were 16.33 m, 17.25 cm, 0.21 m3, 6.68 m, 2.15 m, and 45.45°, respectively. The most important sources of variance for the tree height, DBH, V, and HCB were the block and family, whereas the location, family, and family × location had significant impacts on the tree height, DBH, V, and HCB (p < 0.01). The family heritability for each trait was 0.35, 0.38, 0.62, and 0.19, respectively. Excellent families with strong adaptability and genetic stability were identified using BLUP–GGE biplots for single and multiple traits. The results provide a theoretical basis for the efficient cultivation of C. hystrix in South China.
1. Introduction
The genotype × environment interaction effect (G × E) can conceal the authenticity of genetics, reducing selection efficiency and accuracy [1]. Moreover, the G × E can evaluate the genetic stability and adaptability of tree genotypes in various locations [2,3]. Therefore, the G × E has become an essential factor in the selection and popularization of forest tree genotypes. In addition to focusing on the genetic variations of the tested traits, breeders should also pay more attention to the genetic stability and adaptability of genotypes in various environments. Doing so will help them effectively match germplasms and sites. In view of their research, the growth and morphological traits of plant species are not only regulated by their genotypes and the environment but also affected by the interaction between the two [4,5,6]. Due to the G × E, these traits exhibit varying rank and scale effects on tree traits in different locations [1]. Such variances affect the estimation of genetic parameters such as heritability and genetic gains [1,7], which, in turn, affect the formulation of breeding strategies [8].
Breeders in China and overseas have developed various statistical models to evaluate the patterns and scales of the G × E, analyze the relationship between genetic variations and the ecological environment, and determine genotype adaptability and stability. These models include regression–based stability analysis [9], B-type genetic correlation [10], AMMI analysis [11], GGE biplots [12], factor analysis [13], and BLUP–GGE analysis [14]. The purpose of a ‘suitable genotype for the appropriate location’ is achieved by selecting the appropriate genotypes for cultivation in different ecological zones. The BLUP–GGE joint analysis method combines the advantages of BLUP and GGE biplots and can effectively estimate genetic parameters, as well as determine the optimal genotypes with stability and interactivity. The method can be used to effectively visualize the G × E. This is applicable for the evaluation and research of genotype stability and adaptability and has been widely used to ascertain a suitable range of elite germplasms [15].
Castanopsis hystrix belongs to the Fagaceae family and Castanopsis genus and is a fast-growing and high-yielding species [16]. Thanks to its reddish hues, beautiful texture, and excellent mechanical properties, C. hystrix heartwood is widely used in interior decoration, craft carving, wooden furniture, and various other industries [16,17]. This tree species has already been extensively planted in South China, such as Fujian, Guangdong, Guangxi, and Hainan, as it has been earmarked as a national strategic and reserve forest species for prioritized timber development [16]. These artificial forests will be felled to provide a large amount of precious wood in 50 years. There is an urgent need, however, to analyze the genetic variations and parameters for its important quantitative traits so that high-quality genotypes can be provided for genetic improvement. The results of two single-location measurements in Pingxiang City in Guangxi and Hua’an County in Fujian revealed serious genetic differentiations in the diameter at breast height (DBH), tree height, and individual plant volume (V) among the different C. hystrix families. These differentiations arose from medium-to-high levels of genetic regulation [18,19]. However, the stability and adaptability of the families at different sites have not yet been elucidated. Further investigation into the G × E is warranted when undertaking the selection and breeding of C. hystrix varieties.
The aim of this study was to perform G × E analyses of the growth and morphological traits of C. hystrix families to assist forestry producers in identifying high-quality genotypes exhibiting strong adaptability and genetic stability, thereby maximizing genetic gains. The subjects of this study were 20-year-old C. hystrix experimental forests, and the BLUP–GGE joint analysis method was used to carry out genetic variation and stability analyses of the growth and morphological traits. The main aims were (i) to evaluate the family, location, and G × E of 21 half-sib families at three locations; and (ii) to screen and select the single-location optimal families and families with the best comprehensive growth and morphological performance at multiple locations. The findings will provide a basis for the genetic improvement of valuable tree species such as C. hystrix, as well as scientific support for plantation improvements.
2. Materials and Methods
2.1. C. hystrix Families and Layout of the Experimental Forests
Yulin (YL), Liuzhou (LZ), and Pingxiang (PX) are located in the southeast, southwest, and north of the Guangxi Zhuang Autonomous Region of China. The above experimental bases were arranged for the progeny test of C. hystrix half-sib families, among which 43, 24, and 32 families were, respectively, planted in March 2002. A completely random block design was adopted, with 3 repeats and a row spacing of 2 × 3 m. Each site had 8 plants in each plot. A total of 21 families were planted in common across the three sites to analyze the G × E. These families were acquired from five geographical sources (Bobai, Donglan, Pubei, Rong County, and PX) in the Guangxi Zhuang Autonomous Region. The geographic and climatic information of their origins is shown in Table 1.

