Spatial Analysis of Dense LiDAR Point Clouds for Tree Species Group Classification Using Individual Tree Metrics
Abstract
1. Introduction
2. Materials and Methods
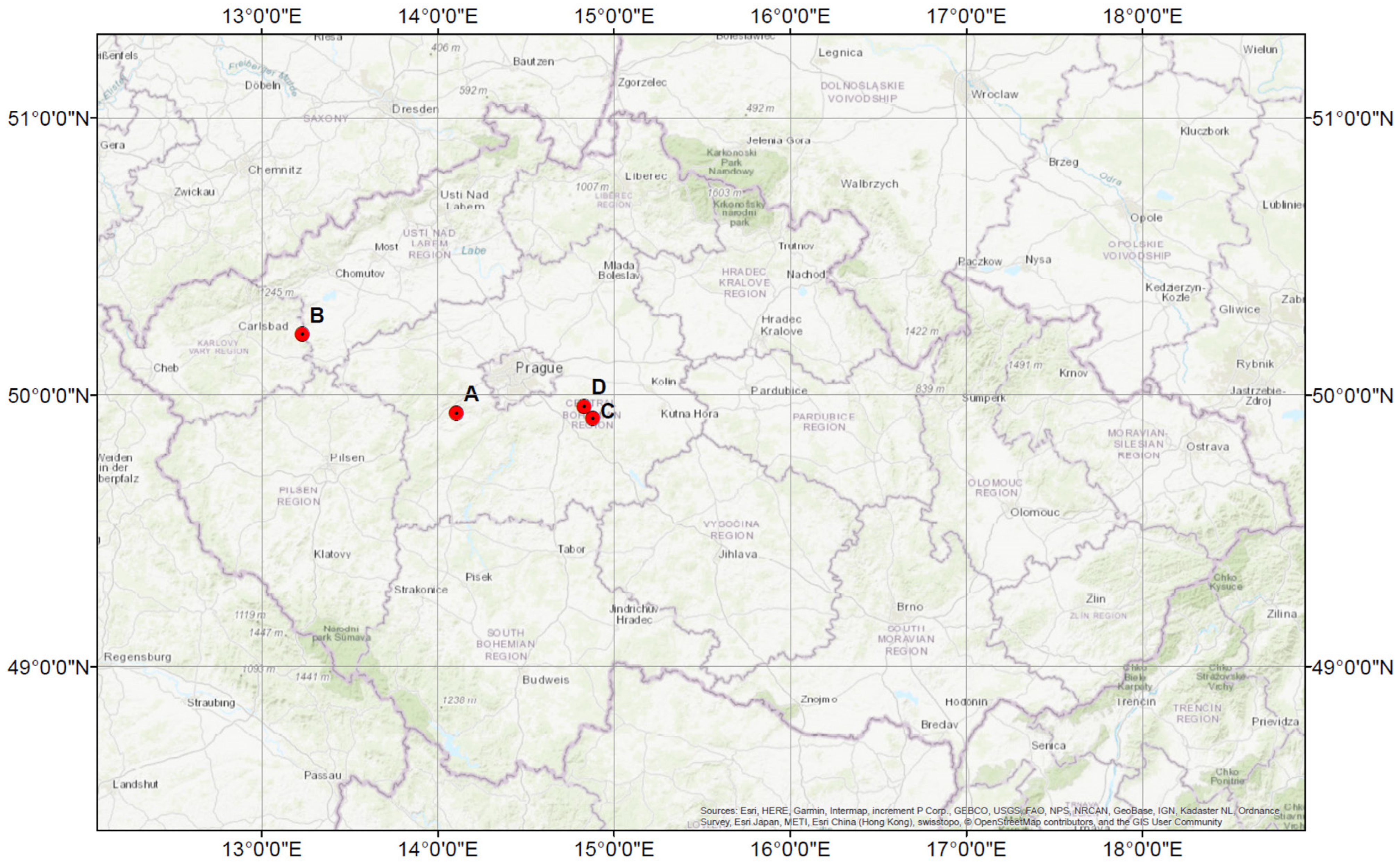
2.1. Study Area
2.2. UAV-Borne LiDAR
2.3. LiDAR Data Acquisition
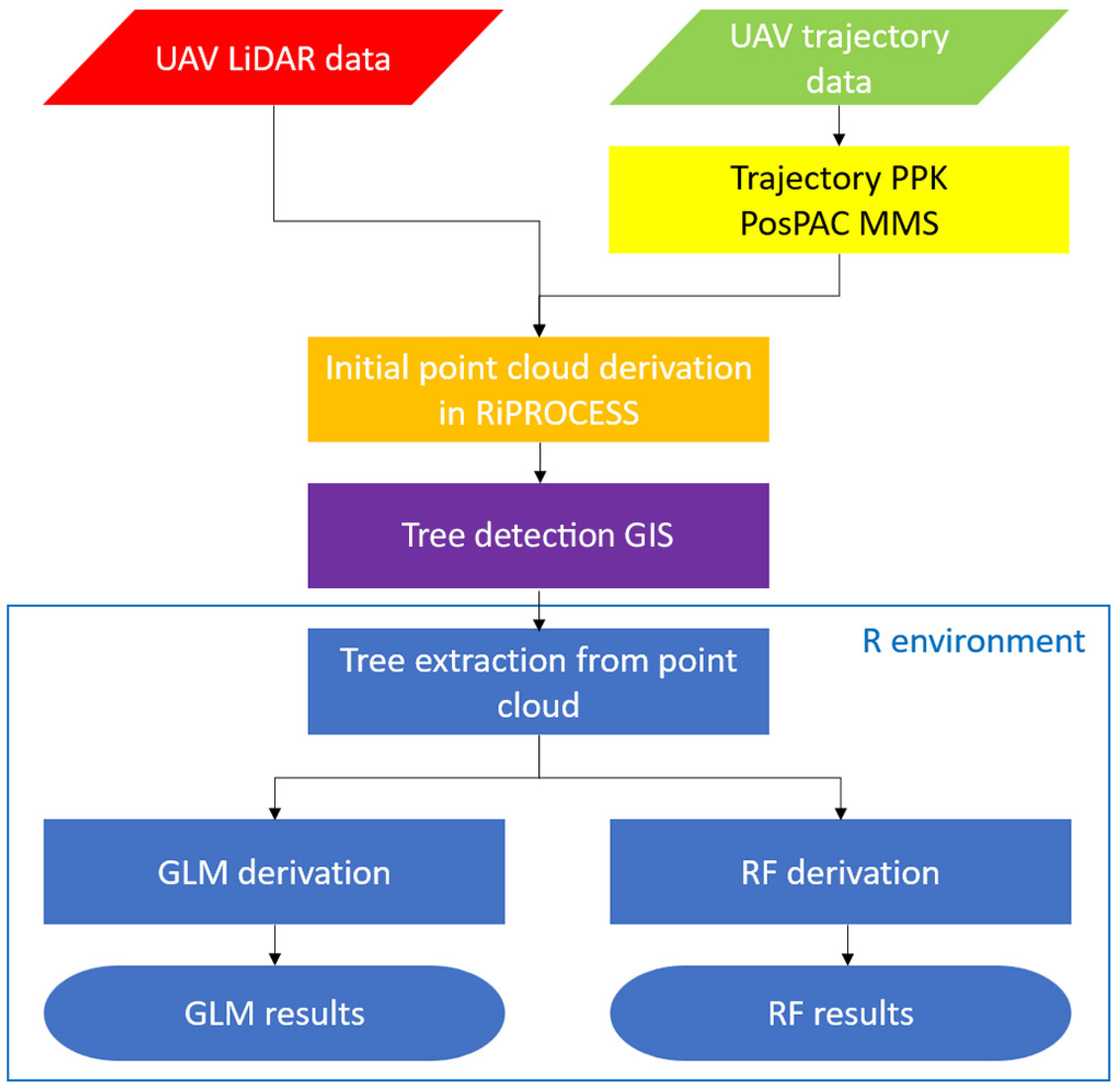
2.4. Data Processing
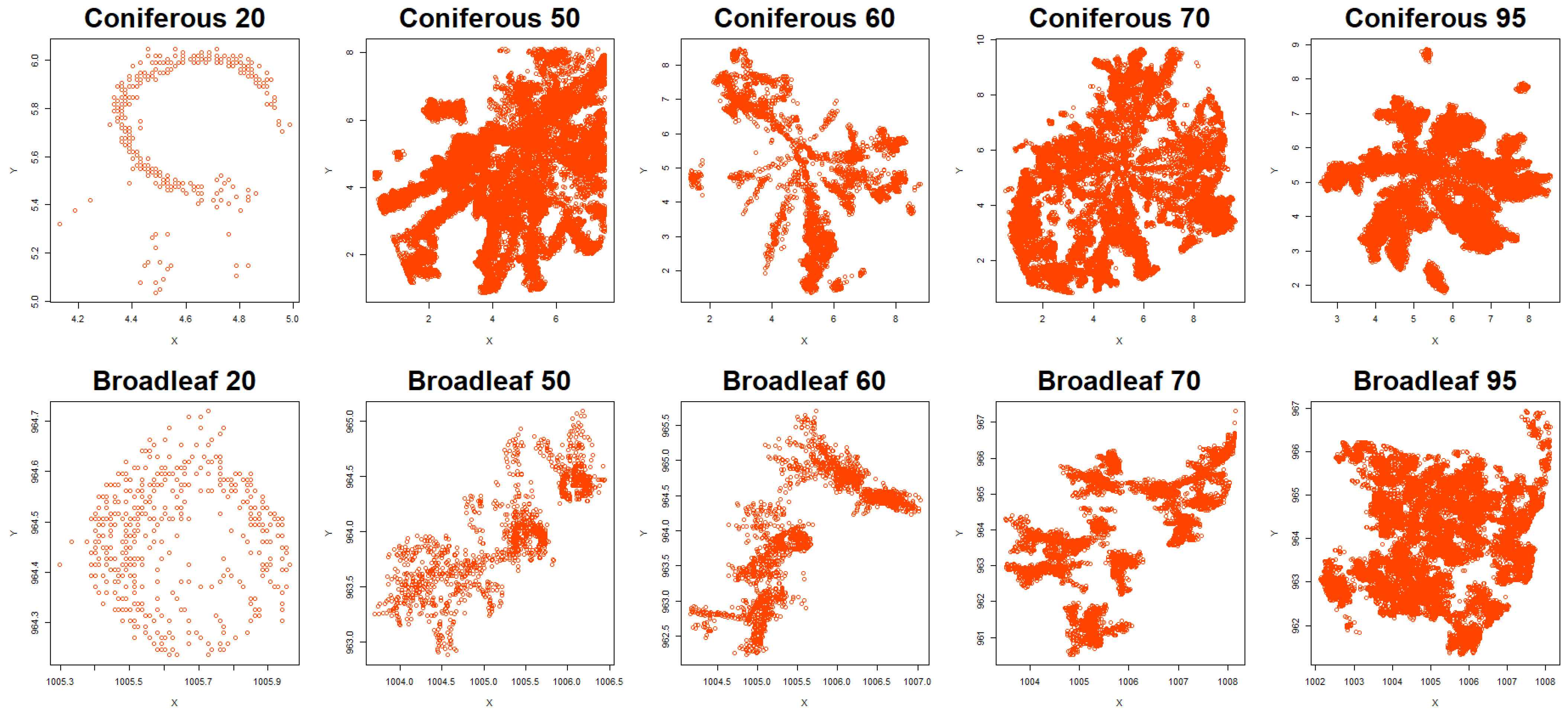
Clark–Evans Spatial Aggregation Index
3. Results
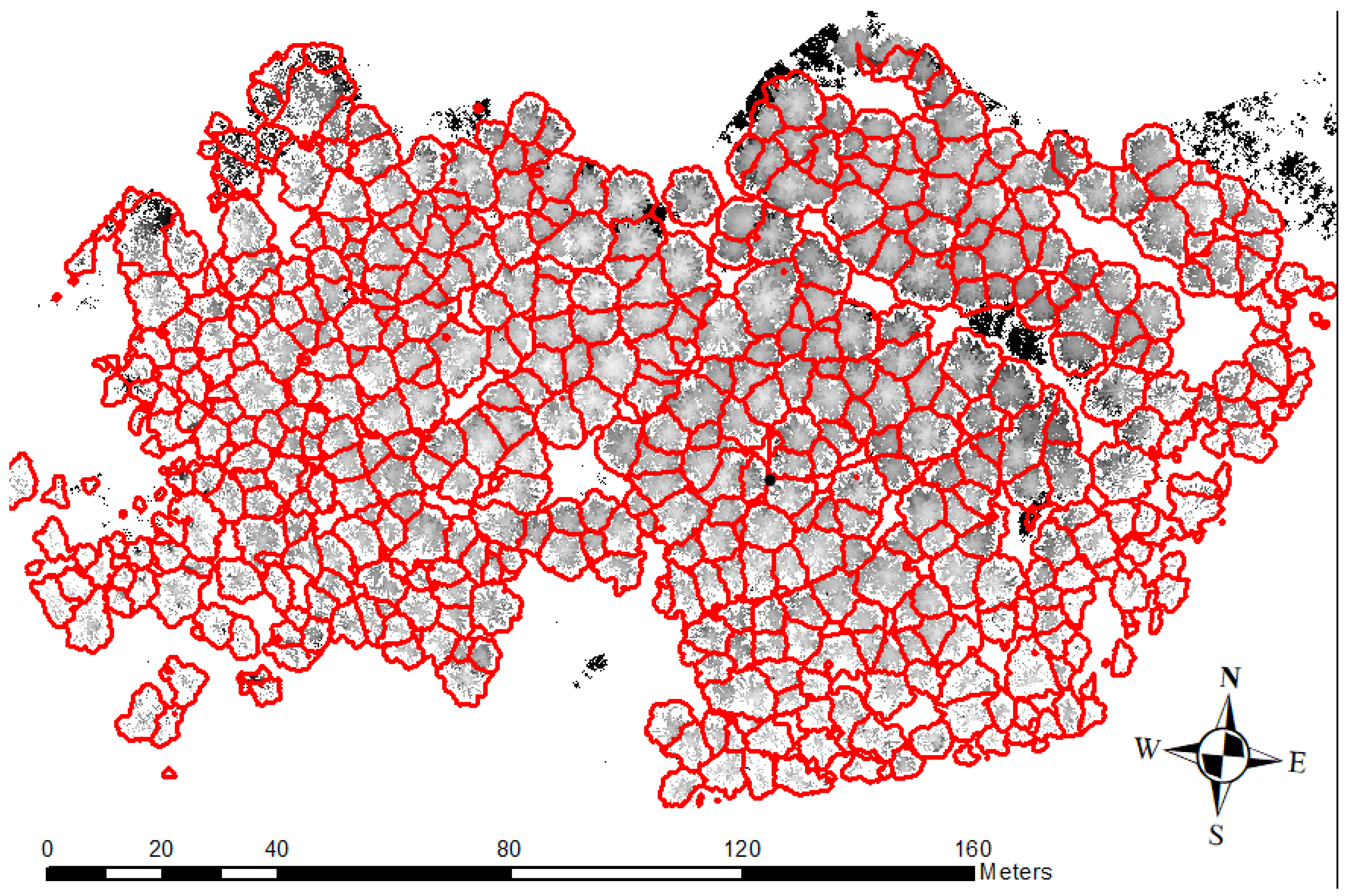
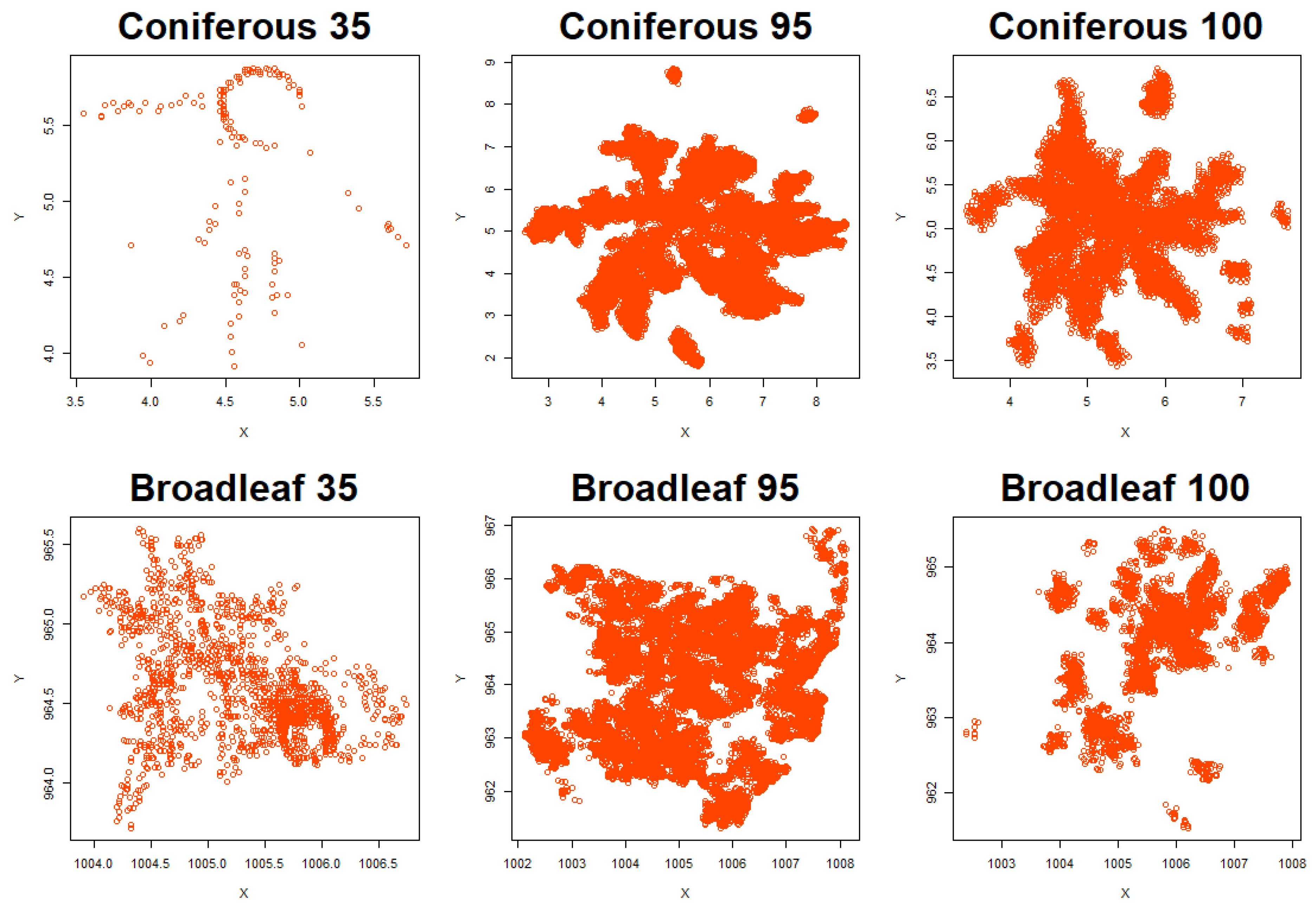
3.1. Tree Delineation
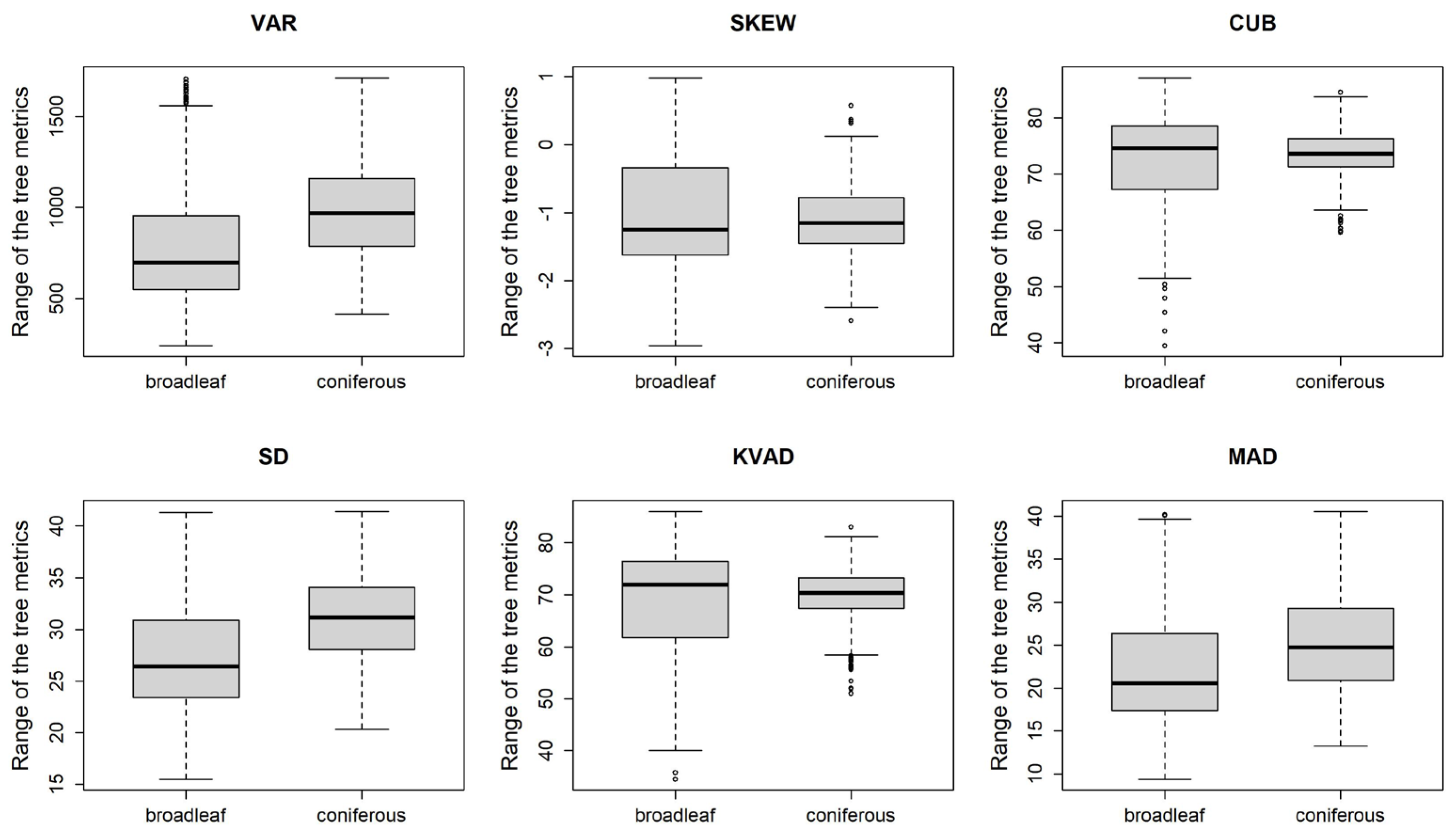
3.2. Generalized Linear Model
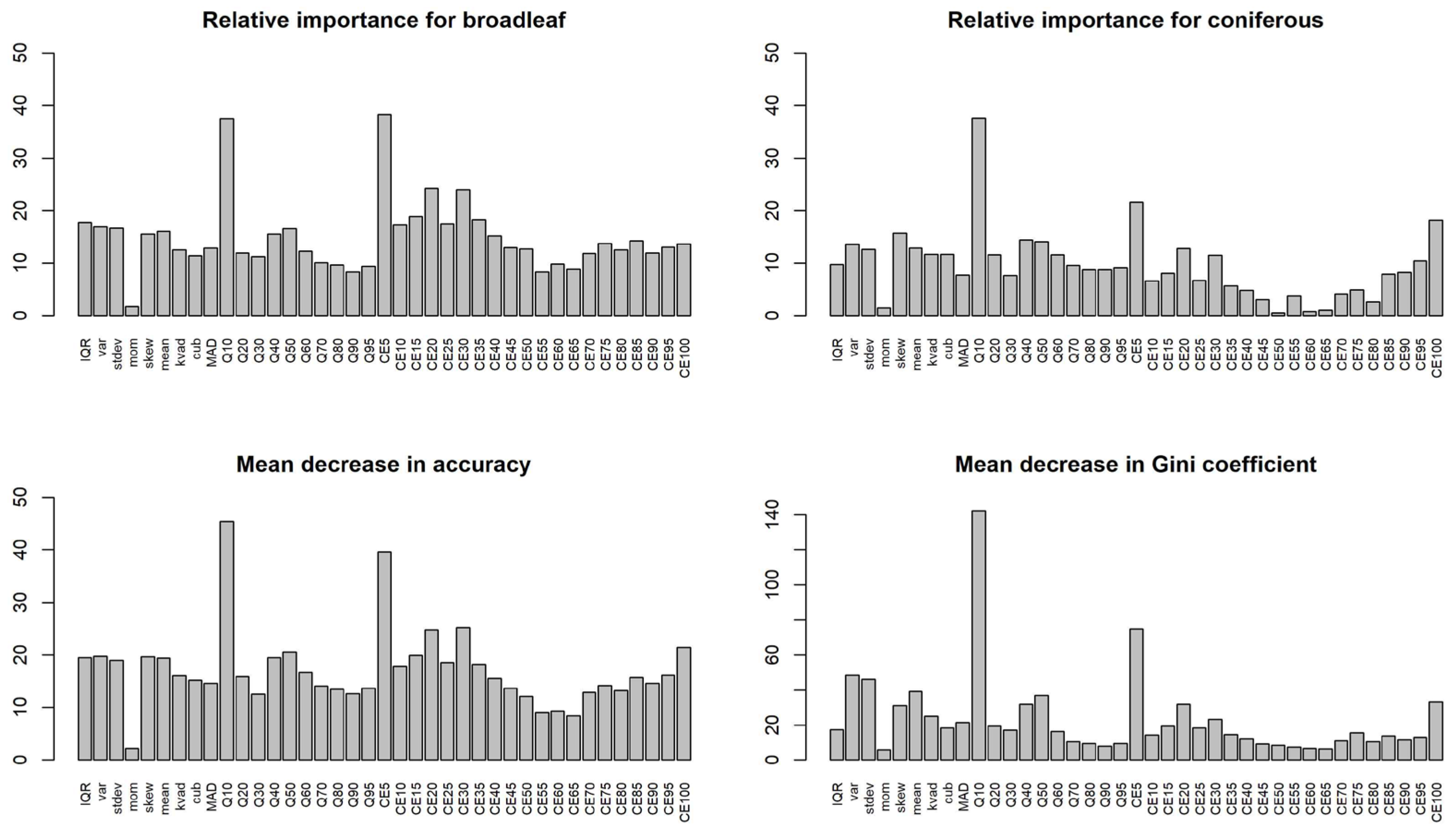
3.3. Random Forest
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Patacca, M.; Lindner, M.; Lucas-Borja, M.E.; Cordonnier, T.; Fidej, G.; Gardiner, B.; Hauf, Y.; Jasinevičius, G.; Labonne, S.; Linkevičius, E.; et al. Significant increase in natural disturbance impacts on European forests since 1950. Glob. Chang. Biol. 2023, 29, 1359–1376. [Google Scholar] [CrossRef] [PubMed]
- Hlásny, T.; Zimová, S.; Merganičová, K.; Štěpánek, P.; Modlinger, R.; Turčáni, M. Devastating outbreak of bark beetles in the Czech Republic: Drivers, impacts, and management implications. For. Ecol. Manag. 2021, 490, 119075. [Google Scholar] [CrossRef]
- Næsset, E.; Gobakken, T. Estimation of Above- and below-Ground Biomass across Regions of the Boreal Forest Zone Using Airborne Laser. Remote Sens. Environ. 2008, 112, 3079–3090. [Google Scholar] [CrossRef]
- Lefsky, M.; Harding, D.; Blair, J.; Parker, G. Laser altimeter canopy height profiles: Methods and validation for closed-canopy, broadleaf forests. Remote Sens. Environ. 2001, 76, 283–297. [Google Scholar]
- Wang, Y.; Lehtomäki, M.; Liang, X.; Pyörälä, J.; Kukko, A.; Jaakkola, A.; Liu, J.; Feng, Z.; Chen, R.; Hyyppä, J. Is Field-Measured Tree Height as Reliable as Believed—A Comparison Study of Tree Height Estimates from Field Measurement, Airborne Laser Scanning and Terrestrial Laser Scanning in a Boreal Forest. ISPRS J. Photogramm. Remote Sens. 2019, 147, 132–145. [Google Scholar] [CrossRef]
