Identification of Late Flavonoid Biosynthesis Genes of Moso Bamboo Reveals the Potential Function of PeANR4 Involved in Osmotic and Salt Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification, Characteristics, and Phylogenetic Analysis of LFBGs in Moso Bamboo
2.2. Expression Analysis of LFBGs in Moso Bamboo
2.3. Transcription Factor Binding Site Analysis in the Promoter of Above Identified Genes
2.4. Correlation Analysis and Co-Expression Network Construction
2.5. Yeast One-Hybrid Assays
2.6. Arabidopsis thaliana Transformation
2.7. Fresh and Dry Weight Measurement of Transgenic Plants
2.8. Measurement of PAs and DMACA Staining
2.9. Treatments of Osmotic and Salt Stress
2.10. Statistical Analysis
3. Results
3.1. Characterization of LFBGs in Moso Bamboo
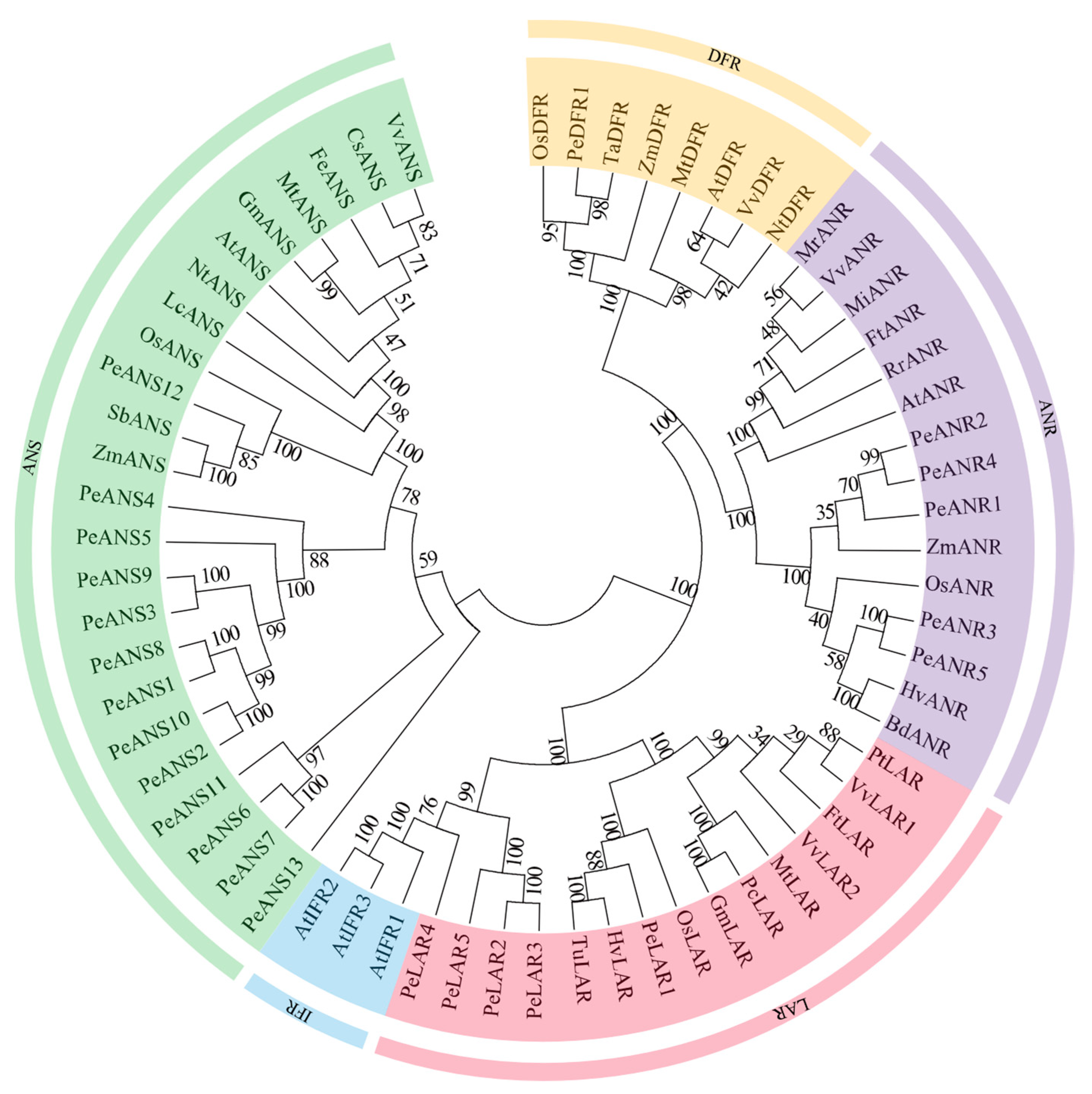
3.2. Phylogenetic Analysis of ANS, DFR, ANR, and LAR
3.3. Expression Patterns of LFBGs in Different Tissues of Moso Bamboo
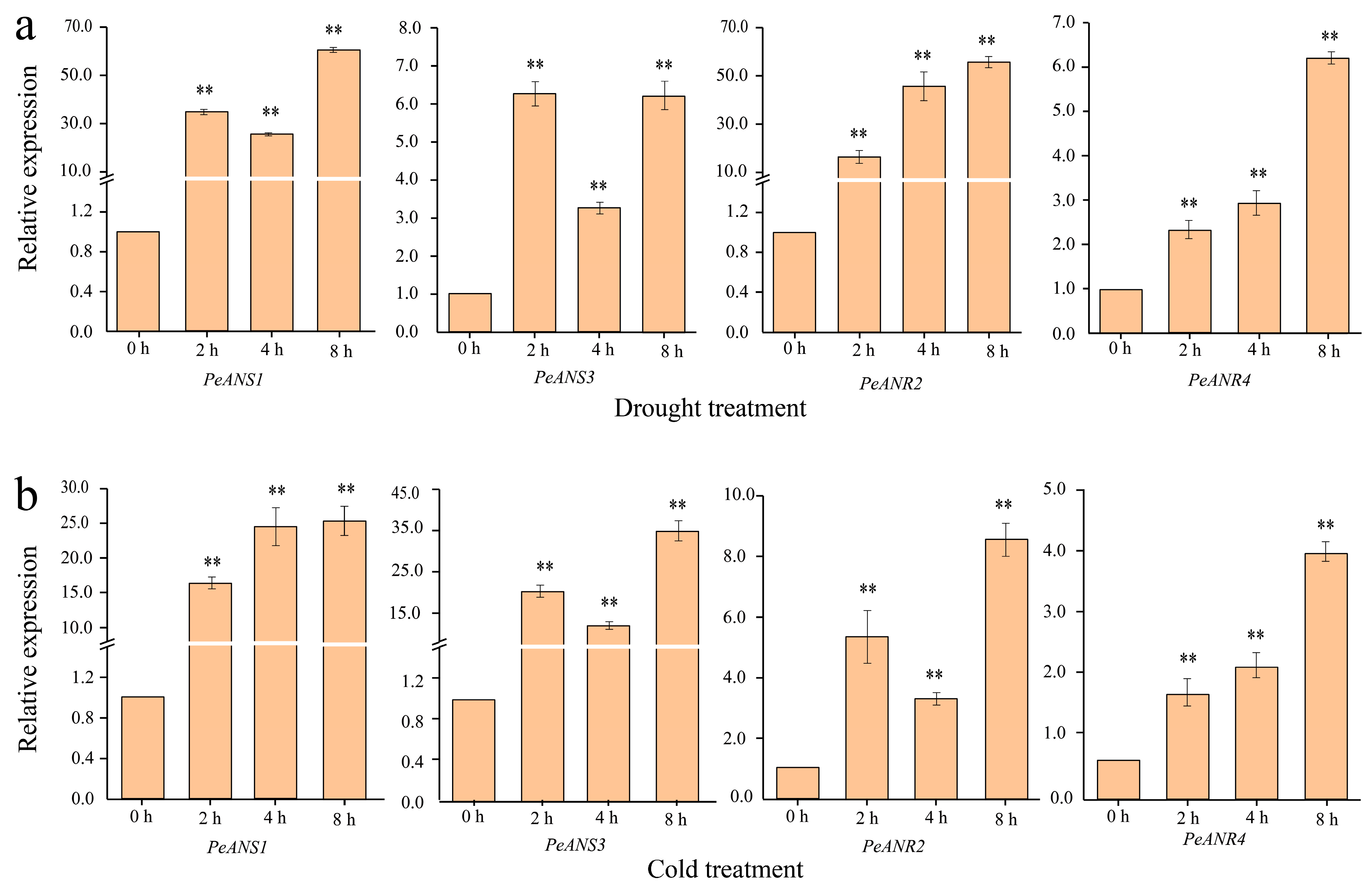
3.4. Expression Patterns of LFBGs in Bamboo Leaves under Drought and Cold Stress
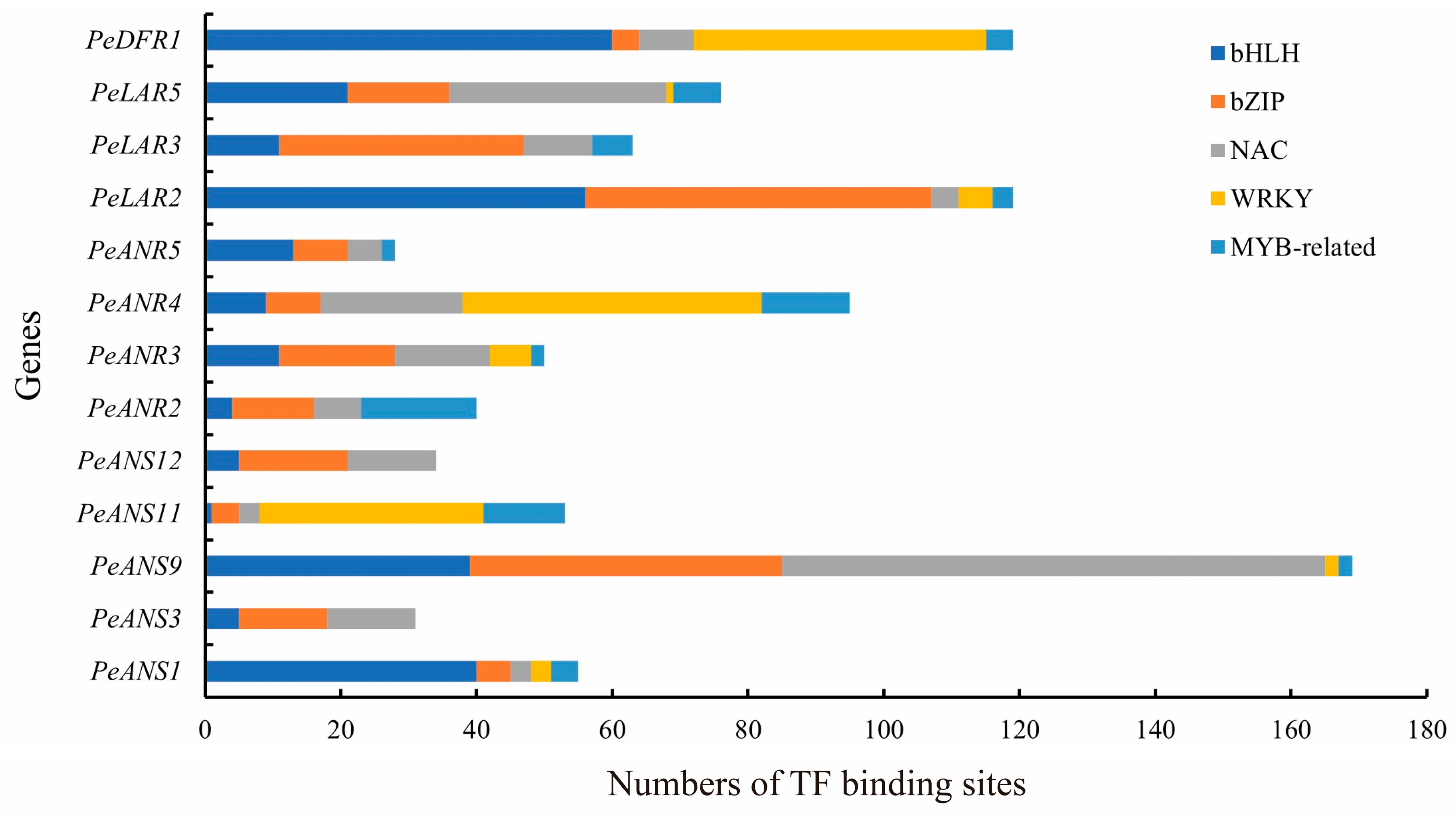
3.5. TF Binding Sites in LFBG Promoters of Moso Bamboo
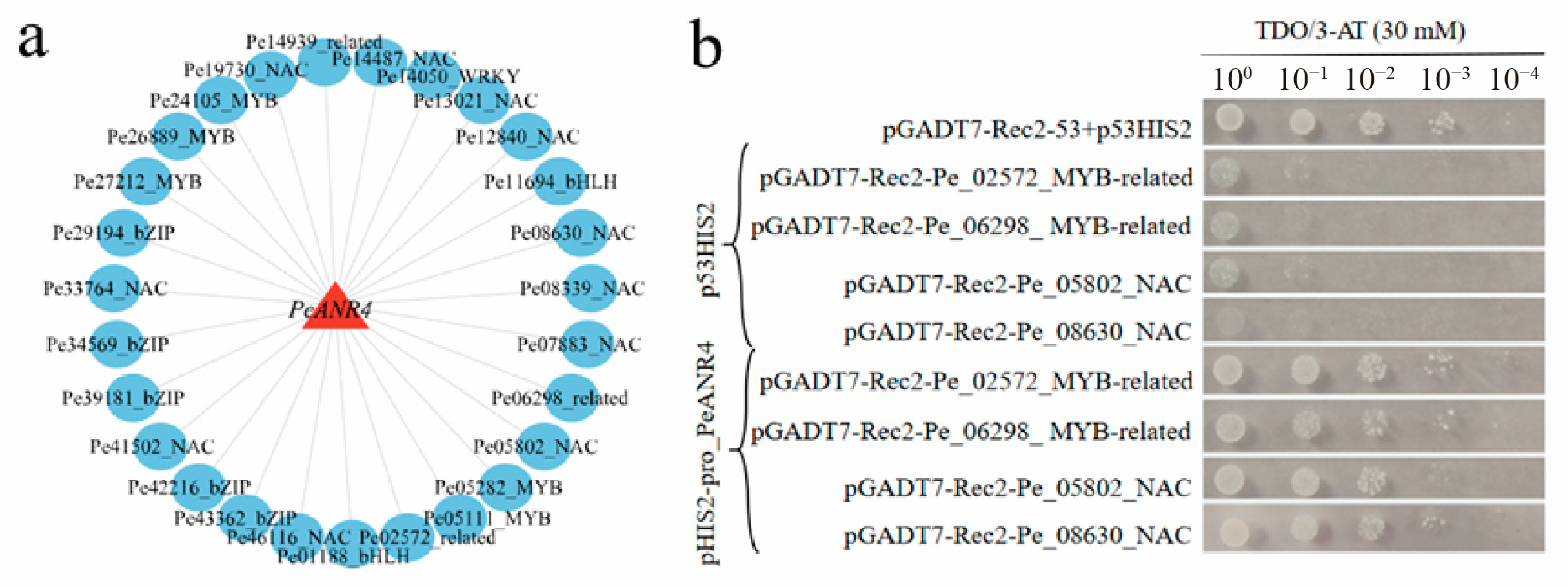
3.6. Co-Expression Network Construction and Validation by Y1H
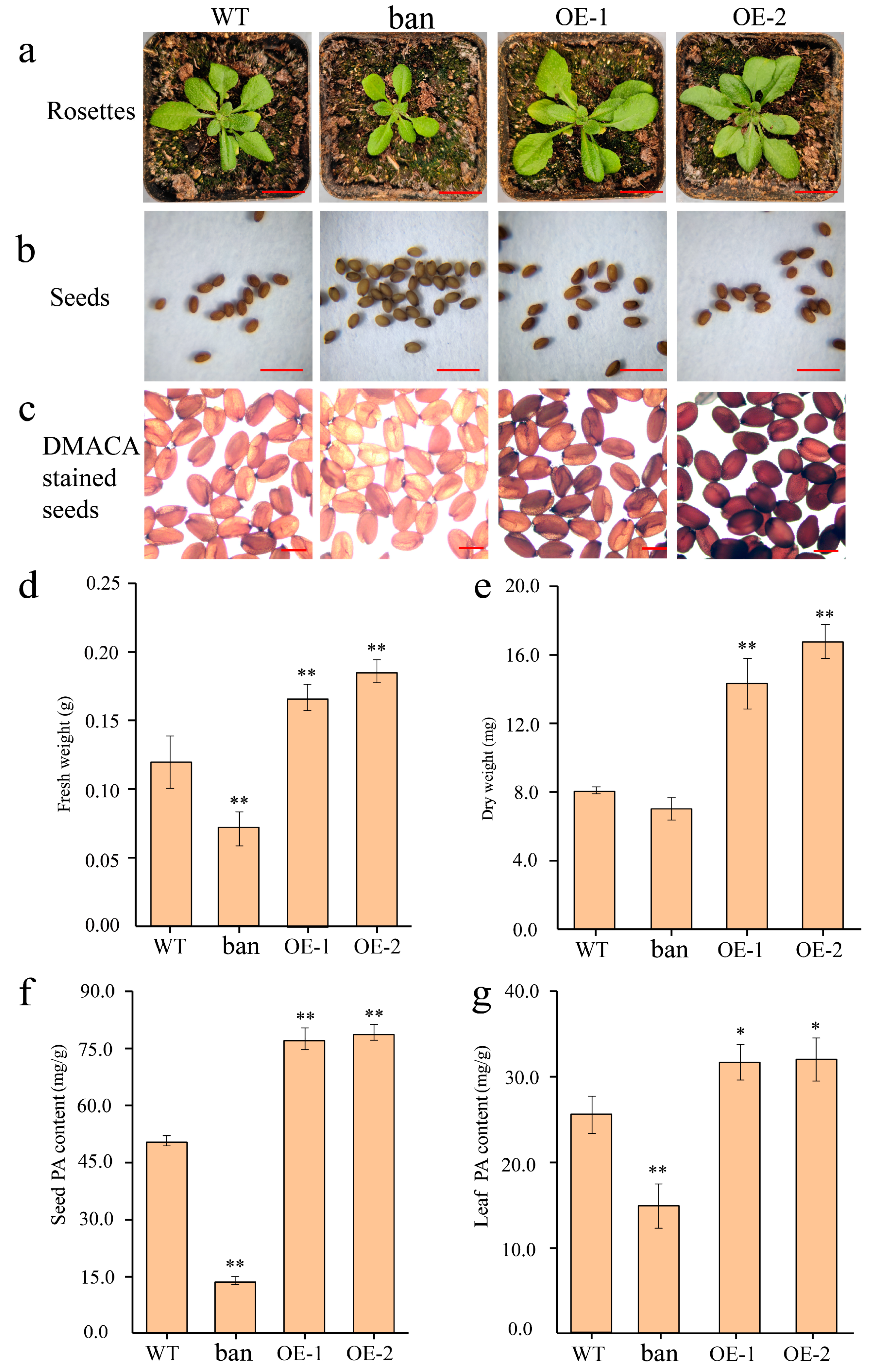
3.7. Ectopic Expression of PeANR4 in Arabidopsis
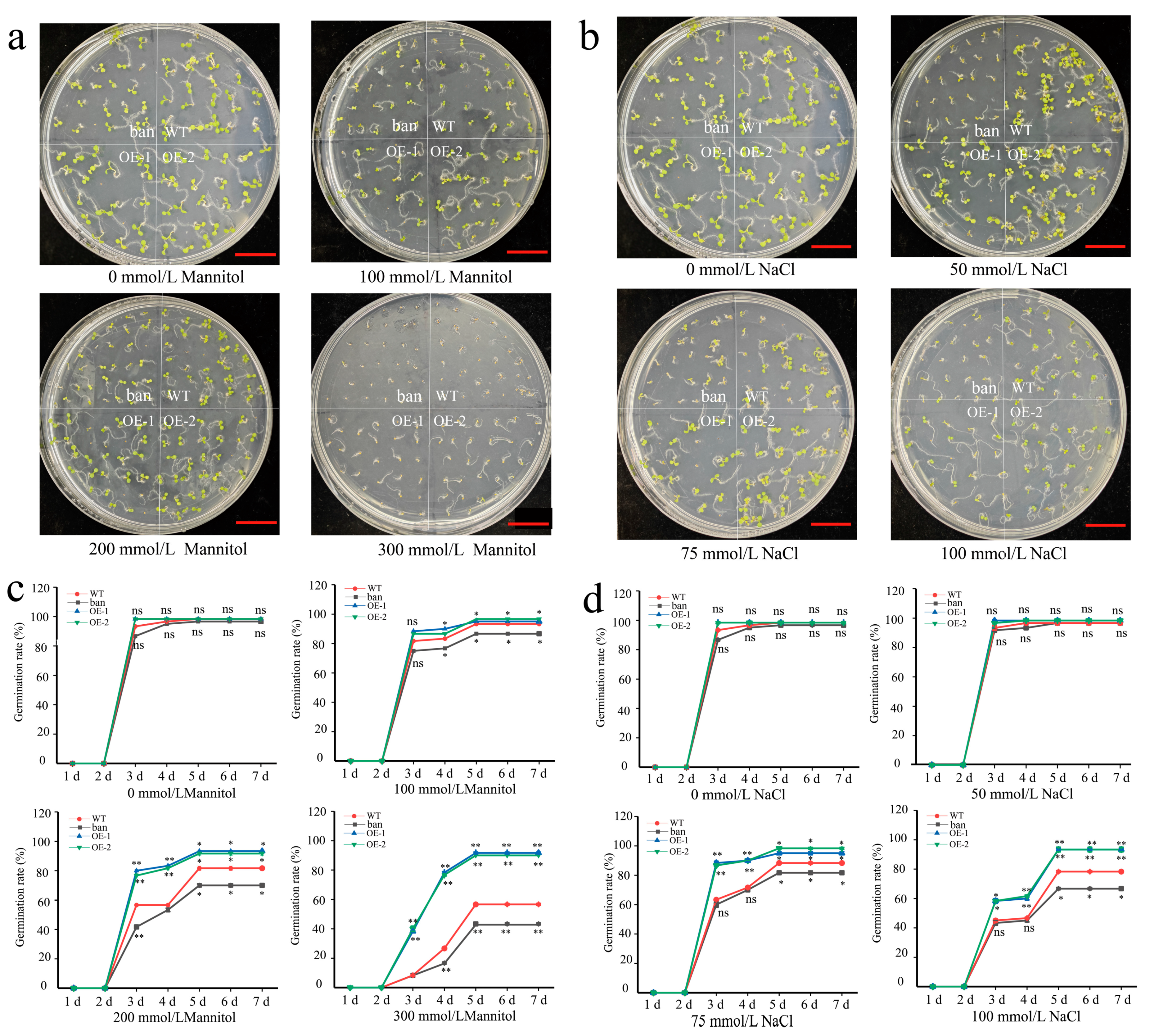
3.8. Effects of Osmotic and Salt Stress on PeANR4 Transgenic Arabidopsis
4. Discussion
4.1. Evolution of Flavonoid Biosynthesis
4.2. Regulation of Flavonoid Biosynthesis
4.3. PAs Synthesis Affecting Plant Growth and Stress Tolerance
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| ANR | Anthocyanidin reductase |
| CDS | Coding sequence |
| DEGs | Differentially expressed genes |
| DETFs | Differentially expressed transcription factors |
| DFR | Dihydroflavonol 4-reductase |
