1. Introduction
Urban trees provide several ecosystem services, one of them being a carbon storage [
1,
2,
3]. During the last decades, a lot of research has been undertaken to bring light to the role of urban forests for carbon sequestration (e.g., [
4,
5,
6,
7,
8] and many others). In comparison to traditional forests, the tree density in urban forests is much lower, but nevertheless urban trees represent a relevant reservoir of carbon and eventually a sink of carbon dioxide in the urban context (e.g., [
9,
10]). While urban tree management aims at maintaining or even enhancing the more obvious urban tree functions (e.g., shading, aesthetics, recreation, see also Konijnendijk et al. [
1]), it might additionally focus on increasing the amount of stored carbon when it comes to climate protection. This requires methods for determining the current carbon storage to be able to evaluate the development of this reservoir.
Knowledge about the urban carbon reservoir and possibly the associated fluxes helps in understanding the role of urban trees in carbon dioxide emission mitigation for single trees (e.g., [
11]), on the local [
8,
10,
12,
13,
14,
15,
16,
17,
18], or nation-wide scale [
2,
4,
6,
19,
20], to deduce the total net balance for a specific area [
9,
21,
22], to conduct demand–supply analysis [
21,
23,
24] for reporting in the context of the United Nations Framework Convention for Climate Change (UNFCCC) [
6], to assess their potential in carbon credit markets (e.g., [
25]), to draw comparisons to other urban (e.g., [
17]) or traditional forests (e.g., [
19]), build storage maps [
13,
26] or to develop management options [
26,
27] with regard to climate change and carbon dioxide mitigation.
For the estimation of stored carbon in urban space, several methods were already developed and applied. On the one hand, there are approaches using remote sensing [
6,
8,
10,
12,
13,
15,
17], either based on airborne photogrammetry and/or airborne lidar. Ground-based lidar, i.e., terrestrial laser scanning (TLS), was applied to estimate single trees biomass (e.g., [
5,
7,
23,
28,
29]). On the other hand, the well-established method of allometric biomass functions is used to estimate tree biomass, which might be upscaled to the desired spatial level. In some cases, predictors were deduced from remote sensing analysis [
10,
13,
15,
17]. Other studies developed specific biomass functions (e.g., [
30,
31] for small trees and Korea, respectively), used equations from nearby locations (e.g., [
32]), or used forest tree biomass function for comparison (e.g., [
16]). One obvious use case for biomass functions is to estimate biomass based on data and attributes stored in a tree inventory, if respective predictors and functions are available. For such a case, we want to develop biomass functions and apply these models to already available data. With that, we can enhance existing management tools by easy means. Remote sensing information is not easily available and accessible in a permanent manner for such management systems, yet.
Since the amount of stored biomass and, hence, carbon (being approximately half of the biomass) cannot be easily determined using existing forest tree biomass functions (e.g., [
29,
33] and also shown by this study), there is a need to develop specific functions for urban trees. Likewise, McPherson et al. [
34] (p. 8ff) even point out that several factors might influence tree growth within and between cities. In this context, the authors highlight management practice, which also depend on regulatory rules. They also state that urban trees exhibit higher variability in habitus than rural trees. Russo et al. [
14] and Strohbach and Haase [
13] also advise to check applicability and comparability, respectively, when existing methods are applied to new areas. Hence, if new biomass functions for urban trees are to be developed, this guidance should be kept in mind and a trade-off made between generality and specificity of the situation at hand.
Biomass functions have already been used for a long time, and an extensive body of literature exists (see, e.g., [
35,
36,
37]). Usually, biomass functions relate easy-to-measure variables, such as diameter in breast height (dbh) and tree height to total or component mass. Due to this relation, such biomass functions are also termed allometric biomass functions (see also [
38]). In urban areas, one of the main difficulties in developing urban tree biomass functions is the scarcity of actual measured biomass data since cutting urban trees is usually not desired. In this study, we aim to develop a set of urban tree biomass functions to estimate the total above-ground biomass (agb) to be able to calculate stored carbon in urban tree stands using single tree attributes from well-established management tools, like communal tree inventories. Additionally, we show a modeling approach capable of improving these functions by including not only urban tree data but also forest tree data. In the following, we describe the two data sets for model building and present the applied regression models. Several alternative models are tested, and the leave-one-out cross-validation results favor the cross-classified mixed model. We discuss the different approaches and apply the proposed cross-classified mixed model on example data from a tree inventory in Munich.
