Impact of Root Rot Disease of Zanthoxylum armatum on Rhizosphere Soil Microbes and Screening of Antagonistic Bacteria
Abstract
1. Introduction
2. Materials and Methods
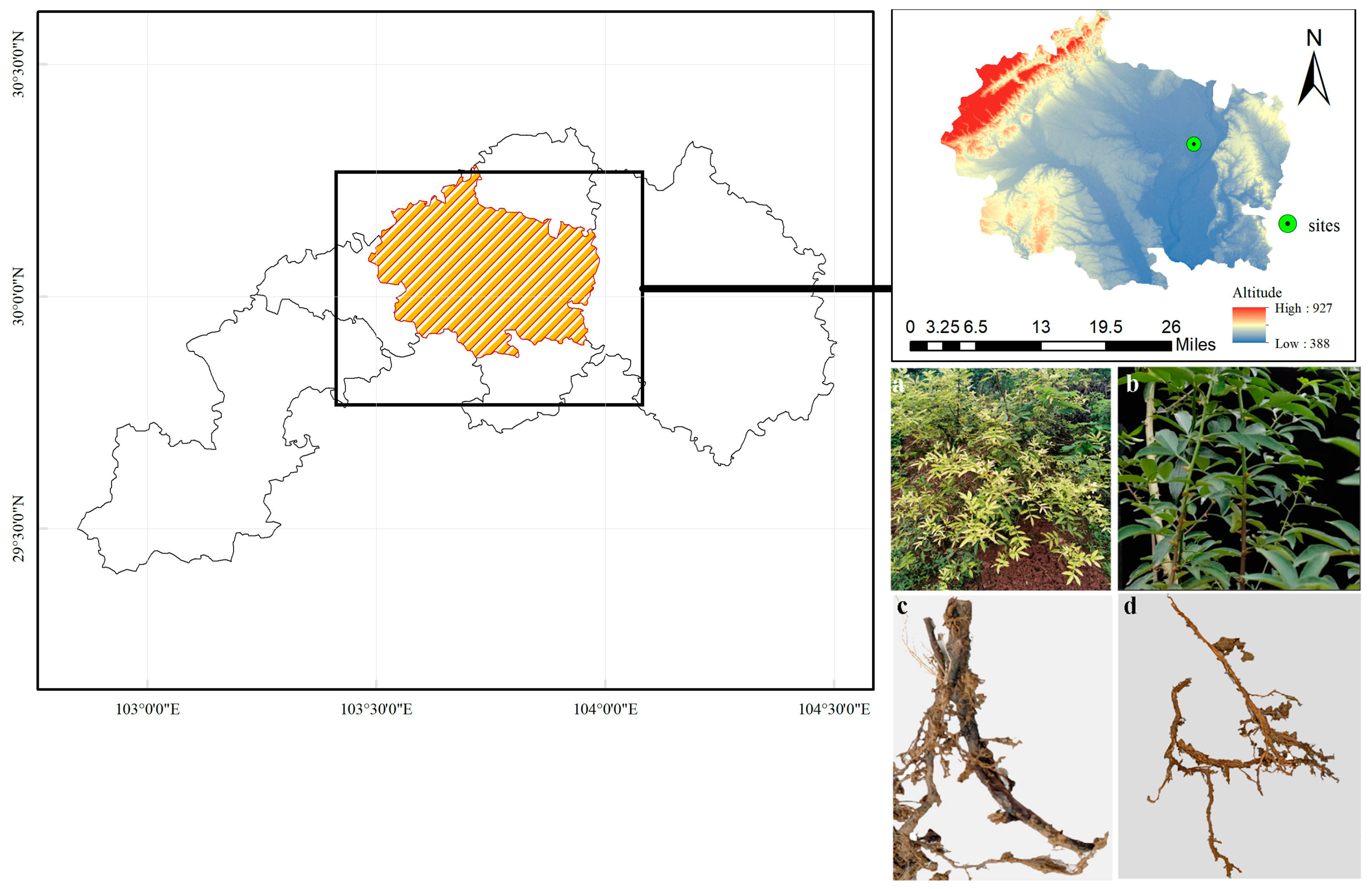
2.1. Experimental Design
2.2. Comparing the Physicochemical Characteristics of Diseased and Healthy Plants’ Rhizosphere Soil
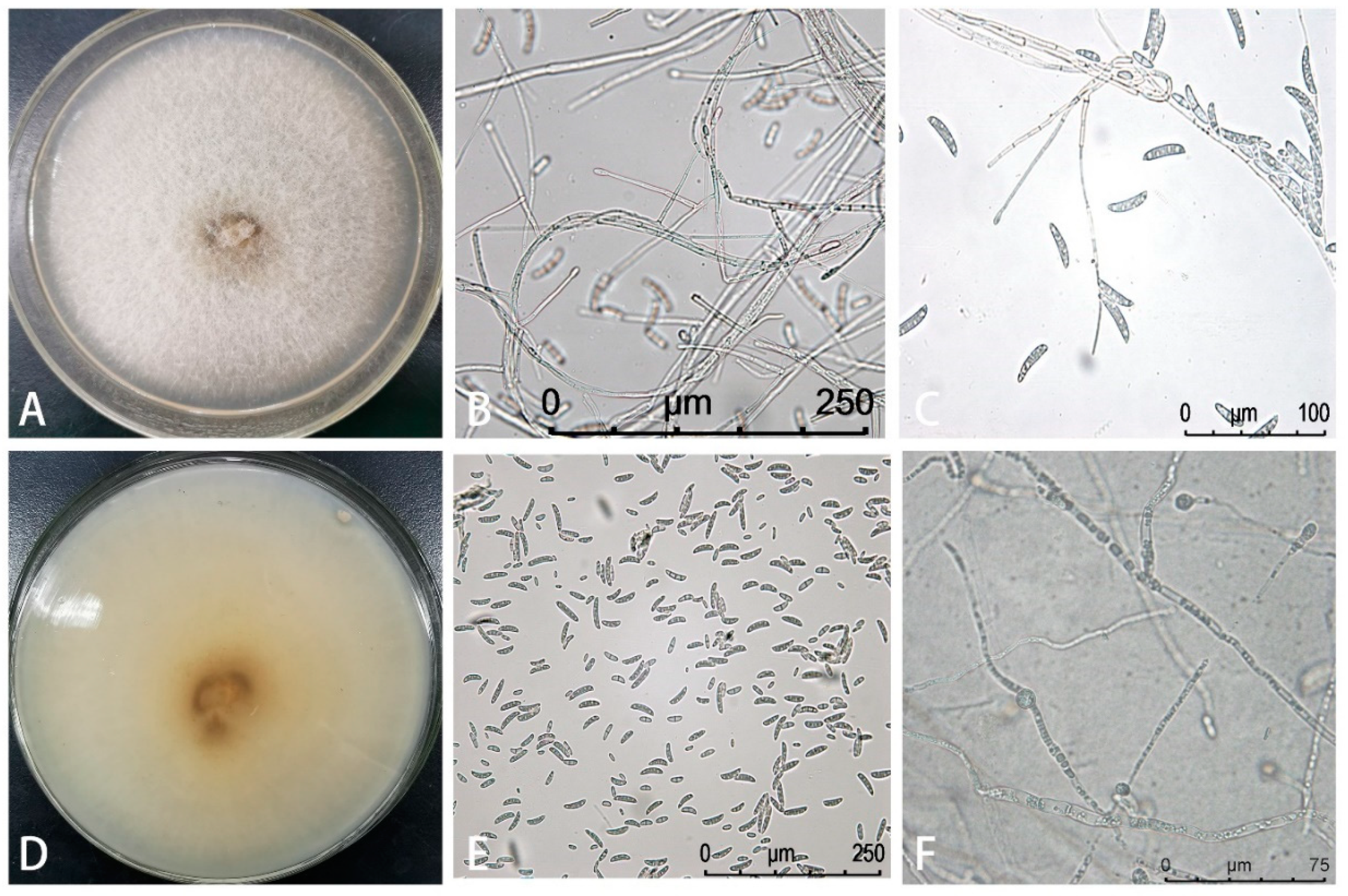
2.3. Isolating and Characterizing the Pathogen Causing Root Rot Disease in Z. armatum
2.4. Characterization of Rhizosphere Soil Microbial Microbiome
2.5. Bioinformatics Analysis
2.6. Isolation of Cultivable Bacterial and Fungal Strains from Rhizosphere Soil
2.7. Screening and Determination of Antagonistic Effects of Antagonistic Bacteria
2.8. Identification of Purified Microbial Strains with Potential Antagonistic Activity
2.9. Statistical Analysis
3. Results
3.1. Isolation and Identification of the Pathogen Causing Root Rot in Z. armatum
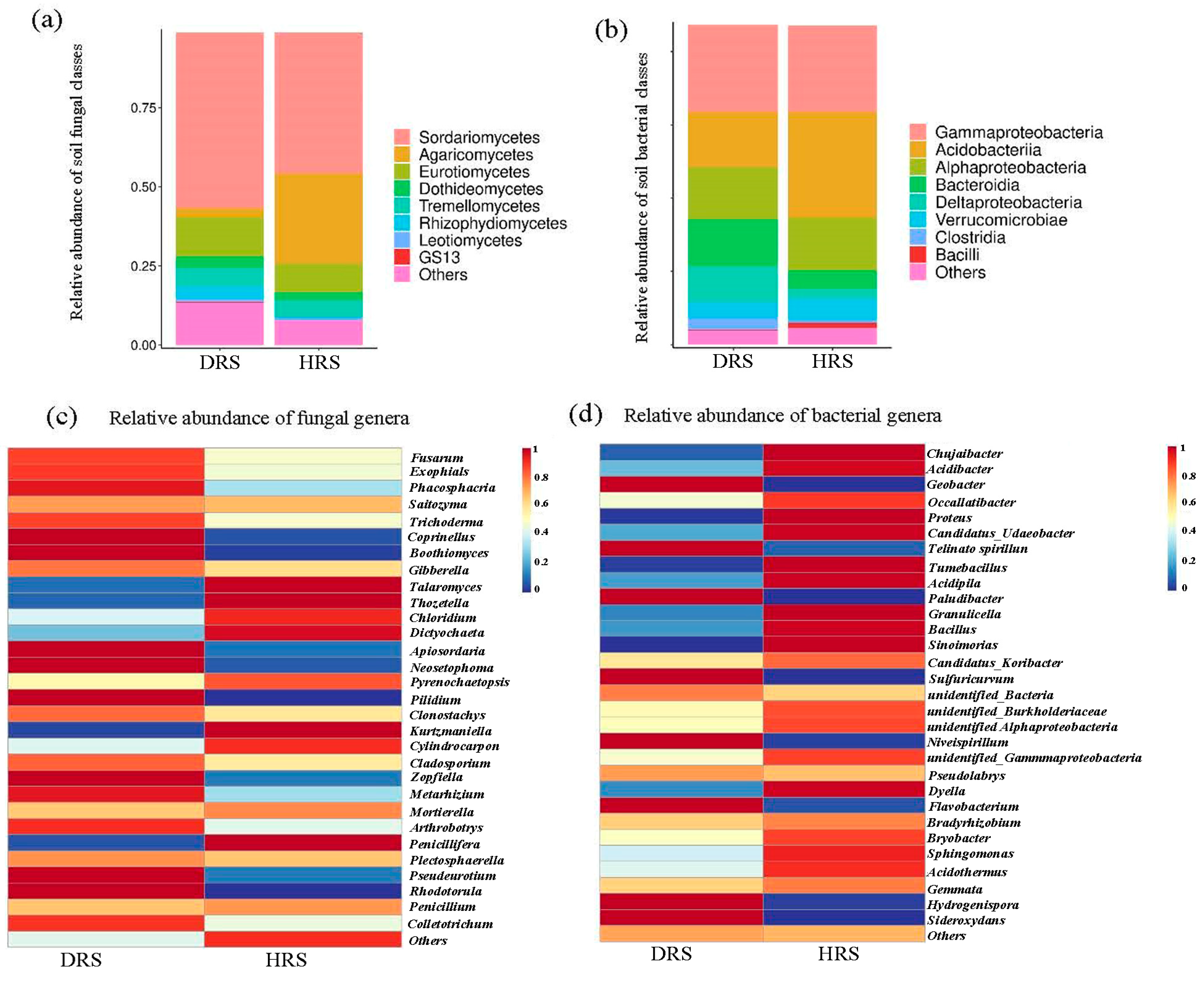
3.2. Impact of Fusarium Abundance on the Overall Taxonomic Distribution of Rhizosphere Soil Microbial Communities
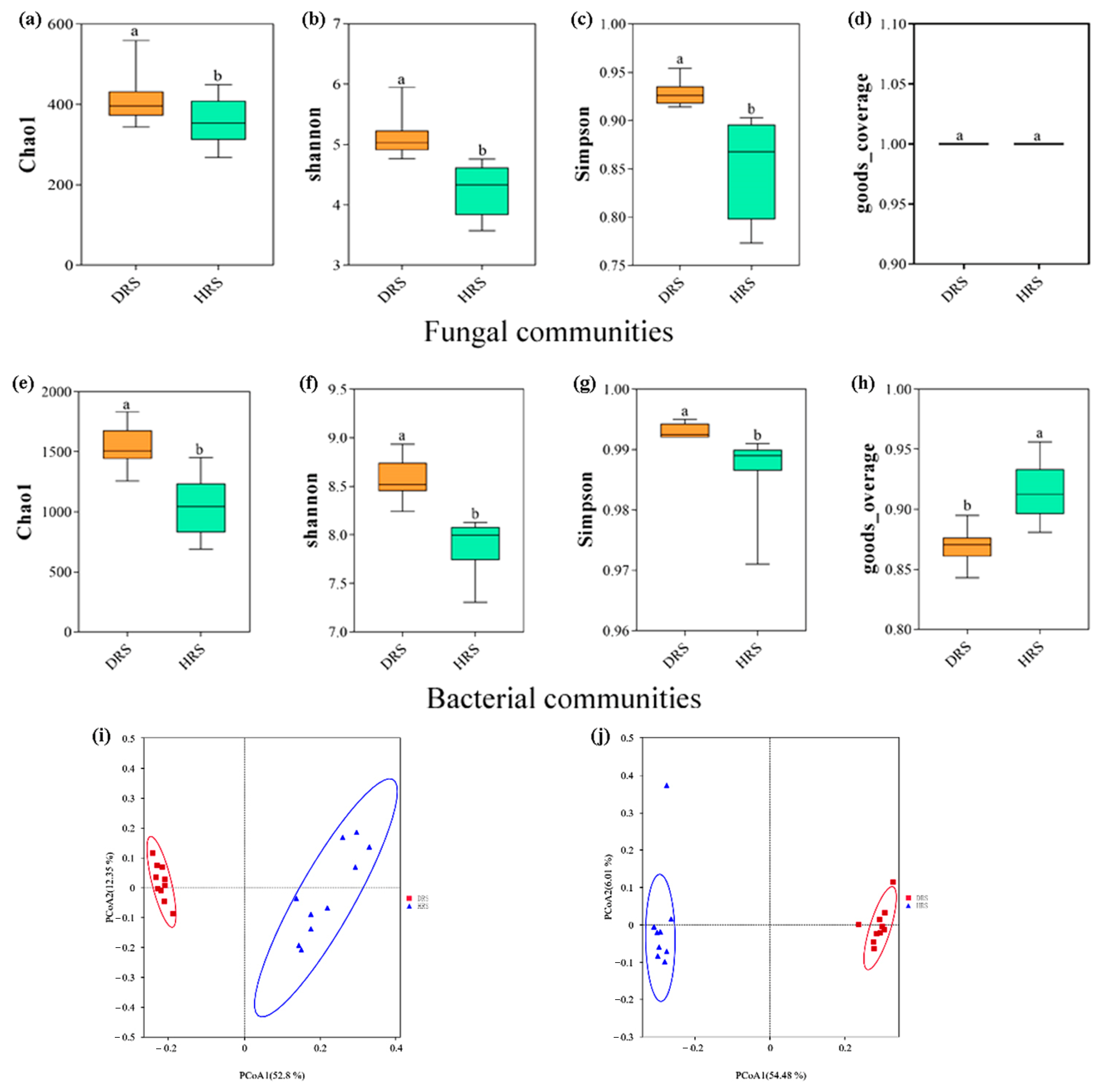
3.3. Study on the Diversity of Rhizosphere Soil Microbial Communities
3.4. Analysis of Rhizosphere Soil Microbial Community Abundance in Z. armatum
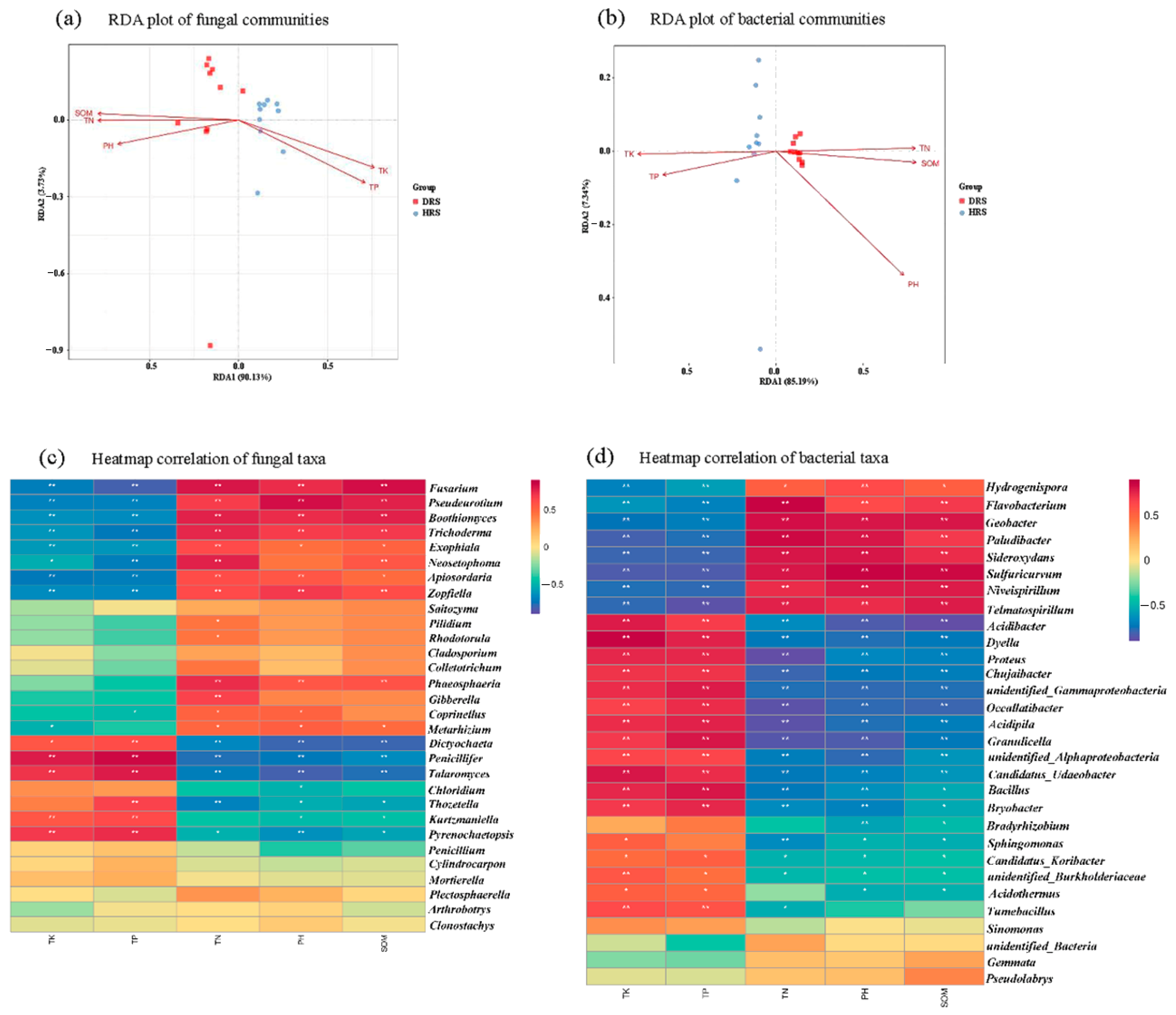
3.5. Relationship between Soil Physicochemical Properties and Microbial Community Composition
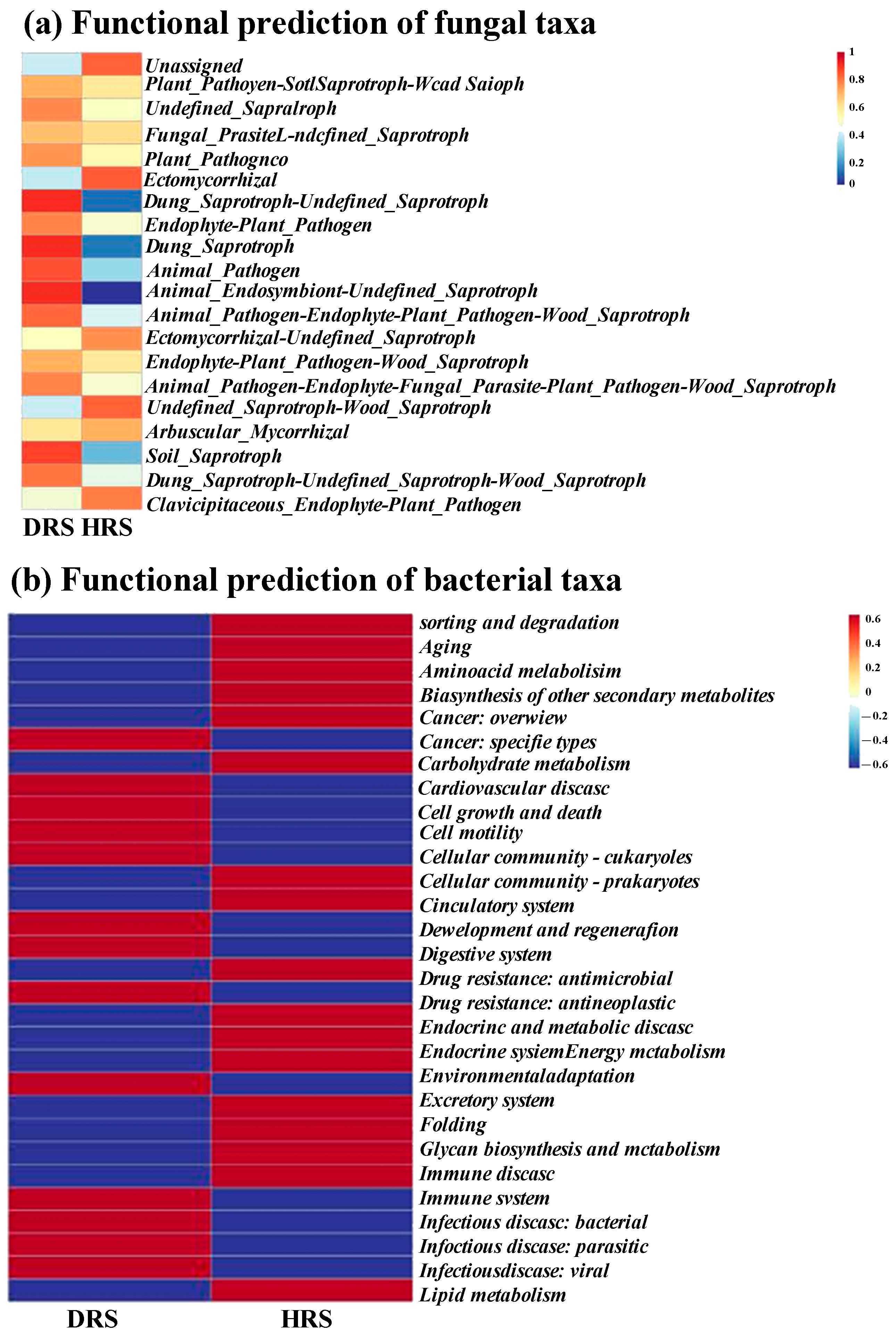
3.6. Functional Prediction of Fungal and Bacterial Taxa
3.7. Screening of Antagonistic Bacteria and Determination of Their Antagonistic Effects
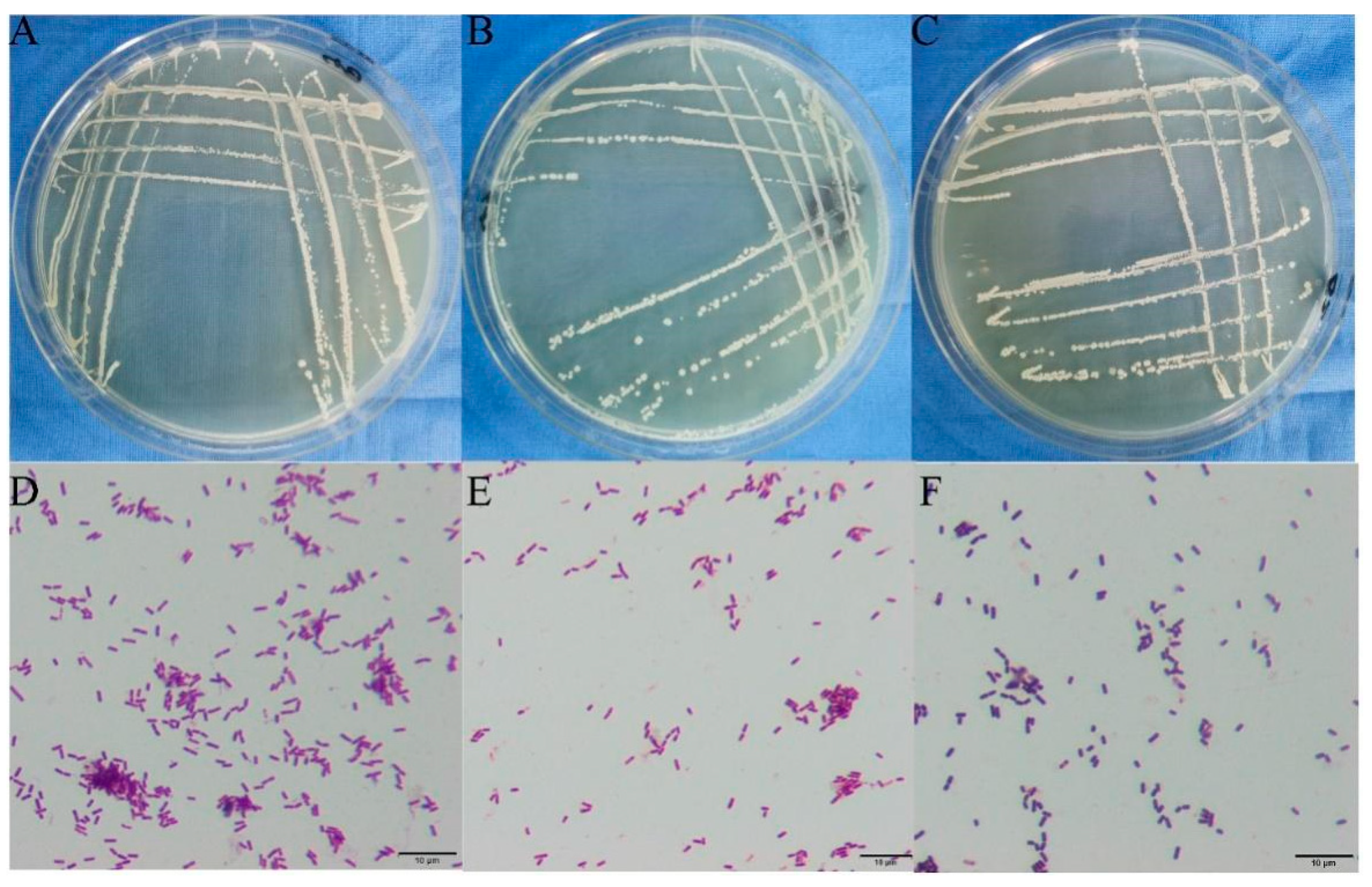
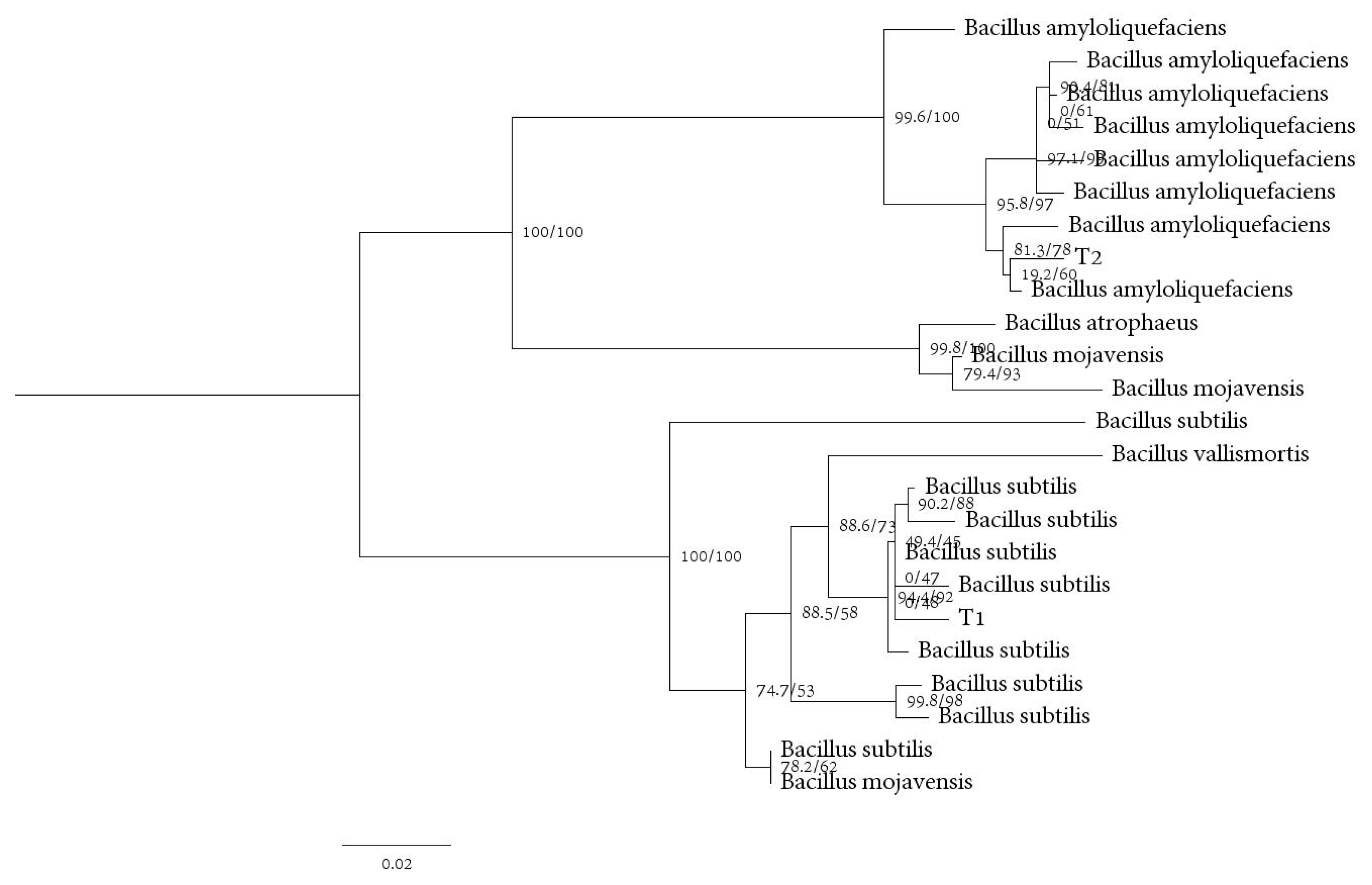
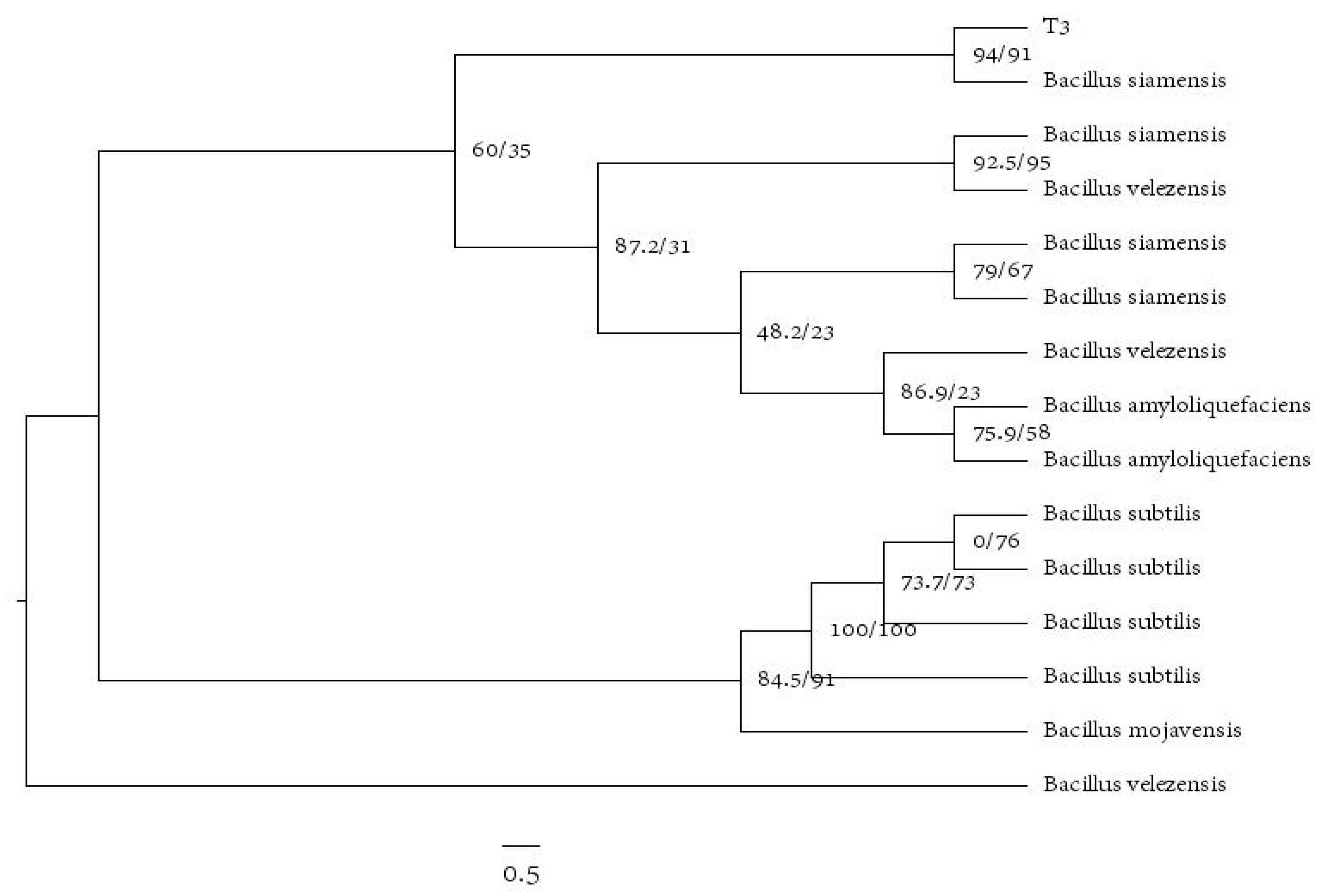
3.8. Identification of Antagonistic Bacteria