- Panagiotidis, D.; Abdollahnejad, A.; Slavík, M. Assessment of Stem Volume on Plots Using Terrestrial Laser Scanner: A Precision Forestry Application. Sensors 2021, 21, 301. [Google Scholar] [CrossRef]
- Terryn, L.; Calders, K.; Disney, M.; Origo, N.; Malhi, Y.; Newnham, G.; Raumonen, P.; kerblom, M.Å.; Verbeeck, H. Tree Species Classification Using Structural Features Derived from Terrestrial Laser Scanning. ISPRS J. Photogramm. Remote Sens. 2020, 168, 170–181. [Google Scholar] [CrossRef]
- Calders, K.; Adams, J.; Armston, J.; Bartholomeus, H.; Bauwens, S.; Bentley, L.P.; Chave, J.; Danson, F.M.; Demol, M.; Disney, M.; et al. Terrestrial Laser Scanning in Forest Ecology: Expanding the Horizon. Remote Sens. Environ. 2020, 251, 112102. [Google Scholar] [CrossRef]
- Gollob, C.; Ritter, T.; Nothdurft, A. Forest Inventory with Long Range and High-Speed Personal Laser Scanning (PLS) and Simultaneous Localization and Mapping (SLAM) Technology. Remote Sens. 2020, 12, 1509. [Google Scholar] [CrossRef]
- Jaakkola, A.; Hyyppä, J.; Kukko, A.; Yu, X.; Kaartinen, H.; Lehtomäki, M.; Lin, Y. A low-cost multi-sensoral mobile mapping system and its feasibility for tree measurements. ISPRS J. Photogramm. Remote Sens. 2010, 65, 514–522. [Google Scholar] [CrossRef]
- Wallace, L.; Lucieer, A.; Watson, C.; Turner, D. Development of a UAV-LiDAR system with application to forest inventory. Remote Sens. 2012, 4, 1519–1543. [Google Scholar] [CrossRef]
- Ayrey, E.; Fraver, S.; Kershaw, J.A.; Kenefic, L.S.; Hayes, D.; Weiskittel, A.R.; Roth, B.E. Layer Stacking: A Novel Algorithm for Individual Forest Tree Segmentation from LiDAR Point Clouds. Can. J. Remote Sens. 2017, 43, 16–27. [Google Scholar] [CrossRef]