3. Results
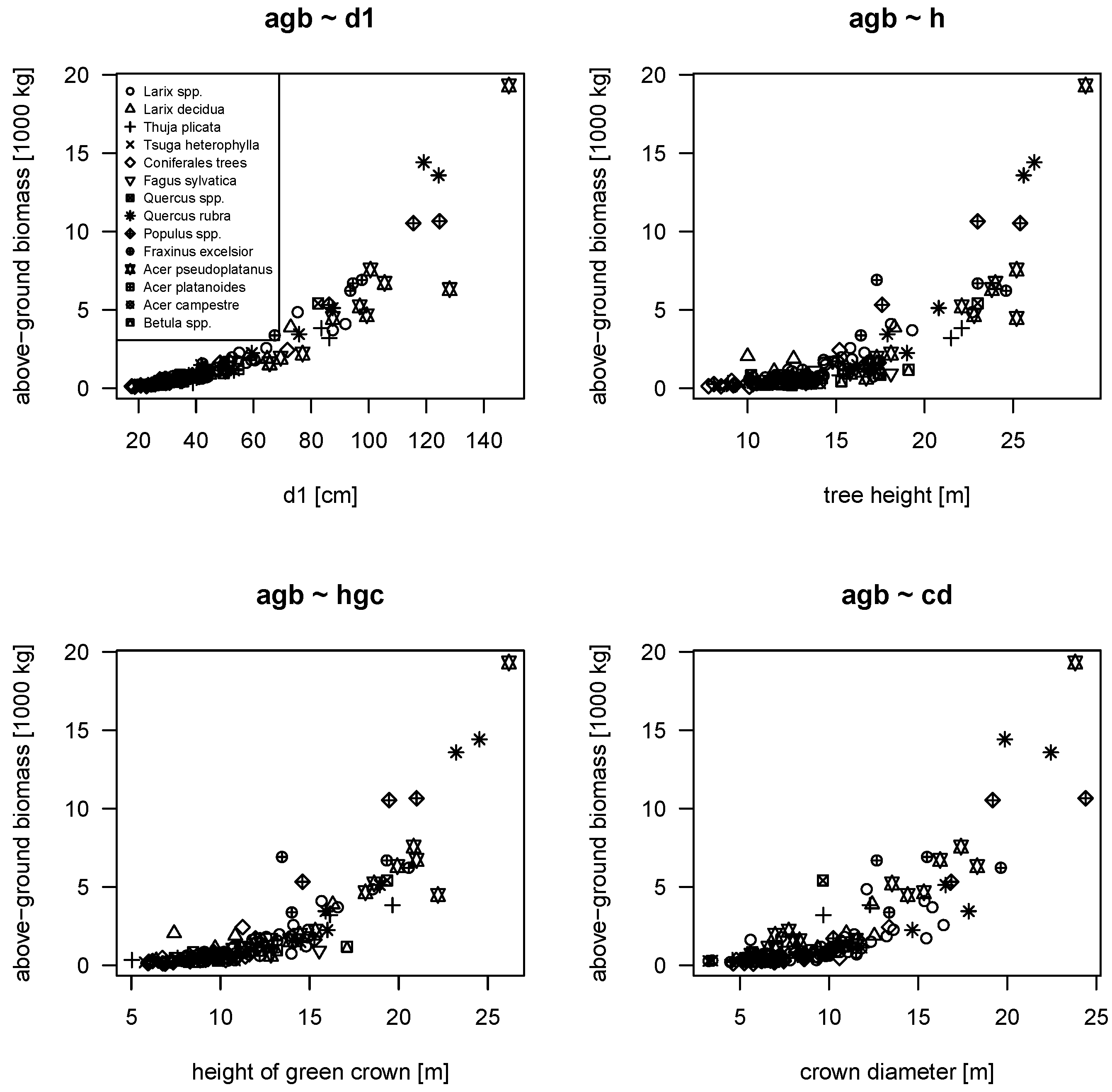
In biomass studies,
is probably the most used predictor. But tree height is also frequently used, as it positively correlates with biomass storage (e.g., [
37]). Trees of same diameter keep more biomass if larger in tree height [
39,
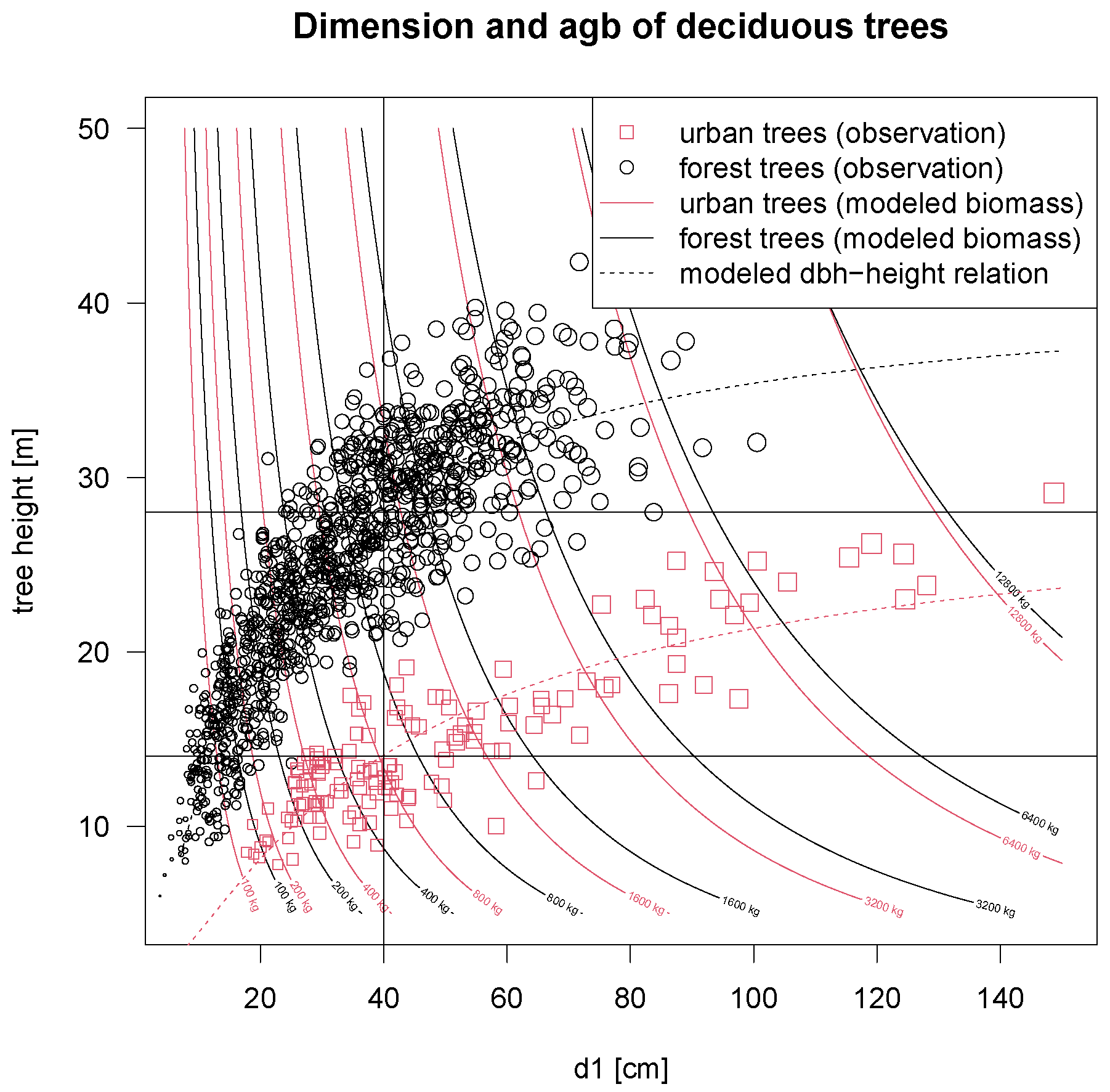
48]. When comparing urban to forest trees, urban trees show different patterns of biomass accumulation with size (see
Figure 2 for the relations between
d1 and
). This finding is even more pronounced if tree height is also taken into account. At the same time, tree height is very different for deciduous urban trees by a given diameter in comparison to forest trees. Based on our data, we can state that the measured deciduous urban trees are 10 to 15 m smaller than their forest counterpart (
Figure 3). Interestingly, in our data, there is actually no overlap in tree height between both origins for all diameter classes. Simultaneously, applying a simple biomass model considering predictors
d1 and
, we find that deciduous urban trees contain more biomass for the same dimensions than forest trees. This is also shown in
Figure 3 by the modeled contour lines (see also the Figure caption for an explanation and example). In consequence, we can state that deciduous urban trees usually hold less biomass than forest trees if only the diameter is considered, but if the comparison includes both diameter and height, deciduous urban trees accumulate more biomass. This seemingly contradictory result is resolved by the fact that urban trees (in our data) are usually smaller than forest trees of the same diameter class and show a different morphology. The observed difference in biomass diminishes as trees get larger and almost vanishes for trees with diameters above 100 cm (
d1). This at least is shown by our data, although only a small share of such large trees is present. Hence, for modeling and application, it is important to include a height measure into the biomass models.