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xu, D.; Zhuo, Z.; Wang, R.; Ye, M.; Pu, B. Modeling the Distribution of Zanthoxylum armatum in China with MaxEnt Modeling. Glob. Ecol. Conserv. 2019, 19, e00691. [Google Scholar]
- Dušan, K.; Branka, L. Modern Trends in the Development of Agriculture and Demands on Plant Breeding and Soil Management. Genetika 2012, 44, 201–216. [Google Scholar]
- Cao, Z.M.; Tian, C.M.; Liang, Y. Investigation on Zanthoxylum Prickly Ash Diseases in Shaanxi and Gansu Provinces. J. Northwest For. Univ. 1994, 9, 39–43. (In Chinese) [Google Scholar]
- Niu, B.; Wang, W.; Yuan, Z.; Sederoff, R.R.; Sederoff, H.; Chiang, V.L.; Borriss, R. Microbial Interactions Within Multiple-Strain Biological Control Agents Impact Soil-Borne Plant Disease. Front. Microbiol. 2020, 11, 585404. [Google Scholar]
- Fira, D.; Dimkić, I.; Berić, T.; Lozo, J.; Stanković, S. Biological Control of Plant Pathogens by Bacillus Species. J. Biotechnol. 2018, 285, 44–55. [Google Scholar]
- Urbina, H.; Breed, M.F.; Zhao, W.; Gurrala, K.L.; Rosling, A. Specificity in Arabidopsis thaliana Recruitment of Root Fungal Communities from Soil and Rhizosphere. Fungal Biol. 2018, 122, 231. [Google Scholar]
- Sanguin, H.; Sarniguet, A.; Gazengel, K.; Moenne-Loccoz, Y.; Grundmann, G.L. Rhizosphere Bacterial Communities Associated with Disease Suppressiveness Stages of Take-All Decline in Wheat Monoculture. New Phytol. 2019, 184, 694–707. [Google Scholar]
- Rashid, M.I.; Mujawar, L.H.; Shahzad, T.; Almeelbi, T.; Ismail, I.M.I.; Oves, M. Bacteria and Fungi Can Contribute to Nutrients Bioavailability and Aggregate Formation in Degraded Soils. Microbiol. Res. 2016, 183, 26–41. [Google Scholar]
- Arici, S.E.; Demirtas, A.E. The Effectiveness of Rhizosphere Microorganisms to Control Verticillium Wilt Disease Caused by Verticillium dahliae Kleb. in Olives. Arab. J. Geosci. 2019, 12, 781. [Google Scholar] [CrossRef]