Table 1.
Profiles for the C. hystrix families in the experiments.
2.2. Overview of the Experimental Sites
The three sites used in this study, YL, LZ, and PX, are all on terrains with low hills. The soil is acidic red loam, and the soil layer is more than 1 m deep. The three locations experience a subtropical monsoon climate, which is warm, humid, and with precipitation and heat occurring at the same time. There is little difference between the annual average and the extreme high temperatures, although the extreme low temperatures show large variations. The extreme low temperature at PX is >0 °C, and thus there is no freezing damage throughout the year, whereas at LZ, it is −8 °C, with water freezing easily in the winter, and at YL, freezing occurs occasionally (Table 2).

Table 2.
Summary data for the experimental sites.
2.3. Determination of Traits
In August 2022, the Vertex IV ultrasonic altimeter (Haglöf IV, Stockholm, Sweden; accuracy 0.1 m) was used to measure the tree height (H) and the vertical height to the lowest branch from the ground to the tree crown (HCB). The DBH was measured with a DBH ruler (accuracy 0.1 cm) 1.3 m from the ground. The crown width was calculated as the half-sum of the four crown radii, which were measured as the horizontal distances from the center of the tree bole to the greatest extent of the crown from the tree bole [20,21]. When the growth survey and determination of traits were carried out at each location, three plants from each plot in each block were measured, and the measurements were repeated thrice. This was because the preservation rates of the families in YL and PX were <0%. In general, 63 plants were surveyed in each block, and 189 plants survived at every site. A total of 567 tree-growth and morphological-trait data were collected.
The equation for calculating the individual plant V is as follows:
where V is the individual plant volume, H is the tree height, DBH is the diameter at breast height, and f is a form factor whose value is 0.421 [22].
2.4. Data Analysis
2.4.1. Estimation of Genetic Parameters
Microsoft Excel 2010 was used for the statistical compilation of the data. Genstat 18.0 software was used to estimate the variance components and genetic parameters of the growth and morphological traits of the participating families.
Estimations of Single-Location Genetic Variance and Heritability
The single-location genetic statistical model is as follows:
where Yij is the average value of the plot for the jth family in the ith block; is the average value of the group; Bi is the value of the effect for the ith block; Fj is the value of the effect for the jth family; Bi × Fj is the block × family interaction effect; and Eij is a random error.
The equation for the single-location family heritability () is as follows:
where , , and denote the variance component of the family, variance component of the block × family interaction effect, and variance component of the environment, respectively; and n and b denote the number of families and blocks, respectively.
Estimation of Multiple-Location Genetic Variance and Heritability
The multiple-location genetic statistical model is as follows:
where Yijk is the average value of the plot for the jth family of the ith block at the kth location; is the average value of the group; Sk is the value of the effect at the kth location; Sk × Bj is the value of the effect of the jth block at the kth location; Fj is the value of the effect of the jth family; Sk × Fj is the value of the interaction effect between the kth location and the jth family; Sk × Bi × Fj is the value of the interaction effect between the ith block and the jth family at the kth location; and Eijk is a random error.
The equation for the multiple-location family heritability () is as follows:
where is the variance component of the family; is the variance of the block × family effect at a location; is the variance of the location × family effect; is the variance component of the environment; and n, b, and s represent the number of families, blocks, and locations, respectively.
Estimation of the Genetic Coefficient of Variances and Environmental Coefficient Variances
The genetic coefficient of variance (CVg) is as follows:
The environmental coefficient of variance (CVe) is:
where and represent the variance component of the family and environment, respectively; and is the average value of the traits.
Estimation of Genetic Gains
The equation for genetic gains (ΔG) is as follows:
where si is the selection intensity; δp is the standard deviation; is the family heritability; and is the average value of the traits.
2.4.2. BLUP–GGE Analysis of the Growth and Morphological Traits
The best linear unbiased prediction method was used to estimate the breeding values of the growth and morphological traits of the C. hystrix families. The theoretical model used was as follows:
where Z is the breeding value of the quantitative traits; μ is the overall average value; Si is the fixed effect of the environment i; SGij is the random interaction effect between the jth family and the ith location; and eij is a random error.
Genstat 18.0 software was used to prepare the BLUP–GGE biplots. Specifically, the impacts of the planting environment on the growth and morphological performance of the C. hystrix families were eliminated using analysis of variance (ANOVA). The main effects of the genotype and G × E were retained. The single-value decomposition method was then used to elucidate the principal components.