We tested different approaches for fitting urban tree biomass functions. The first set of models are based on the 144 urban trees, and the second set encompasses 2205 trees, including forest trees. We evaluated these models using AIC (first set of models [
57]), model residual error, leave-one-out cross-validated root mean squared error (RMSE) and mean error (BIAS).
The first set of models, fitted using the urban tree data only, show that all four selected variables (
d1,
,
and
) contribute to explain the above-ground biomass of urban trees. This is coherent since all variables describe trees in their volumetric extent (see also
Figure 1). The fitted mixed models had the random effect terms for factor
species usually placed on the parameter
belonging to
d1. Heteroscedasticity (c.f.
Section 2.2) was best treated also by this covariate, except for model mm4, which makes use of the model predictions to weight the errors (see
Table 3). The best model by AIC (mm4, AIC = 1905.4) makes use of
d1,
and
for the fixed effects with random effects set on the exponent of
d1. Considering the other fit statistics, i.e., residual standard error (
) and cross-validated RMSE and BIAS (see
Table 3 and
Table 4), other models are not much worse, or even better.
In terms of predictive performance, mm5 shows the lowest group-level cross-validated RMSE and BIAS (see
Table 4, index “g” and “cv”). When it comes to applicability, the model mm1 seems to be a good choice as well because it only requires easy-to-measure quantities (
d1 and
, no laborious crown measurements) and exhibits only a slightly higher bias than mm5. The model results also indicate that the height of the green crown (
) might offer more information than does the tree height (
)—at least for model fitting. Comparing cross-validated results, it seems that using
instead of
delivers more accurate predictions on average (models mm1 and mm5). The model gm6, a species-independent generalized nonlinear least squares model, was fitted as a reference without an hierarchical approach and, indeed, shows the smallest residual standard error. But all other fit statistics do not show any advantage of that model.
As mentioned above, our full data clearly indicate that there is a difference in the above-ground biomass between urban and forest trees of the same diameter class. Modeling the deviation between urban and forest biomass to be able to use well-approved forest biomass functions using an additive or multiplicative correction factor does not lead to satisfactory results: this type of model, based on adjusting forest tree biomass functions, turns out to have the highest RMSE
cv values of all tested models and moderately high BIAS
cv values (see
Table 5). The additive model shows lower RMSE
cv but higher BIAS
cv than the multiplicative model.
The CCMM model of Equation (
4), an extension of the well-suited mm1 model (with predictors
d1 and
) with both urban and forest trees, shows best values for both RMSE
cv and BIAS
cv. Lastly, the simplified FM model, implementing Equation (
5) using a binary variable encoding forest and urban origin, exhibits a BIAS
cv high as −90.6 kg (almost nine times higher than the best model in absolute values) and moderately high RMSE
cv (see
Table 5).