- Vauhkonen, J.; Korpela, I.; Maltamo, M.; Tokola, T. Imputation of Single-Tree Attributes Using Airborne Laser Scanning-Based Height, Intensity, and Alpha Shape Metrics. Remote Sens. Environ. 2010, 114, 1263–1276. [Google Scholar] [CrossRef]
- Aubry-Kientz, M.; Dutrieux, R.; Ferraz, A.; Saatchi, S.; Hamraz, H.; Williams, J.; Coomes, D.; Piboule, A.; Vincent, G. A Comparative Assessment of the Performance of Individual Tree Crowns Delineation Algorithms from ALS Data in Tropical Forests. Remote Sens. 2019, 11, 1086. [Google Scholar] [CrossRef]
- Kellner, J.R.; Armston, J.; Birrer, M.; Cushman, K.C.; Duncanson, L.; Eck, C.; Falleger, C.; Imbach, B.; Král, K.; Krůček, M.; et al. New Opportunities for Forest Remote Sensing Through Ultra-High-Density Drone Lidar. Surv. Geophys. 2019, 40, 959–977. [Google Scholar] [CrossRef]
- Yan, W.; Guan, H.; Cao, L.; Yu, Y.; Li, C.; Lu, J.Y. A Self-Adaptive Mean Shift Tree-Segmentation Method Using UAV LiDAR Data. Remote Sens. 2020, 12, 515. [Google Scholar] [CrossRef]
- Krůček, M.; Král, K.; Cushman, K.C.; Missarov, A.; Kellner, J.R. Supervised Segmentation of Ultra-High-Density Drone Lidar for Large-Area Mapping of Individual Trees. Remote Sens. 2020, 12, 3260. [Google Scholar] [CrossRef]
- Bruggisser, M.; Hollaus, M.; Otepka, J.; Pfeifer, N. Influence of ULS Acquisition Characteristics on Tree Stem Parameter Estimation. ISPRS J. Photogramm. Remote Sens. 2020, 168, 28–40. [Google Scholar] [CrossRef]
- Kuželka, K.; Slavík, M.; Surový, P. Very High Density Point Clouds from UAV Laser Scanning for Automatic Tree Stem Detection and Direct Diameter Measurement. Remote Sens. 2020, 12, 1236. [Google Scholar] [CrossRef]
- Wieser, M.; Mandlburger, G.; Hollaus, M.; Otepka, J.; Glira, P.; Pfeifer, N. A case study of UAS borne laser scanning for measurement of tree stem diameter. Remote Sens. 2017, 9, 1154. [Google Scholar] [CrossRef]
- Brede, B.; Lau, A.; Bartholomeus, H.M.; Kooistra, L. Comparing RIEGL RiCOPTER UAV LiDAR Derived Canopy Height and DBH with Terrestrial LiDAR. Sensors 2017, 17, 2371. [Google Scholar] [CrossRef]