3. Results
3.1. Genetic Variances in the Growth Traits of C. hystrix
The results of the single-location ANOVA for the growth traits of 20-year-old C. hystrix are shown in Table 3. The block and family effects for the tree height, DBH, and V at each location were either significant (p < 0.05) or extremely significant (p < 0.01). The block × family effects of the three traits were extremely significant at the LZ site (p < 0.01). However, there were no significant differences at PX, and the tree height was only found to have a significant interaction effect at YL (p < 0.05). The interaction effects of the DBH and V were not significant.

Table 3.
Analysis of the variances in growth traits of C. hystrix at the YL, LZ, and PX locations.
Analyses of the performances of all families showed that the average annual growth in the height, DBH, and V for the C. hystrix trees at 20 years of age was the smallest at YL, with values of 0.64 m, 0.73 cm, and 0.006 m3, respectively. This was followed by LZ and then PX, with values of 0.74 m, 0.92 cm, and 0.0075 m3, and 0.94 m, 1.12 cm, and 0.018 m3, respectively (Table 4). Among the three experimental forests, the families with the largest growth in tree height were 21, 1, and 9, with values of 18.41, 20.80, and 19.93 m, respectively. The DBH growth for family 21 at YL and LZ was the largest at 20.53 and 16.06 cm, respectively, whereas family 20 had the highest value at PX (25.50 cm). The families with the largest individual plant V were 21, 1, and 18, with values of 0.29 m3, 0.24 m3, and 0.51 m3, respectively (Table 4).

Table 4.
Descriptive statistical analysis of the growth traits of C. hystrix.
The results of the triple-location ANOVA for the growth traits of C. hystrix are shown in Table 5. The results indicate extremely significant differences (p < 0.01) among families and locations for the tree height, DBH, and V, as well as for the family × location effect. These results indicate that the growth performances of the same family under different site conditions were significantly different. The growth performances of different families within the same site were also inconsistent, but there were obvious family × site effects.

Table 5.
Analysis of the variances in the growth traits of C. hystrix at the three locations.
3.2. Genetic Variances in the Morphological Traits of C. hystrix
The results of the single-location ANOVA for the morphological traits of the 20-year-old C. hystrix are shown in Table 6. The block and family were determined to be the main sources of high HCB variances. However, the block × family effect was only extremely significant at the YL site (p < 0.01). The main sources of CW variances among the C. hystrix families at YL were the block, family, and block × family. Their contributions to the CW variance, however, were not significant at LZ and PX.

Table 6.
Analysis of the variances in the morphological traits of C. hystrix at the YL, LZ, and PX locations.
A descriptive statistical analysis of the morphological traits of the 20-year-old C. hystrix families is shown in Table 7. From the perspective of the families’ overall performance, the average HCB, CW, and branch angle were 6.68 m, 2.15 m, and 45.45°, respectively. The average values for the morphological traits at the PX pilot location were higher than the group average, indicating that the PX pilot plants had the strongest natural adjustment abilities. The CW was relatively small after the large branches from the lower layer were pruned, leading to the maintenance of good crown shapes. The HCB of the C. hystrix families at LZ was the lowest. The thick branches at the sides were not pruned in time, which affected the crown shapes.

Table 7.
Descriptive statistical analysis of the morphological traits of C. hystrix.
The ANOVA results for the morphological traits of the families at the three locations showed that the location, family, and family × location effects were all extremely significant (p < 0.01). This indicated that the pruning abilities of trees in the same family were significantly different under different site conditions, the pruning strength of different families at the same location was different, and the pruning abilities of the various families at the three locations lacked stability. The CW and branch angle were only significantly different between locations (p < 0.01). Variances among families and the family × location effect did not reach a significant or extremely significant level, indicating that site conditions were the main factors Table 8.

Table 8.
Analysis of the variances in the morphological traits of C. hystrix at the three locations.
3.3. Estimating the Genetic Parameters for the Growth and Morphological Traits of C. hystrix
The ANOVA results for the CW and branch angle were not significant, and the variance components could not be resolved. Furthermore, the variance components of the family in terms of the tree height, DBH, V, and HCB ranged from 0.00001 to 3.59, indicating that these four traits were all genetically regulated. Relatively speaking, the genetic coefficients of variance for V (ranging from 17.88% to 34.21%) were greater than those for the other traits, indicating that the genetic stability of the individual plant V was weak. The DBH, tree height, V, and HCB of C. hystrix had strong heritabilities. The family heritability was 0.41–0.83, indicating that these traits were strongly regulated by genetics (Table 9).