- Kennedy, A.C.; Smith, K.L. Soil Microbial Diversity and the Sustainability of Agricultural Soils. Plant Soil 1995, 170, 75–86. [Google Scholar]
- Luo, L.; Guo, C.; Wang, L.; Zhang, J.; Deng, L.; Luo, K.; Huang, H.; Huang, H.; Liu, Y.; Liu, Y.; et al. Negative Plant-Soil Feedback Driven by Re-Assemblage of the Rhizosphere Microbiome with the Growth of Panax notoginseng. Front. Microbiol. 2019, 10, 1597. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, H.; Sun, L.; Qi, G.; Chen, S.; Zhao, X. Microbial Community Composition Is Related to Soil Biological and Chemical Properties and Bacterial Wilt Outbreak. Sci. Rep. 2017, 7, 343. [Google Scholar] [CrossRef]
- Zhou, X.; Wu, F. Dynamics of the Diversity of Fungal and Fusarium Communities during Continuous Cropping of Cucumber in the Greenhouse. FEMS Microbiol. Ecol. 2012, 80, 469–478. [Google Scholar] [CrossRef]
- Cha, J.Y.; Han, S.; Hong, H.J.; Cho, H.; Kim, D.; Kwon, Y.; Kwon, S.K.; Crüsemann, M.; Bok Lee, Y.; Kim, J.F.; et al. Microbial and Biochemical Basis of a Fusarium Wilt-Suppressive Soil. ISME J. 2016, 10, 119–129. [Google Scholar] [CrossRef]
- Li, H.; Ma, X.; Tang, Y.; Yan, C.; Hu, X.; Huang, X.; Lin, M.; Liu, Z. Integrated Analysis Reveals an Association between the Rhizosphere Microbiome and Root Rot of Arecanut palm. Pedosphere 2021, 31, 725–735. [Google Scholar] [CrossRef]
- Pieterse, C.M.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.; Bakker, P.A. Induced Systemic Resistance by Beneficial Microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The Importance of the Microbiome of the Plant Holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef]
- Islam, M.A.; Nain, Z.; Alam, M.K.; Banu, N.A.; Islam, M.R. In Vitro Study of Biocontrol Potential of Rhizospheric Pseudomonas aeruginosa against Fusarium oxysporum f. sp. Cucumerinum. Egypt. J. Biol. Pest Control 2018, 28, 90. [Google Scholar] [CrossRef]