- Hyyppä, E.; Yu, X.; Kaartinen, H.; Hakala, T.; Kukko, A.; Vastaranta, M.; Hyyppä, J. Comparison of Backpack, Handheld, under-Canopy UAV, and above-Canopy UAV Laser Scanning for Field Reference Data Collection in Boreal Forests. Remote Sens. 2020, 12, 3327. [Google Scholar] [CrossRef]
- Hyyppä, E.; Hyyppä, J.; Hakala, T.; Kukko, A.; Wulder, M.A.; White, J.C.; Pyörälä, J.; Yu, X.; Wang, Y.; Virtanen, J.P.; et al. Under-Canopy UAV Laser Scanning for Accurate Forest Field Measurements. ISPRS J. Photogramm. Remote Sens. 2020, 164, 41–60. [Google Scholar] [CrossRef]
- Puliti, S.; Breidenbach, J.; Astrup, R. Estimation of Forest Growing Stock Volume with UAV Laser Scanning Data: Can It Be Done without Field Data? Remote Sens. 2020, 12, 1245. [Google Scholar] [CrossRef]
- Liu, K.; Shen, X.; Cao, L.; Wang, G.; Cao, F. Estimating forest structural attributes using UAV-LiDAR data in Ginkgo plantations. ISPRS J. Photogramm. Remote Sens. 2018, 146, 465–482. [Google Scholar] [CrossRef]
- Michałowska, M.; Rapí, J.; Cao, L.; Fernández-Manso, A. A Review of Tree Species Classification Based on Airborne LiDAR Data and Applied Classifiers. Remote Sens. 2021, 13, 353. [Google Scholar] [CrossRef]
- Pereira Martins-Neto, R.; Garcia Tommaselli, A.M.; Imai, N.N.; Honkavaara, E.; Miltiadou, M.; Saito Moriya, E.A.; David, H.C. Tree Species Classification in a Complex Brazilian Tropical Forest Using Hyperspectral and LiDAR Data. Forests 2023, 14, 945. [Google Scholar] [CrossRef]
- Holmgren, J.; Persson, Å. Identifying Species of Individual Trees Using Airborne Laser Scanner. Remote Sens. Enviton. 2004, 90, 415–423. [Google Scholar] [CrossRef]
- Belgiu, M.; Drăgu, L. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Vapnik, V. The support vector method of function estimation. In Nonlinear Modeling; Springer: Berlin/Heidelberg, Germany, 1998; pp. 55–85. [Google Scholar]