Table 9.
Single-location genetic parameters for the growth and morphological traits of C. hystrix.
The environmental coefficients of variance for the tree height, DBH, individual plant V, and HCB were greater than the genetic coefficients of variance. Relatively speaking, these traits were more affected by the environment of the sites. The family heritabilities of the tree height, DBH, and V were 0.35, 0.38, and 0.62, respectively. These were thus found to be moderately heritable traits, and their values were greater than that of the HCB, indicating that the genetic stability of the growth traits was stronger than that of the morphological traits (Table 10).

Table 10.
Genetic parameters for the growth and morphological traits of C. hystrix at the three locations.
3.4. BLUP
The YL and PX sites each had five families with positive breeding values for the growth and morphological traits. These were 12, 14, 15, 20, and 21 at YL, and 7, 14, 15, 19, and 20 at PX. The LZ site only had two families (1 and 21) with positive breeding values for the tree height, DBH, V, and HCB. At all three sites, families 10 and 17 had positive breeding values for the tree height; families 14 and 20 had positive breeding values for the individual plant V; and families 20 and 15 had positive breeding values for the DBH and HCB, respectively Table 11.

Table 11.
Breeding values for the growth and morphological traits of C. hystrix.
3.5. GGE-BLUP
The results show that 82.90% of the variance in the tree height was affected by the genotype effect and the G × E (Figure 1a and Figure 2a). Specifically, Principal Components 1 and 2 accounted for 44.87% and 38.3% of the total variances, respectively (Figure 1a). The which–won–where biplots show that the blue circle divided the three pilot sites into two site types: YL and PX were combined into one category and LZ was separate. The C. hystrix families on the outermost edge formed a pentagon based on two-by-two connections and enclosed the rest of the families. The five vertical lines of the pentagon divided the participating families into five different sectors. Among them, family 21 was at the apex of the YL and PX sectors, whereas family 1 was at the apex of the LZ sector. This indicates that family 21 had the greatest growth at YL and PX, whereas family 1 had the greatest growth at LZ (see Figure 1a). The mean vs. stability biplots indicate that the 21 C. hystrix families were divided into three types. Families 10, 14, and 21 were high-yielding but unstable, families 16 and 20 were high-yielding and stable, and families 2, 3, 4, 5, 6, 7, 8, 9, 11, 12, 13, 15, 17, 18, and 19 were low-yielding (see Figure 2a).
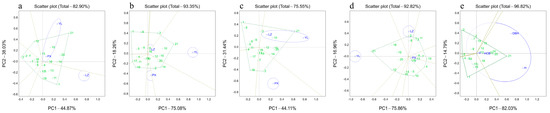
Figure 1.
Which–won–where GGE biplot based on the single traits and comprehensive traits at the three sites. (a) Biplot analysis based on the height BLUP. (b) Biplot analysis based on the DBH BLUP. (c) Biplot analysis based on the V. (d) Biplot analysis based on the HCB BLUP. (e) Biplot analysis based on the all-traits BLUP. LZ, YL, and PX indicate the three sites used for the C. hystrix family trials. Letters followed by numbers indicate the number of families. The green polygon was formed by connecting the outermost families lines, and the gray line was the perpendicular line from the origin to the green polygon.
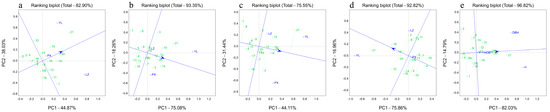
Figure 2.
Mean vs. stability GGE biplot based on the single traits and comprehensive traits at the three sites. (a) Biplot analysis based on the height BLUP. (b) Biplot analysis based on the DBH BLUP. (c) Biplot analysis based on the V. (d) Biplot analysis based on the HCB BLUP. (e) Biplot analysis based on the all-traits BLUP. LZ, YL, and PX indicate the three sites used for the C. hystrix family trials. Letters followed by numbers indicate the number of families. The blue lines with an arrow represented the mean environmental axis. The green vertical dotted lines represent the average yield and stability of each family in all experimental sites.
The GGE biplots for the DBH explained a total of 93.35% of the total variance, of which Principal Components 1 and 2 explained 75.08% and 18.26% of the total variances, respectively (Figure 1b and Figure 2b). Families 21, 2, and 3 had the largest DBH growth at YL, LZ, and PX, respectively (Figure 1b); families 15 and 20 were high-yielding and stable (Figure 2b); families 10, 14, and 21 were high-yielding but unstable; and families 1, 2, 3, 4, 5, 6, 7, 8, 9, 11, 12, 13, 16, 17, 18, and 19 were low-yielding.