| DMACA | p-Dimethylamino cinnamaldehyde |
| DW | Dry weight |
| FPKM | Fragments per kilobase per million |
| FW | Fresh weight |
| LAR | Leucoanthocyanidin reductase |
| LDOX/ANS | (Leuco)anthocyanidin dioxygenase |
| LFBGs | Late flavonoid biosynthesis genes |
| MATE | Multidrug and toxic compound extrusion |
| PAs | Proanthocyanidins or condensed tannins |
| PCC | Pearson correlation coefficient |
| ROS | Reactive oxygen species |
| TF | Transcription factor |
| Y1H | Yeast one-hybrid |
References
- Dixon, R.A.; Xie, D.Y.; Sharma, S.B. Proanthocyanidins—A final frontier in flavonoid research? New Phytol. 2005, 165, 9–28. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.; Prescott, A.; Mackay, S.; Bartlett, J.; Vrijlandt, E. Control of anthocyanin biosynthesis in flowers of Antirrhinum majus. Plant J. 1991, 1, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Saslowsky, D.; Winkel-Shirley, B. Localization of flavonoid enzymes in Arabidopsis roots. Plant J. 2001, 27, 37–48. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 2002, 5, 218–223. [Google Scholar] [CrossRef]
- Xu, Z.C.; Yin, J.; Zhou, B.; Liu, Y.T.; Yu, Y.; Li, G.Q. Grape seed proanthocyanidin protects liver against ischemia/reperfusion injury by attenuating endoplasmic reticulum stress. World J. Gastroenterol. 2015, 21, 7468–7477. [Google Scholar] [CrossRef]
- Bakheet, S.A.; Alhuraishi, A.M.; Al-Harbi, N.O.; Al-Hosaini, K.A.; Al-Sharary, S.D.; Attia, M.M.; Alhoshani, A.R.; Al-Shabanah, O.A.; Al-Harbi, M.M.; Imam, F.; et al. Alleviation of aflatoxin B1-induced genomic damage by proanthocyanidins via modulation of DNA repair. J. Biochem. Mol. Toxicol. 2016, 30, 559–566. [Google Scholar] [CrossRef]
- Lepiniec, L.; Debeaujon, I.; Routaboul, J.M.; Baudry, A.; Pourcel, L.; Nesi, N.; Caboche, M. Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 2006, 57, 405–430. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. It takes a garden. How work on diverse plant species has contributed to an understanding of flavonoid metabolism. Plant Physiol. 2001, 127, 1399–1404. [Google Scholar] [CrossRef]