- Mountrakis, G.; Im, J.; Ogole, C. Support vector machines in remote sensing: A review. ISPRS J. Photogramm. Remote Sens. 2011, 66, 247–259. [Google Scholar] [CrossRef]
- Kranjčić, N.; Medak, D.; Župan, R.; Rezo, M. Support Vector Machine accuracy assessment for extracting green urban areas in towns. Remote Sens. 2019, 11, 655. [Google Scholar] [CrossRef]
- Pal, M. Random forest classifier for remote sensing classification. Int. J. Remote Sens. 2005, 26, 217–222. [Google Scholar] [CrossRef]
- RIEGL Data Sheet, RIEGL VUX-1UAV. 2019. Available online: http://www.riegl.com/products/unmanned-scanning/riegl-vux-1uav22/ (accessed on 20 July 2023).
- Panagiotidis, D.; Abdollahnejad, A.; Surový, P.; Chiteculo, V. Determining tree height and crown diameter from high-resolution UAV imagery. Int. J. Remote Sens. 2017, 38, 2392–2410. [Google Scholar] [CrossRef]
- Team, R.C. An introduction to dplR. Ind. Commer Train. 2019, 10, 11–18. [Google Scholar] [CrossRef]
- Næsset, E.; Gobakken, T. Estimating forest growth using canopy metrics derived from airborne laser scanner data. Remote Sens. Environ. 2005, 96, 453–465. [Google Scholar] [CrossRef]
- Kamińska, A.; Lisiewicz, M.; Stereńczak, K.; Kraszewski, B.; Sadkowski, R. Species-related single dead tree detection using multi-temporal ALS data and CIR imagery. Remote Sens. Environ. 2018, 219, 31–43. [Google Scholar] [CrossRef]
- Pekár, S.; Brabec, M. Modern Analysis of Biological Data: Generalized Linear Models in R; Masarykova Univerzita: Brno, Czechia, 2016; ISBN 8021081066. [Google Scholar]
- Breiman, L.; Cutler, A.; Liaw, A.; Wiener, M. Package ‘RandomForest’. 2018, p. 29. Available online: https://cran.r-hub.io/web/packages/randomForest/index.html (accessed on 20 July 2023).
- Han, H.; Guo, X.; Yu, H. Variable selection using Mean Decrease Accuracy and Mean Decrease Gini based on Random Forest. In Proceedings of the 2016 7th IEEE International Conference on Software Engineering and Service Science (ICSESS), Beijing, China, 26–28 August 2016; pp. 219–224. [Google Scholar]