The sources of 75.55% of the V variance were the genotype effect and the G × E. Principal Components 1 and 2 accounted for 44.11% and 31.44% of the total variance, respectively (Figure 1c and Figure 2c). YL and LZ belonged to the same site type and family 21 had the largest V growth, whereas PX was a separate site type in which family 18 had the largest V growth (Figure 1c). Families 14, 15, and 20 were high-yielding and stable; families 2 and 22 were high-yielding but unstable; and families 1, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 16, 17, 18, and 19 were low-yielding (Figure 2c).
The genetic effect and G × E explained 92.82% of the variance in the HCB (Figure 1d and Figure 2d). The ratio of Principal Components 1 to 2 was approximately 4:1. Families 21, 2, and 19 had the best pruning abilities at YL, LZ, and PX, respectively (Figure 1d), whereas families 15, 18, and 20 had strong and stable pruning abilities; families 2, 12, and 21 had strong but unstable pruning abilities; and families 1, 3, 4, 5, 6, 7, 8, 9, 10, 11, 13, 14, 16, 17, and 19 had poor pruning abilities (Figure 2d).
Principal Components 1 and 2 reflected 82.03% and 14.79% of the total variance, respectively (Figure 1e and Figure 2e). Both principal components reflected 96.85% of the genotype effect and the G × site interaction effect. Family 21 had the best overall performance in terms of the tree height, DBH, individual plant V, and HCB (Figure 1e). Four families (10, 14, 20, and 21) had excellent and stable comprehensive performances (Figure 2e), whereas the remaining 27 families had poor growth and morphological performances overall. The average genetic gains for their growth traits—tree height (H), diameter at breast height (DBH), and volume (V)—and morphological traits—height to live crown base (HCB)—were 21.29%, 22.92%, 21.25%, and −0.14%, respectively (Table 12).

Table 12.
Selected families and estimated genetic gains.
4. Discussion
Among the C. hystrix families investigated in this study, there were significant differences in their growth and morphological traits. These differences indicate that the traits have great genetic potential among the tested families, and excellent families could potentially be selected. The examined traits were also all found to exhibit significant effects based on the respective site, suggesting that C. hystrix has strong phenotypic plasticity. These findings have also been confirmed in numerous previous studies [15,23,24,25]. The results of this study, however, have further revealed that the environmental coefficients of variance of the tested traits were greater than the genetic coefficients of variance, and the site factors had a substantial contribution rate to the growth and morphology of the C. hystrix families. These facts indicate that the environmental factor of the planting sites had a stronger impact on the variances in the corresponding traits than the G factor. This phenomenon is also consistent with previous research on conifers (such as Larix kaempferi and Pinus taeda) and broad-leaved species (such as Betula alnoides and B. platyphylla) [6,24,26,27].
According to a survey on natural factors in the original habitat of C. hystrix, the soil pH value ranged from 4.36 to 6.23 [28], which was consistent with the soil acidity among the three sites. Previous studies showed that the correlation between the growth traits of C. hystrxi and the soil pH was weak [29]. So, it can be inferred that the growth differences were basically unrelated to the soil pH in the trials. The growth performance of C. hystrix at the LZ and YL sites was worse than that at the PX site. This was because the first two sites are located close to the Tropic of Cancer and have lower cumulative temperatures, whereas the PX site is located further south with a higher cumulative temperature. Differences in latitude and altitude often lead to corresponding changes in the climate, soil chemical property, and other factors, which, in turn, affects tree growth [30]. In addition, the C. hystrix families at PX had the largest tree-height values, which were 5.90 and 1.29 m taller than those at YL and LZ, respectively. Even though the stand at the PX site maintained a high growth rate, its natural pruning abilities were also the strongest, which promoted the natural pruning of the thick branches. Consequently, the average HCBs of the C. hystrix families at PX were 2.49 and 2.76 m higher than those at YL and LZ, respectively.
The tree height, DBH, V, and HCB of the C. hystrix families presented rank and scale effects at the three investigated sites [1]. This phenomenon was similarly identified during the popularization and utilization of loblolly pine and other tree species [23,24,25,26,27] and is essentially caused by the G × E. When breeding forest trees, it is crucial to determine which environmental factors are the key driving factors for the G × E. Doing so provides an important reference for the division of suitable planting regions to help improve tree species. The geographical differences between YL, LZ, and PX led to differences in the cumulative temperature, precipitation, and soil nutrients of the forest land. For a specific genotype set, the greater the site heterogeneity, the higher the G × E amplitude [31]. In addition, differences in the responses to environmental changes among genotypes constitute the internal driving force generated by the G × E [1]. Therefore, the analysis of the G × E helps to estimate the real genetic parameters of environmental quantitative traits in multiple locations. These analyses also provide a basis for the selection of genotypes with excellent phenotypes and stable genetics, as well as optimizing environmental evaluations. By selecting the best genotypes for the appropriate site, the aim of increasing the production and efficiency of artificial timber forests can be achieved.