Summing up, the best-performing model for urban trees is the CCMM. To assure that the CCMM also reproduces observed biomass of forest trees equally well, we compared the bias-adjusted predictions with the predictions of the German NFI functions [
48] for Norway spruce, Scots pine, Douglas fir, European beech, oak and sycamore. Results indicate slightly higher RMSE values for all species except Douglas fir and sycamore (−26.9% to +17.8%) and smaller absolute BIAS values for all species, except for Norway spruce (NFI: +8.0 kg, CCMM: −12.8 kg). Highest (in absolute terms) relative BIAS of the CCMM model is −3.2%.
When evaluating the different models for particular species, results might show different patterns (see
Table A1 and
Table A2 in
Appendix A for absolute and relative cross-validated RMSE and BIAS). In this case, no model exhibits best results for all of the 14 considered deciduous tree species. The “best” models show smallest BIAS
cv values for only four species and smallest RMSE
cv values for only five species, respectively. Still, the CCMM shows very good results: it is the only model with all relative BIAS values below 10% and relative RMSE ranges between 10 and 36% (cf.
Table A2). Similarly, mm1 and mm5 models also perform well, but the largest BIAS is higher (for sycamore and birch, respectively). Only for black locust is the result of mm1 better than that of CCMM considering RMSE and BIAS. But both models (mm1 and mm5) exceed 10% BIAS for 3 and 4 tree species, respectively. Moreover, both models can only predict deciduous trees. This is an advantage of the CCMM model because coniferous trees in urban areas can be represented by the integration of coniferous tree species data and the methodology of cross-classified mixed models. An independent evaluation of the performance for these additional tree species in the city is not possible with our data. But it has been shown that the model performs well in all testable aspects (deciduous urban trees as well as coniferous and deciduous forest trees).
In summary, the finally proposed CCMM model (Equation (
4)) uses only
d1 and
as predictors due to data limitations, but includes independent random effects given
species for the intercept and the two parameters. Random effects based on the
origin (traditional forest vs. urban area) were significant only for the intercept (
), making it a scaling parameter (this finding lead to the development of the factor model of Equation (
5)). The estimated parameters for the CCMM model are given in
Table 6, and the random effects for different species are given in
Table A3 in
Appendix B. The factor for correcting bias is estimated to be 1.012081 (see [
51,