- Bogs, J. Proanthocyanidin synthesis and expression of genes encoding leucoanthocyanidin reductase and anthocyanidin reductase in developing grape berries and grapevine leaves. Plant Physiol. 2005, 139, 652–663. [Google Scholar] [CrossRef]
- Pang, Y.; Peel, G.J.; Sharma, S.B.; Tang, Y.; Dixon, R.A. A transcript profiling approach reveals an epicatechin-specific glucosyltransferase expressed in the seed coat of Medicago truncatula. Proc. Natl. Acad. Sci. USA 2008, 105, 14210–14215. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Dixon, R.A. MATE transporters facilitate vacuolar uptake of epicatechin 3′-O-glucoside for proanthocyanidin biosynthesis in Medicago truncatula and Arabidopsis. Plant Cell 2009, 21, 2323–2340. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Pang, Y.; Dixon, R.A. The mysteries of proanthocyanidin transport and polymerization. Plant Physiol. 2010, 153, 437–443. [Google Scholar] [CrossRef]
- Xie, D.Y.; Sharma, S.B.; Paiva, N.L.; Ferreira, D.; Dixon, R.A. Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 2003, 299, 396–399. [Google Scholar] [CrossRef]
- Devic, M.; Guilleminot, J.; Debeaujon, I.; Bechtold, N.; Bensaude, E.; Koornneef, M.; Pelletier, G.; Delseny, M. The BANYULS gene encodes a DFR-like protein and is a marker of early seed coat development. Plant J. 1999, 19, 387–398. [Google Scholar] [CrossRef]
- Pelletier, M.K.; Murrell, J.R.; Shirley, B.W. Characterization of flavonol synthase and leucoanthocyanidin dioxygenase genes in Arabidopsis. Further evidence for differential regulation of “early” and “late” genes. Plant Physiol. 1997, 113, 1437–1445. [Google Scholar] [CrossRef]