In addition to genetic regulations, the impacts of the environmental effects and G × E on the genetic parameter estimations for the measured traits cannot be ignored [32]. The site conditions of the single-location experiments were consistent and the block variances were relatively small, resulting in the overestimation of the heritabilities of the tested traits. Among them, the heritabilities of the tree height, DBH, and V were 0.41–0.72, 0.50–0.83, and 0.54–0.74, respectively. These values revealed genetic regulations at the moderate level or above, which was consistent with the results of Zhu et al. [18] and Fang [19]. However, the ANOVA results of the three locations showed that the high environmental variances and variances in the family × site effects had diluted the genetic potential of the traits. In contrast to the results of this study, the single- and multiple-location heritabilities of the full-sib families in a third-generation horsetail pine seed garden were found to be similar [33]. This was mainly related to the genetic background of the tested families. In addition, the family heritabilities for the tree height, DBH, and V were higher than that of the HCB, indicating that growth traits were subjected to stronger genetic regulations. This was also consistent with the test results for B. alnoides [6].
Although the economic benefits of timber forests are dependent on their growth traits, the contributions of their morphological traits to their economic value cannot be ignored [32]. Improvements in morphological heritability are thus also considered an important component in the breeding of valuable tree species. The goal of forest tree breeding has thus changed from the selection of single traits to the comprehensive selection of multiple traits. However, owing to the inconsistent performances of the growth and morphological traits, it is often challenging to simultaneously improve the two trait types [4,6]. In the present study, multiple-trait BLUP–GGE biplots were used to select four families with excellent comprehensive performances. These families not only maintained their fast-growing characteristics but their morphological traits also performed well, such that their breeding efficiencies were in line with expectations. Considering that the tested stand was 20 years old and had reached half of the rotation period, the measured traits tended to be stable, and the measured results were accurate and reliable. The final families that were selected can thus be directly promoted and applied by forestry production units. These findings provide a theoretical basis for the efficient cultivation of C. hystrix in commercial forests in South China.
5. Conclusions
The genetic parameters for various traits of 21 C. hystrix families at 20 years of age located at three sites in China were estimated based on a comprehensive growth (tree height, DBH, and V) and morphological (HCB, CW, and branch angle) trait analysis. The aim was to clarify the impacts of the G × E. The results showed that the tree height, DBH, and average annual growth in the individual plant V of the C. hystrix families was 0.64–0.94 m, 0.73–1.12 cm, and 0.006–0.018 m3, respectively. Genetic testing at YL, LZ, and PX revealed that the tested traits were affected by the family, site, and family × site effects. The heritabilities of the tree height, DBH, and V were 0.35, 0.38, and 0.62, respectively, indicating moderate genetic regulation; HCB was under low-intensity genetic regulation with a heritability of 0.19. Considering the extreme significance of the G × E, the targeted cultivation of C. hystrix timber forests should aim to optimize the breeding objectives of improved varieties. Specifically, families 10, 14, 20, and 21 were selected as the four ideal families with comprehensive multiple traits, making them suitable for popularization and planting in various locations. The average genetic gains of the tree height, DBH, V, and HCB were 15.81%, 12.67%, 8.2%, and −0.14%, respectively. The GGE biplots based on BLUP were effectively applied to evaluate the C. hystrix families and their suitable planting sites, thereby providing a theoretical basis for the effective establishment and management of C. hystrix commercial forests in South China.
Author Contributions
G.L., Z.T., H.J., W.S. and J.X. designed the experiment; G.L., Z.L., L.T. and H.Z. performed the experiments and collected the data; G.L. and W.S. analyzed the data; G.L. and J.X. contributed to writing the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Guangxi Natural Science Foundation Project (Project No.: 2023GXNSFAA026497) and the Experimental Center of Tropical Forestry Scientific Fund under the Chinese Academy of Forestry (Project No. RL-2020-01).
Data Availability Statement
When needed, data can be uploaded.