- Iban, M.C.; Sekertekin, A. Machine learning based wildfire susceptibility mapping using remotely sensed fire data and GIS: A case study of Adana and Mersin provinces, Turkey. Ecol. Inform. 2022, 69, 101647. [Google Scholar] [CrossRef]
- Clark, P.J.; Evans, F.C. Distance to Nearest Neighbor as a Measure of Spatial Relationships in Populations. Ecology 1954, 35, 445–453. [Google Scholar] [CrossRef]
- Fabrika, M.; Pretsch, H. Analysis and Modelling of Forest Ecosystems; Technical University in Zvolen: Zvolen, Slovakia, 2011; pp. 1–599. [Google Scholar]
- Reitberger, J.; Krzystek, P.; Stilla, U. Analysis of full waveform LIDAR data for the classification of deciduous and coniferous trees. Int. J. Remote Sens. 2008, 29, 1407–1431. [Google Scholar] [CrossRef]
- Ørka, H.O.; Næsset, E.; Bollandsås, O.M. Classifying species of individual trees by intensity and structure features derived from airborne laser scanner data. Remote Sens. Environ. 2009, 113, 1163–1174. [Google Scholar] [CrossRef]
- Brandtberg, T. Classifying individual tree species under leaf-off and leaf-on conditions using airborne lidar. ISPRS J. Photogramm. Remote Sens. 2007, 61, 325–340. [Google Scholar] [CrossRef]
- Liang, X.; Matikainen, L. Deciduous-Coniferous Tree Classification Using Difference Between First and Last Pulse Laser Signatures. IAPRS 2007, 36, 253–257. [Google Scholar]
- Kim, S.; McGaughey, R.J.; Andersen, H.E.; Schreuder, G. Tree species differentiation using intensity data derived from leaf-on and leaf-off airborne laser scanner data. Remote Sens. Environ. 2009, 113, 1575–1586. [Google Scholar] [CrossRef]
- Vetrivel, A.; Gerke, M.; Kerle, N.; Vosselman, G. Identification of damage in buildings based on gaps in 3D point clouds from very high resolution oblique airborne images. ISPRS J. Photogramm. Remote Sens. 2015, 105, 61–78. [Google Scholar] [CrossRef]