- Pang, Y.; Peel, G.J.; Wright, E.; Wang, Z.; Dixon, R.A. Early steps in proanthocyanidin biosynthesis in the model legume Medicago truncatula. Plant Physiol. 2007, 145, 601–615. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhao, S.; Fei, B.; Liu, H.; Yang, H.; Dai, H.; Wang, D.; Jin, W.; Tang, F.; Gao, Q.; et al. Announcing the genome atlas of bamboo and gattan (GABR) project: Promoting research in evolution and in economically and ecologically beneficial plants. Gigascience 2017, 6, 1–7. [Google Scholar] [CrossRef]
- Wedler, J.; Daubitz, T.; Schlotterbeck, G.; Butterweck, V. In vitro anti-inflammatory and wound-healing potential of a Phyllostachys edulis leaf extract--identification of isoorientin as an active compound. Planta Med. 2014, 80, 1678–1684. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Xia, D.; Huang, J.; Ge, Q.; Mao, J.; Liu, S.; Zhang, Y. Functional components of bamboo shavings and bamboo leaf extracts and their antioxidant activities in vitro. J. Med. Food 2015, 18, 453–459. [Google Scholar] [CrossRef]
- Shu, G.; Kong, F.; Xu, D.; Yin, L.; He, C.; Lin, J.; Fu, H.; Wang, K.; Tian, Y.; Zhao, X. Bamboo leaf flavone changed the community of cecum microbiota and improved the immune function in broilers. Sci. Rep. 2020, 10, 12324. [Google Scholar] [CrossRef]
- Yu, Y.; Li, Z.; Cao, G.; Huang, S.; Yang, H. Bamboo leaf flavonoids extracts alleviate oxidative stress in HepG2 cells via naturally modulating reactive oxygen species production and Nrf2-mediated antioxidant defense responses. J. Food Sci. 2019, 84, 1609–1620. [Google Scholar] [CrossRef]
- Li, H.; Chen, Y.; Peng, Q.; Tan, X.; Chen, G.; Zhou, H.; Yan, R. Flavonoids from bamboo leaves improve the stability of unsaturated fatty acids in the lipids of walnut emulsions. Ind. Crops Prod. 2022, 178, 114609. [Google Scholar] [CrossRef]