Acknowledgments
We would like to express our sincere appreciation to Jie Zeng for his guidance on the research proposal and manuscript revisions, and to Chunsheng Wang for his guidance on data processing.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Li, Y.J.; Suontama, M.; Burdon, R.D.; Dungey, H.S. Genotype by environment interactions in forest tree breeding: Review of methodology and perspectives on research and application. Tree Genet. Genomes 2017, 13, 60. [Google Scholar]
- Lee, K.; Kim, I.S.; Lee, S.W. Estimation of genetic parameters on growth characteristics of a 35-year-old Pinus koraiensis progeny trial in South Korea. J. For. Res. 2020, 32, 2227–2236. [Google Scholar]
- Munhoz, L.; Biernaski, F.; Peres, F.; Dias, A.; Tabarussi, E. Predicted genetic gains for growth traits and genotype × environment interaction in Pinus greggii: New perspectives for genetic improvement in Brzzil. An. Da Acad. Bras. De Ciencisa 2021, 93 (Suppl. S3), e20101486. [Google Scholar]
- Baltunis, B.S.; Brawner, J.T. Clonal stability in Pinus radiata across New Zealand and Australia 1: Growth and form traits. New For. 2010, 40, 305–322. [Google Scholar]
- Suontama, M.; Low, C.B.; Stovold, G.T.; Miller, M.A.; Fleet, K.R.; Li, Y.; Dungey, H.S. Genetic parameters and genetic gains across three breeding cycles for growth and form traits of Eucalyptus regnans in New Zealand. Tree Genet. Genomes 2015, 11, 133. [Google Scholar]
- Yin, M.Y.; Guo, J.J.; Wang, C.S.; Zhao, Z.G.; Zeng, J. Genetic parameter estimates and genotype × environment interactions of growth and quality traits for Betula alnoides Buch.-Ham. Ex, D. Don in four provenance-family trials in Southern China. Forests 2019, 10, 1036. [Google Scholar]
- Shelbourne, C.J. Genotype-Environment Interaction: Its Study and Its Implications in Forest Tree Improvement; The IUFRO Genetics and SABRAO Joint Symposium: Tokyo, Japan, 1972; pp. 1–28. [Google Scholar]
- Zas, R.; Merlo, E.; Fernández-López, J. Genotype by environment interaction in maritime pine families in Galicia, Northwest Spain. Silvae Genet. 2004, 53, 175–182. [Google Scholar]
- Skrøppa, T. A critical evaluation of methods available to estimate the genotype × environment interaction. Stud. For. Suec. 1982, 166, 3–14. [Google Scholar]
- Burdon, R.D. Genetic correlation as a concept for studying genotype-environment interaction in forest tree breeding. Silvae Genetica. 1977, 26, 168–175. [Google Scholar]
- Richard, C. Statistical analysis of regional yield trials: AMMI analysis of factorial designs. Agric. Syst. 1996, 51, 242–244. [Google Scholar]
- Yan, W.K.; Hunt, L.A.; Sheng, Q.L.; Szlavnics, Z. Cultivar Evaluation and Mega-Environment Investigation Based on the GGE Biplot. Crop Sci. 2000, 40, 597–605. [Google Scholar]
- Cullis, B.R.; Smith, A.B.; Coombes, N.E. On the design of early generation variety trials with correlated data. J. Agric. Biol. Environ. Stat. 2006, 11, 381–393. [Google Scholar]
- Ling, J.; Xiao, Y.; Hu, J.; Wang, F.; Ouyang, F.; Wang, J.; Weng, Y.; Zhang, H. Genotype by environment interaction analysis of growth of Picea koraiensis families at different sites using BLUP-GGE. New For. 2021, 52, 113–127. [Google Scholar] [CrossRef]
- Li, Y.X.; Zhang, X.X.; Zhang, Q.H.; Jiang, L.P.; Han, R.; Sun, S.Q.; Hu, X.Q.; Pei, X.N.; Zhan, C.L.; Zhao, X.Y. G × E analysis of growth traits of Betula platyphylla clones at three separated sites in Northeastern China. Phyton-Int. J. Exp. Bot. 2022, 91, 2055–2068. [Google Scholar]
- Li, N.; Yang, Y.M.; Xu, F.; Chen, X.Y.; Wei, R.Y.; Li, Z.Y.; Pan, W.; Zhang, W.H. Genetic diversity and population structure analysis of Castanopsis hystrix and construction of a core collection using phenotypic traits and molecular markers. Genes 2022, 13, 2383. [Google Scholar] [CrossRef]
- Jiang, J.H.; Lv, J.X. Preliminary research of Castanopsis hystrix and Betula alnoides plantation wood applied in furniture and decoration. Sci. Silvae Sin. 2008, 44, 136–140, (In Chinese with English Abstract). [Google Scholar]