- Álvarez, B.; Biosca, E.G. Bacteriophage-Based Bacterial Wilt Biocontrol for an Environmentally Sustainable Agriculture. Front. Plant Sci. 2017, 8, 1218. [Google Scholar] [CrossRef]
- Cazorla, F.M.; Mercado-Blanco, J. Biological Control of Tree and Woody Plant Diseases: An Impossible Task? BioControl 2016, 61, 233–242. [Google Scholar]
- Yuan, J.; Chaparro, J.M.; Manter, D.K.; Zhang, R.; Vivanco, J.M.; Shen, Q. Roots from distinct plant developmental stages are capable of rapidly selecting their own microbiome without the influence of environmental and soil edaphic factors. Soil Biol. Biochem. 2015, 89, 206–209. [Google Scholar]
- Frey-Klett, P.; Burlinson, P.; Deveau, A.; Barret, M.; Tarkka, M.; Sarniguet, A. Bacterial-Fungal Interactions: Hyphens between Agricultural, Clinical, Environmental, and Food Microbiologists. Microbiol. Mol. Biol. Rev. 2011, 75, 583–609. [Google Scholar]
- Hassani, M.A.; Durán, P.; Hacquard, S. Microbial Interactions within the Plant Holobiont. Microbiome 2018, 6, 58. [Google Scholar]
- Kemen, E. Microbe–Microbe Interactions Determine Oomycete and Fungal Host Colonization. Curr. Opin. Plant Biol. 2014, 20, 75–81. [Google Scholar] [CrossRef]
- Hong, Y.; Chen, S.; Zhang, Y.; Chen, Y.; Yu, L.; Liu, Y.; Liu, Y.; Cheng, H.; Liu, Y. Rapid Identification of Soil Organic Matter Level via Visible and Near-Infrared Spectroscopy: Effects of Two-Dimensional Correlation Coefficient and Extreme Learning Machine. Sci. Total Environ. 2018, 644, 1232–1243. [Google Scholar]
- Bremner, J.M. Determination of Nitrogen in Soil by the Kjeldahl Method. J. Agric. Sci. 1960, 55, 11–33. [Google Scholar]