- Wang, J.; Yue, Y.D.; Tang, F.; Sun, J. TLC screening for antioxidant activity of extracts from fifteen bamboo species and identification of antioxidant flavone glycosides from leaves of Bambusa. textilis McClure. Molecules 2012, 17, 12297–12311. [Google Scholar] [CrossRef]
- Wang, H.; Lu, P.; Li, Z.; Yuan, C.; Liu, H.; Zhao, J.; Lu, W.; Wang, J. Oligomeric proanthocyanidins and bamboo leaf flavonoids improve the quality of bull semen cryopreservation. Molecules 2022, 27, 1144. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Y.; Cheng, J.; Zhao, J.; Shi, R.; He, L.; Li, Q.; Chen, Y. Efficient purification of flavonoids from bamboo shoot residues of Phyllostachys edulis by macroporous resin and their hypoglycemic activity. Food Chem. X 2022, 16, 100505. [Google Scholar] [CrossRef]
- Sun, H.; Wang, S.; Zhu, C.; Yang, K.; Liu, Y.; Gao, Z. A new biotechnology for in-planta gene editing and its application in promoting flavonoid biosynthesis in bamboo leaves. Plant Methods 2023, 19, 20. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Gao, Z.; Wang, L.; Wang, J.; Wang, S.; Fei, B.; Chen, C.; Shi, C.; Liu, X.; Zhang, H.; et al. Chromosome-level reference genome and alternative splicing atlas of moso bamboo (Phyllostachys edulis). Gigascience 2018, 7, giy115. [Google Scholar] [CrossRef]
- Huang, Z.; Jin, S.H.; Guo, H.D.; Zhong, X.J.; He, J.; Li, X.; Jiang, M.Y.; Yu, X.F.; Long, H.; Ma, M.D.; et al. Genome-wide identification and characterization of TIFY family genes in moso bamboo (Phyllostachys edulis) and expression profiling analysis under dehydration and cold stresses. Peer J. 2016, 4, e2620. [Google Scholar] [CrossRef]
- Yang, K.; Li, L.; Lou, Y.; Zhu, C.; Li, X.; Gao, Z. A regulatory network driving shoot lignification in rapidly growing bamboo. Plant Physiol. 2021, 187, 900–916. [Google Scholar] [CrossRef]