- Shen, X.; Cao, L. Tree-species classification in subtropical forests using airborne hyperspectral and LiDAR data. Remote Sens. 2017, 9, 1180. [Google Scholar] [CrossRef]
- Krzystek, P.; Serebryanyk, A.; Schnörr, C.; Červenka, J.; Heurich, M. Large-scale mapping of tree species and dead trees in Sumava National Park and Bavarian Forest National Park using lidar and multispectral imagery. Remote Sens. 2020, 12, 661. [Google Scholar] [CrossRef]
- Immitzer, M.; Neuwirth, M.; Böck, S.; Brenner, H.; Vuolo, F.; Atzberger, C. Optimal input features for tree species classification in Central Europe based on multi-temporal Sentinel-2 data. Remote Sens. 2019, 11, 2599. [Google Scholar] [CrossRef]



| Estimate | Std. Error | z Value | p Value | ||
|---|---|---|---|---|---|
| Intercept | −109.149 | 16.950 | −6.439 | 1.20 × 10−10 | *** |
| VAR | −0.059 | 0.0114 | −5.130 | 2.90 × 10−7 | *** |
| SD | 2.887 | 0.737 | 3.917 | 8.95 × 10−5 | *** |
| SKEW | −5.504 | 2.541 | −2.166 | 0.0303 | * |
| KVAD | 4.322 | 1.163 | 3.717 | 0.0002 | *** |
| CUB | −3.774 | 1.211 | −3.116 | 0.0018 | ** |
| MAD | 2.016 | 0.505 | 3.991 | 6.57 × 10−5 | *** |
| Q20 | 0.116 | 0.035 | 3.325 | 0.0009 | *** |
| Q50 | −0.156 | 0.031 | −5.034 | 4.79 × 10−7 | *** |
| Q60 | −0.229 | 0.062 | −3.689 | 0.0002 | *** |
| Q70 | −0.254 | 0.119 | −2.152 | 0.0314 | * |
| Q95 | 0.404 | 0.196 | 2.064 | 0.0391 | * |
| CE35 | 1.944 | 1.040 | 1.869 | 0.0616 | |
| CE95 | 5.049 | 2.190 | 2.305 | 0.0212 | * |
| CE100 | 9.993 | 1.857 | 5.383 | 7.34 × 10−8 | *** |
| Estimate | Std. Error | z Value | p Value | ||
|---|---|---|---|---|---|
| Intercept | −1.053 × 102 | 14.190 | −7.421 | 1.17 × 10−13 | *** |
| VAR | −4.742 × 10−2 | 0.009 | −5.442 | 5.27 × 10−8 | *** |
| SD | 2.001 | 0.644 | 3.121 | 0.0018 | |
| KVAD | 4.633 | 0.954 | 4.856 | 1.20 × 10−6 | *** |