- Qiao, J.; Zhu, Y.; Jia, X.; Huang, L.; Shao, M. Vertical Distribution of Soil Total Nitrogen and Soil Total Phosphorus in the Critical Zone on the Loess Plateau, China. Catena 2018, 166, 310–316. [Google Scholar]
- Zebec, V.; Rastija, D.; Lončarić, Z.; Bensa, A.; Popović, B.; Ivezić, V. Comparison of Chemical Extraction Methods for Determination of Soil Potassium in Different Soil Types. Eurasian Soil Sci. 2017, 50, 1420–1427. [Google Scholar] [CrossRef]
- Fang, Z.D. Plant Disease Research Methods, 3rd ed.; China Agricultural Press: Beijing, China, 1998. [Google Scholar]
- Schuck, S.; Weinhold, A.; Luu, V.T.; Baldwin, I.T. Isolating Fungal Pathogens from a Dynamic Disease Outbreak in a Native Plant Population to Establish Plant-Pathogen Bioassays for the Ecological Model Plant Nicotiana Attenuata. PLoS ONE 2014, 9, e102915. [Google Scholar]
- O’Donnell, K. Progress towards a Phylogenetic Classification of Fusarium. Sydowia 1996, 48, 57–70. [Google Scholar]
- Wu, X.; Zhang, H.; Chen, J.; Shang, S.; Wei, Q.; Yan, J.; Tu, X. Comparison of the Fecal Microbiota of Dholes High-Throughput Illumina Sequencing of the V3–V4 Region of the 16S RRNA Gene. Appl. Microbiol. Biotechnol. 2016, 100, 3577–3586. [Google Scholar] [CrossRef]
- Xiong, W.; Zhao, Q.; Xue, C.; Xun, W.; Zhao, J.; Li, R.; Wu, H. Comparison of Fungal Community in Black Pepper-Vanilla and Vanilla Monoculture Systems Associated with Vanilla Fusarium Wilt Disease. Front. Microbiol. 2016, 7, 117. [Google Scholar] [CrossRef]
- Liu, S.; Li, P.; Van Zwieten, L.; Tu, J.; Gan, W.; Lu, S.; Wang, H.; Wu, L. Edaphic Variables Influence Soil Bacterial Structure under Successive Fertilization of Paulownia Plantation Substituting Native Vegetation. J. Soils Sediments 2021, 21, 2922–2937. [Google Scholar] [CrossRef]