55]). As long as predictions correspond to the group level, i.e., refer to a particular species and location as in our example, the given approach for bias correction by
is valid and corrects for the bias of transformation of the common within-group error. For prediction on population level (i.e., setting the random effects to zero), the uncertainty of the between-group error must be incorporated as well (e.g., see Appendix 1 of [
58]).
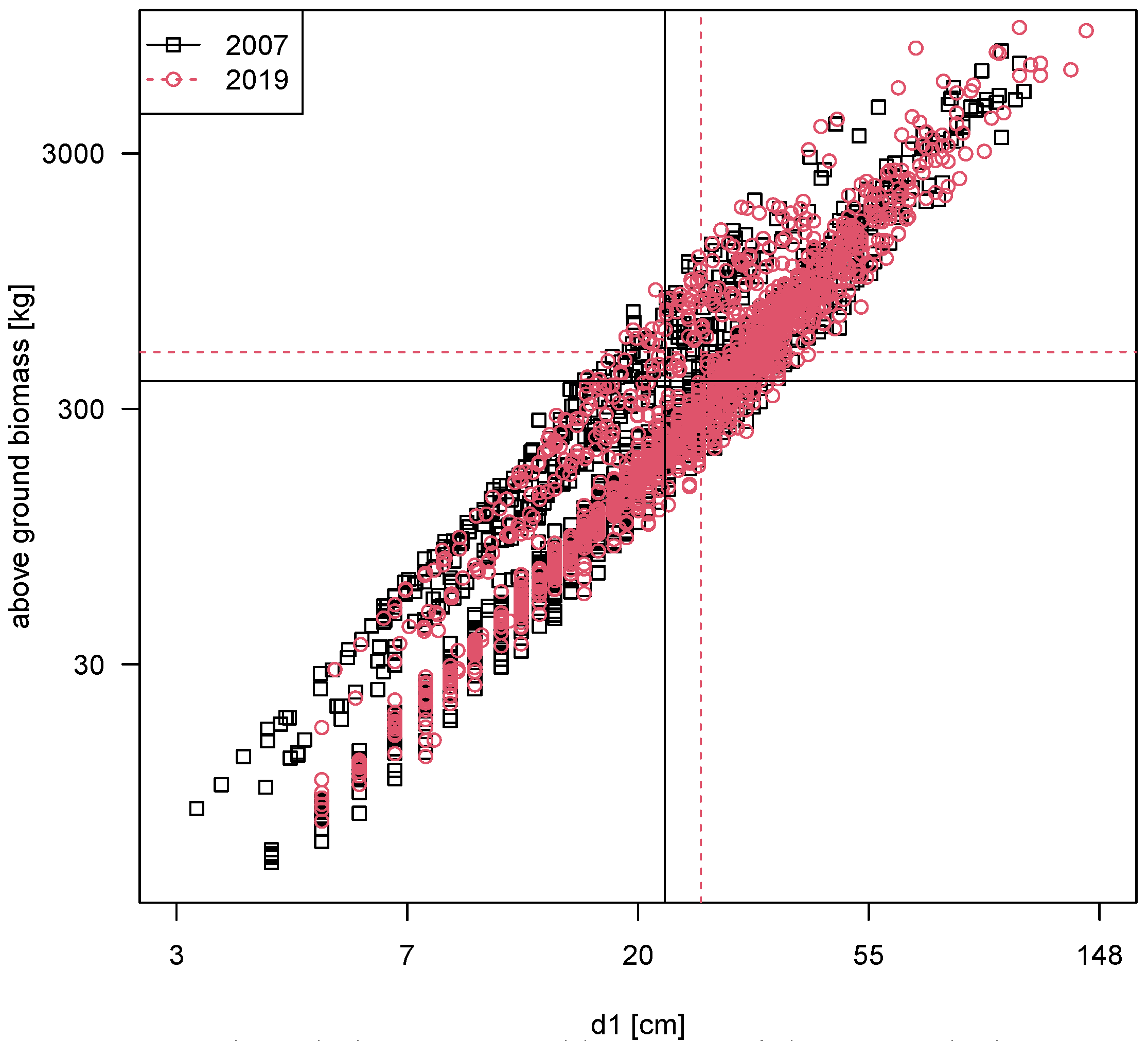
Sample Application
To show the applicability of the developed model, we applied the CCMM with iSiMan5 tree management software [
59] to a small subset of data collected during a regular inventory by TreeConsult Brudi & Partner in a residential area in the city of Munich, Germany. The model estimates the stored above-ground biomass
of the urban trees, which can also be easily translated into stored amount of carbon by applying the respective carbon content factor (see, e.g., [
60,
61]). If data for multiple points in time are available, in this case for 2007 and 2019 (see
Table 7), it even is possible to calculate the respective net carbon fluxes.
The results for our example data show that in 2019, the urban trees stored about 941 tons of biomass, i.e., about 453 tons of carbon (c.f.
Figure 4). This amount of storage was achieved by 2260 trees, meaning an average amount of stored biomass of 416 kg per tree. Compared to 2007, this is an increase of about 86 tons of biomass or a plus of 10.2%. The average sink capacity for this set of trees is thus approximately 3.5 tons of carbon per year, even though 503 trees were lost at the same time (−18.2%). By an increased average diameter and constant tree height, the loss in numbers could be overcompensated in terms of carbon storage. This highlights again the importance of managing, tending and conserving especially middle-aged and old urban trees, which not only act as a carbon storage but also serve further ecosystem services (social, economic, ecological, climatic and aesthetical, see further, e.g., [
1,
3]). Since the developed equation of CCMM takes into account the different tree species and their sizes, further analyses in combination with the data from the tree inventory are possible.
4. Discussion
There are several other, supposedly more modern, approaches to determine the biomass and C-sink potential of urban trees (e.g., aerial and terrestrial laser scanning). But the use of the close allometric relationships between simple-to-measure tree attributes and the target variables biomass and carbon storage, especially in combination with regular and repeated inventory data, remains an accurate and low-cost method to determine urban tree carbon storage. In addition, the method enables more modern techniques (e.g., terrestrial laser scanning) to expand the data basis for model building.