| CUB | −3.748 | 1.016 | −3.689 | 0.0002 | *** |
| MAD | 2.059 | 0.429 | 4.802 | 1.57 × 10−6 | *** |
| Q20 | 0.096 | 0.030 | 3.144 | 0.0017 | ** |
| Q30 | 0.042 | 0.024 | 1.783 | 0.0746 | * |
| Q50 | −0.135 | 0.030 | −4.516 | 6.31 × 10−6 | *** |
| Q60 | −0.232 | 0.059 | −3.915 | 9.02 × 10−5 | *** |
| Q70 | −0.284 | 0.084 | −3.382 | 0.0007 | *** |
| Q90 | −0.403 | 0.221 | −1.825 | 0.0679 | |
| Q95 | 0.824 | 0.233 | 3.536 | 0.0004 | *** |
| Broadleaf | Coniferous | Class. Error | |
|---|---|---|---|
| Broadleaf | 263 | 27 | 9.3% |
| Coniferous | 27 | 728 | 3.6% |
| Broadleaf | Coniferous | Class. Error | |
|---|---|---|---|
| Broadleaf | 252 | 38 | 13.1% |
| Coniferous | 39 | 716 | 5.2% |
| Broadleaf | Coniferous | Class. Error | |
|---|---|---|---|
| Broadleaf | 260 | 30 | 10.3% |
| Coniferous | 21 | 734 | 2.8% |
| Broadleaf | Coniferous | Class. Error | |
|---|---|---|---|
| Broadleaf | 256 | 34 | 11.7% |
| Coniferous | 30 | 725 | 4% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Slavík, M.; Kuželka, K.; Modlinger, R.; Surový, P. Spatial Analysis of Dense LiDAR Point Clouds for Tree Species Group Classification Using Individual Tree Metrics. Forests 2023, 14, 1581. https://doi.org/10.3390/f14081581
Slavík M, Kuželka K, Modlinger R, Surový P. Spatial Analysis of Dense LiDAR Point Clouds for Tree Species Group Classification Using Individual Tree Metrics. Forests. 2023; 14(8):1581. https://doi.org/10.3390/f14081581
Chicago/Turabian StyleSlavík, Martin, Karel Kuželka, Roman Modlinger, and Peter Surový. 2023. "Spatial Analysis of Dense LiDAR Point Clouds for Tree Species Group Classification Using Individual Tree Metrics" Forests 14, no. 8: 1581. https://doi.org/10.3390/f14081581
APA StyleSlavík, M., Kuželka, K., Modlinger, R., & Surový, P. (2023). Spatial Analysis of Dense LiDAR Point Clouds for Tree Species Group Classification Using Individual Tree Metrics. Forests, 14(8), 1581. https://doi.org/10.3390/f14081581