- Zhu, J.Y.; Shen, W.H.; Jiang, Y.; Lu, L.H.; Tan, Y.B.; Liu, X. Genetic variation and superior family selection of Castanopsis hystrix families. J. Trop. Subtrop. Bot. 2014, 22, 270–280, (In Chinese with English Abstract). [Google Scholar]
- Fang, B.J. Analysis and selection of genetic variation on growth traits among provenances/families of Castanopsis hystrix. Subtrop. Agric. Res. 2021, 17, 22–29, (In Chinese with English Abstract). [Google Scholar]
- Fu, L.Y.; Sharma, R.P.; Hao, K.J.; Tang, S.Z. A generalized interregional nonlinear mixed-effects crown width model for Prince Rupprecht larch in northern China. For. Ecol. Manag. 2017, 389, 364–373. [Google Scholar]
- Fu, L.Y.; Sun, H.; Sharma, R.P.; Lei, Y.C.; Zhang, H.R.; Tang, S.Z. Nonlinear mixed-effects crown width models for individual trees of Chinese fir (Cunninghamia lanceolata) in south-central China. For. Ecol. Manag. 2013, 302, 210–220. [Google Scholar]
- Pang, L.F.; Ma, Y.P.; Sharma, R.P.; Rice, S.; Song, X.; Fu, L. Developing an improved parameter estimation method for the segmented taper equation through combination of constrained two-dimensional optimum seeking and least square regression. Forests 2016, 7, 194. [Google Scholar] [CrossRef]
- Bian, L.M.; Shi, J.S.; Zheng, R.H.; Chen, J.H.; Wu, H.X. Genetic parameters and genotype-environment interactions of Chinese fir (Cunninghamia lanceolata) in Fujian province. Can. J. For. Res. 2014, 44, 582–592. [Google Scholar] [CrossRef]
- Zhao, X.Y.; Xia, H.; Wang, X.W.; Wang, C.; Liang, D.Y.; Li, K.L.; Liu, G.F. Variance and stability analysis of growth characters in half-sib Betula platyphylla families at three different sites in China. Euphytica 2016, 208, 173–186. [Google Scholar] [CrossRef]
- Murillo, O.; Resende, V.; Badilla, Y.; Gamboa, J.P. Genotype by environment interaction and teak (Tectona grandis L.) selection in Costa Rica. Silvae Genet. 2019, 68, 116–121. [Google Scholar] [CrossRef]
- Diao, S.; Hou, Y.M.; Xie, Y.H.; Sun, X.M. Age trends of genetic parameters, early selection and family by site interactions for growth traits in Larix kaempferi open-pollinated families. BMC Genet. 2016, 17, 104–115. [Google Scholar] [CrossRef]
- Souza, B.M.; Aguiar, A.V.D.; Dambrat, H.M.; Galucha, S.C.; Tambarussi, E.V.; Sestrem, M.S.C.D.S.; Tomigian, D.S.; Freitas, M.L.M.; Venson, I.; Torres-Dini, D. Effects of previous land use on genotype-by-environment interactions in two loblolly pine progeny tests. For. Ecol. Manag. 2022, 503, 119762. [Google Scholar] [CrossRef]
- Jiang, Y.; Xiao, Y.Q. Survey on natural factor in original habitat of Castanopsis hystrix of native forest in Guangxi. Guangxi For. Sci. 2007, 36, 202–205, (In Chinese with English Abstract). [Google Scholar]
- Liu, G.J.; Jia, H.Y.; Xu, J.M.; Niu, C.H.; Zeng, J.; Lan, G.; Zhu, M.F.; Li, W.Z. Effects of soil physicochemical properties on growth and heartwood formation of Castanopsis hystrix plantation. For. Res. 2021, 34, 88–97, (In Chinese with English Abstract). [Google Scholar]
- Krishnamoorthy, M.; Palanisamy, K.; Francis, A.P.; Girrrsan, K. Impact of environmental factors and altitude on growth and reproductive characteristics of teak (Tectona grandis Linn. f.) in Southern India. J. For. Environ. Sci. 2016, 32, 353–366. [Google Scholar] [CrossRef]
- Li, Y.; Xue, J.; Clinton, P.W.; Dungey, H.S. Genetic parameters and clone by environment interactions for growth and foliar nutrient concentrations in radiate pine on 14 widely diverse New Zealand sites. Tree Genet. Genomes 2015, 11, 10. [Google Scholar] [CrossRef]
- Vargas-Hernandez, J.J.; Adams, W.T.; Joyce, D.G. Quantitative genetic structure of stem form and branching traits in Douglas fir seedlings and implications for early selection. Silvae Genatica 2003, 52, 36–44. [Google Scholar]
- Yuan, C.Z.; Zhang, Z.; Jin, G.Q.; Zheng, Y.; Zhou, Z.C.; Sun, L.S.; Tong, H.B. Genetic parameters and genotype by environment interactions influencing growth and productivity in Masson pine in east and central China. For. Ecol. Manag. 2021, 487, 118991. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).