- Ma, X.; Zhao, H.; Xu, W.; You, Q.; Yan, H.; Gao, Z.; Su, Z. Co-expression gene network analysis and functional module identification in bamboo growth and development. Front. Genet. 2018, 9, 574. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Li, Z.; Zhu, C.; Liu, Y.; Sun, H.; Li, X.; Gao, Z. Identification of KFB family in moso bamboo reveals the potential function of PeKFB9 involved in stress response and lignin polymerization. Int. J. Mol. Sci. 2022, 23, 12568. [Google Scholar] [CrossRef]
- Fan, C.; Ma, J.; Guo, Q.; Li, X.; Wang, H.; Lu, M. Selection of reference genes for quantitative real-time PCR in bamboo (Phyllostachys edulis). PloS ONE 2013, 8, e56573. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Liu, Y.; Shi, Z.; Maximova, S.; Payne, M.J.; Guiltinan, M.J. Proanthocyanidin synthesis in Theobroma cacao: Genes encoding anthocyanidin synthase, anthocyanidin reductase, and leucoanthocyanidin reductase. BMC Plant Biol. 2013, 13, 202. [Google Scholar] [CrossRef] [PubMed]
- Stafford, H.A. Flavonoid evolution: An enzymic approach. Plant Physiol. 1991, 96, 680–685. [Google Scholar] [CrossRef]
- Ma, Q.; Xia, Z.; Cai, Z.; Li, L.; Cheng, Y.; Liu, J.; Nian, H. GmWRKY16 enhances drought and salt tolerance through an ABA-mediated pathway in Arabidopsis thaliana. Front. Plant Sci. 2019, 9, 1979. [Google Scholar] [CrossRef] [PubMed]
- Koes, R.E.; Quattrocchio, F.; Mol, J.N.M. The flavonoid biosynthetic pathway in plants: Function and evolution. BioEssays 1994, 16, 123–132. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, L.; Jiang, X.; Dai, X.; Xu, L.; Li, T.; Xing, D.; Li, Y.; Li, M.; Gao, L.; et al. Evolutionary and functional characterization of leucoanthocyanidin reductases from Camellia sinensis. Planta 2018, 247, 139–154. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.Z.; Zhu, Y.; Liu, Z.; Du, C.; Li, K.G.; Xie, D.Y. An integrated approach to demonstrating the ANR pathway of proanthocyanidin biosynthesis in plants. Planta 2012, 236, 901–918. [Google Scholar] [CrossRef]