- Lin, T.; Li, L.; Gu, X.; Owusu, A.M.; Li, S.; Han, S.; Cao, G.; Zhu, T.; Li, S. Seasonal Variations in the Composition and Diversity of Rhizosphere Soil Microbiome of Bamboo Plants as Infected by Soil-Borne Pathogen and Screening of Associated Antagonistic Strains. Ind. Crop. Prod. 2023, 197, 116641. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly Accurate OTU Sequences from Microbial Amplicon Reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Lu, Q.; Zheng, Z.; Li, S.; Li, S.; Liu, Y.; Zhu, T.; Chen, L.; Yang, C.; Han, S. Soil cadmium stress affects the phyllosphere microbiome and associated pathogen resistance differently in male and female poplars. J. Exp. Bot. 2023, 74, 2188–2202. [Google Scholar] [CrossRef] [PubMed]
- Sunar, K.; Chakraborty, U.; Chakraborty, B.N. Exploitation of Native Microflora of Darjeeling Hills for Sclerotial Rot Management and Growth Promotion in Pulses. Indian Phytopathol. 2014, 67, 59–69. [Google Scholar]
- Ritpitakphong, U.; Falquet, L.; Vimoltust, A.; Berger, A.; Métraux, J.P.; L’Haridon, F. The microbiome of the leaf surface of Arabidopsis protects against a fungal pathogen. New Phytol. 2016, 210, 1033–1043. [Google Scholar]
- Masum, M.M.I.; Liu, L.; Yang, M.; Hossain, M.M.; Siddiqa, M.M.; Supty, M.E.; Ogunyemi, S.O.; Hossain, A.; An, Q.; Li, B. Halotolerant Bacteria Belonging to Operational Group Bacillus amyloliquefaciens in Biocontrol of the Rice Brown Stripe Pathogen Acidovorax Oryzae. J. Appl. Microbiol. 2018, 125, 1852–1867. [Google Scholar] [CrossRef]
- Borba, M.P.; Ferrero, A.P.D.S.; de Souza Lameira, R.; Van Der Sand, S.T. The Intricate Molecular Identification of Streptomyces: A Case Study on Antarctic Soil Isolates. Arch. Microbiol. 2022, 204, 476. [Google Scholar] [CrossRef] [PubMed]