Our data analysis shows that a simple transfer of forest biomass functions into urban space needs adjustment because the allometry of urban trees differs strongly from those of forest trees. Although the same tree size in terms of diameter (e.g., d1) can be found in both landscapes, tree heights (and also crown habitus) differ significantly. In our data set, there was actually no overlap in tree heights for any given diameter class, so the two origins should be considered separate entities. Still, both are made up by trees, obeying allometric rules. Differences in biomass between urban and forest trees are more prominent for smaller tree dimensions and diminish as trees mature. This is probably because urban trees grow under less light competition in less confined spaces and therefore grow less tall and form larger crowns right from the very beginning. The larger trees get, both origins resemble a more unconstrained habitus and thus show more comparable biomass and carbon storage. As a clear result, we can state that there is a need for urban tree biomass functions in addition to forest tree biomass functions.
The application of forest biomass models in an urban setting is an extrapolation of those models, which requires a significant correction. Our approaches using the NFI biomass functions, including correction, performed worse in comparison to most other tested models. The models, developed for urban tree biomass, make use of the predictors
d1 and
, which are regularly measured during urban tree inventories. In the case of using a diameter measured in a different height, e.g., in 1.3 m (
) as regularly used in forestry, a simple linear regression can convert between both variables due to high correlation (see, e.g., [
62]). It turns out by model cross validation that additional variables like height of green crown (hgc) or crown diameter (cd)—although significant during model fitting—do not or only slightly improve the models. Actually, our CCMM model shows best overall statistics only requiring
d1 and
.
We propose to use the CCMM model, which fits several species and origins (urban and forest areas) at once. This model shows the best performance compared to models only using data from urban areas. Hence, this model “learns” from forest tree data and improves the modeled relationship. Additionally, it is possible to estimate yet unmeasured crossings (e.g., conifer species in urban areas). Due to the lack of data, this case cannot be validated, but we could show that the model behaves well in all statistically verifiable aspects. It predicts forest biomass equally well as the NFI functions and outperforms the urban tree-only models by better capturing the general allometric relation. But of course, the difference between urban trees and forest trees is contained only in the random effect of the scaling parameter. Thus, a similar difference between these two groups of tree species of the two origins is assumed since both deciduous and coniferous trees have more space and less competition in urban areas than in the traditional forest environment. Hence, crown formation and height growth are likely to be modified in a similar direction for both groups.
The CCMM uses the mixed-effects modeling framework. From a theoretical perspective, the validity of a model is assured, among other things, by checking the assumption of normality for the estimated random effects. This is difficult if the factor variable has only two levels, and often 5–6 factor levels are recommended as a minimum. Nevertheless, the case of less factor levels is not uncommon (e.g., female vs. male), and some authors see no reason to prevent the use of mixed models in such cases (e.g., [
63], p. 247/275f). Our results indicate the convergence and suitable parameter estimates of the CCMM model as well as proper fit statistics so that we can assume the correctness and applicability of the model.
The set of (urban) tree species included reflects the situation in Karlsruhe, Germany in 2011, and hence, it is not necessarily representative for other cities, not even in Germany. Some tree species are represented only by a small number of samples, so it is important to enhance our data by more tree species, especially if future development may be directed toward more suitable tree species in a changing urban climate. There might also be differences within tree species depending on different urban regions (c.f. [
34]), requiring more specific and localized models. Using the mixed-models approach, new tree species (or subspecies with special habitus like
Populus nigra italica) can be added into the model by estimating their random effect even with a few observations only so that local situations can be handled with little effort [
64]. Besides that, collecting data from different cities—here, TLS might be a suitable technique—might help with building a more general model, which includes random effect terms for different cities and makes it possible to expand the model geographically. The data of the forest trees instead originate from different studies (both with regard to the geographic origin and sampling method).
As an example, we applied the CCMM model to a subset of a tree inventory in Munich, Germany. The assignment of the encountered tree species to the model tree species, as well as the regional transfer of the model, eventually leads to a certain inaccuracy of the result. Yet, this is unavoidable, as the corresponding basic data are missing so far.
With the application of the model, and despite its certain inaccuracies in application, it was possible to represent carbon storage at two different points in time and, thus, estimate carbon fluxes over the period under consideration. The closer look at the performance of the stand or even of the individual species or genera, also on different site types, can thereby provide important advice for future tree management in urban areas, taking climate protection and climate adaptation into account.