- Dixon, P. VEGAN, a Package of R Functions for Community Ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Ginestet, C. ggplot2: Elegant Graphics for Data Analysis. J. R. Stat. Soc. Ser. A Stat. Soc. 2011, 174, 245–246. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R. Predictive Functional Profiling of Microbial Communities Using 16S RRNA Marker Gene Sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Qu, Q.; Zhang, Z.; Peijnenburg, W.J.G.M.; Liu, W.; Lu, T.; Hu, B.; Chen, J.; Chen, J.; Lin, Z.; Qian, H. Rhizosphere Microbiome Assembly and Its Impact on Plant Growth. J. Agric. Food Chem. 2020, 68, 5024–5038. [Google Scholar] [CrossRef]
- Luan, F.G.; Zhang, L.L.; Lou, Y.Y.; Wang, L.; Liu, Y.N.; Zhang, H.Y. Analysis of Microbial Diversity and Niche in Rhizosphere Soil of Healthy and Diseased Cotton at the Flowering Stage in Southern Xinjiang. Genet. Mol. Res. 2015, 14, 1602–1611. [Google Scholar] [CrossRef]
- Van Elsas, J.D.; Chiurazzi, M.; Mallon, C.A.; Elhottovā, D.; Krištůfek, V.; Salles, J.F. Microbial Diversity Determines the Invasion of Soil by a Bacterial Pathogen. Proc. Natl. Acad. Sci. USA 2012, 109, 1159–1164. [Google Scholar] [CrossRef]
- Raza, W.; Ling, N.; Zhang, R.; Huang, Q.; Xu, Y.; Shen, Q. Success Evaluation of the Biological Control of Fusarium Wilts of Cucumber, Banana, and Tomato since 2000 and Future Research Strategies. Crit. Rev. Biotechnol. 2016, 26, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Egamberdieva, D. Bacillus Spp.: A Potential Plant Growth Stimulator and Biocontrol Agent under Hostile Environmental Conditions. In Bacilli and Agrobiotechnology; Springer: Cham, Switzerland, 2016; pp. 91–111. [Google Scholar]
- Lee, S.; Gi, K.H.; Cheol, S.G.; Min, R.C. Disruption of Firmicutes and Actinobacteria Abundance in Tomato Rhizosphere Causes the Incidence of Bacterial Wilt Disease. ISME J. 2020, 15, 330–347. [Google Scholar] [CrossRef]
- Rosenzweig, N.; Tiedje, J.M.; Quensen, J.F., III; Meng, Q.; Hao, J.J. Microbial Communities Associated with Potato Common Scab-Suppressive Soil Determined by Pyrosequencing Analyses. Plant Dis. 2012, 96, 718–725. [Google Scholar] [CrossRef] [PubMed]
- Fan, B. Bacillus velezensis FZB42 in 2018: The Gram-Positive Model Strain for Plant Growth Promotion and Biocontrol. Front. Microbiol. 2018, 9, 2491. [Google Scholar] [CrossRef] [PubMed]
- Paul, C.S.; Anton, H.; Gao, X.W.; Rainer, B. Biocontrol Mechanism by Root-Associated Bacillus amyloliquefaciens FZB42—A Review. Front. Microbiol. 2015, 6, 780. [Google Scholar]


| Sample | 16S | gyrA | gyrB |
|---|---|---|---|
| T1 | OP317514 | OQ659395 | OQ659398 |
| T2 | OP324656 | OQ659396 | OQ659399 |
| T3 | OP317360 | OQ659397 | OQ659400 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, S.; Zheng, Z.; Li, S.; Liu, Y.; Li, S.; Yang, C.; Qiao, T.; Lin, T.; Zhu, T. Impact of Root Rot Disease of Zanthoxylum armatum on Rhizosphere Soil Microbes and Screening of Antagonistic Bacteria. Forests 2023, 14, 1561. https://doi.org/10.3390/f14081561
Han S, Zheng Z, Li S, Liu Y, Li S, Yang C, Qiao T, Lin T, Zhu T. Impact of Root Rot Disease of Zanthoxylum armatum on Rhizosphere Soil Microbes and Screening of Antagonistic Bacteria. Forests. 2023; 14(8):1561. https://doi.org/10.3390/f14081561
Chicago/Turabian StyleHan, Shan, Zhenlei Zheng, Shujiang Li, Yinggao Liu, Shuying Li, Chunlin Yang, Tianmin Qiao, Tiantian Lin, and Tianhui Zhu. 2023. "Impact of Root Rot Disease of Zanthoxylum armatum on Rhizosphere Soil Microbes and Screening of Antagonistic Bacteria" Forests 14, no. 8: 1561. https://doi.org/10.3390/f14081561
APA StyleHan, S., Zheng, Z., Li, S., Liu, Y., Li, S., Yang, C., Qiao, T., Lin, T., & Zhu, T. (2023). Impact of Root Rot Disease of Zanthoxylum armatum on Rhizosphere Soil Microbes and Screening of Antagonistic Bacteria. Forests, 14(8), 1561. https://doi.org/10.3390/f14081561