- Verwoert, I.I.; Verbree, E.C.; van der Linden, K.H.; Nijkamp, H.J.; Stuitje, A.R. Cloning, nucleotide sequence, and expression of the Escherichia coli fabD gene, encoding malonyl coenzyme A-acyl carrier protein transacylase. J. Bacteriol. 1992, 174, 2851–2857. [Google Scholar] [CrossRef]
- Jez, J.M.; Bowman, M.E.; Dixon, R.A.; Noel, J.P. Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase. Nat. Struct. Biol. 2000, 7, 786–791. [Google Scholar]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef]
- Xiao, X.; Zhu, C.; Yang, K.; Liu, Y.; Li, Z.; Guo, D.; Gao, Z. Low temperature promotes flavonoid synthesis and expression pattern analysis of the related genes in leaves of Phyllostachys edulis. J. Trop. Subtrop. Bot. 2022. accepted. [Google Scholar]
- Cheng, A.X.; Han, X.J.; Wu, Y.F.; Lou, H.X. The function and catalysis of 2-oxoglutarate-dependent oxygenases involved in plant flavonoid biosynthesis. Int. J. Mol. Sci. 2014, 15, 1080–1095. [Google Scholar] [CrossRef]
- Xie, D.Y.; Sharma, S.B.; Dixon, R.A. Anthocyanidin reductases from Medicago truncatula and Arabidopsis thaliana. Arch. Biochem. Biophys. 2004, 422, 91–102. [Google Scholar] [CrossRef]
- Tanner, G.J.; Francki, K.T.; Abrahams, S.; Watson, J.M.; Larkin, P.J.; Ashton, A.R. Proanthocyanidin biosynthesis in plants. Purification of legume leucoanthocyanidin reductase and molecular cloning of its cDNA. J. Biol. Chem. 2003, 278, 31647–31656. [Google Scholar] [CrossRef]
- Li, H.; Tian, J.; Yao, Y.Y.; Zhang, J.; Song, T.T.; Li, K.T.; Yao, Y.C. Identification of leucoanthocyanidin reductase and anthocyanidin reductase genes involved in proanthocyanidin biosynthesis in Malus crabapple plants. Plant Physiol. Biochem. 2019, 139, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Qi, C.; Wang, C.; Wang, S.; Zhou, C.; Ren, Y.; Cheng, Z.; Zhang, X.; Guo, X.; Zhao, Z.; et al. Determinant factors and regulatory systems for anthocyanin biosynthesis in rice apiculi and stigmas. Rice 2021, 14, 37. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Zhang, Y.; Zhang, L.; Wang, B.; Zhao, Y.; Irfan, M.; Chen, L.; Feng, Y. Regulation of MYB transcription factors of anthocyanin synthesis in lily flowers. Front. Plant Sci. 2021, 12, 761668. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Jia, H.; Xing, M.; Jin, R.; Grierson, D.; Gao, Z.; Sun, C.; Chen, K.; Xu, C.; Li, X. Genome-wide analysis of MYB gene family in chinese bayberry (Morella rubra) and identification of members regulating flavonoid biosynthesis. Front. Plant Sci. 2021, 12, 691384. [Google Scholar] [CrossRef]
- Ma, D.; Reichelt, M.; Yoshida, K.; Gershenzon, J.; Constabel, C.P. Two R2R3-MYB proteins are broad repressors of flavonoid and phenylpropanoid metabolism in poplar. Plant J. 2018, 96, 949–965. [Google Scholar] [CrossRef]
- Zhai, R.; Wang, Z.; Zhang, S.; Meng, G.; Song, L.; Wang, Z.; Li, P.; Ma, F.; Xu, L. Two MYB transcription factors regulate flavonoid biosynthesis in pear fruit (Pyrus bretschneideri Rehd.). J. Exp. Bot. 2016, 67, 1275–1284. [Google Scholar] [CrossRef]
- Xu, P.; Wu, L.; Cao, M.; Ma, C.; Xiao, K.; Li, Y.; Lian, H. Identification of MBW complex components implicated in the biosynthesis of flavonoids in woodland strawberry. Front. Plant Sci. 2021, 12, 774943. [Google Scholar] [CrossRef]
- Yang, J.; Wu, X.; Aucapiña, C.B.; Huang, J.; Hao, Z.; Zhang, Y.; Ren, Y.; Miao, N. NtMYB12 requires for competition between flavonol and (pro)anthocyanin biosynthesis in Narcissus tazetta tepals. Mol. Hortic. 2023, 3, 2. [Google Scholar] [CrossRef]
- An, X.H.; Tian, Y.; Chen, K.Q.; Liu, X.J.; Liu, D.D.; Xie, X.B.; Cheng, C.G.; Cong, P.H.; Hao, Y.J. MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol. 2015, 56, 650–662. [Google Scholar] [CrossRef]
- Hong, M.; Hu, K.; Tian, T.; Li, X.; Chen, L.; Zhang, Y.; Yi, B.; Wen, J.; Ma, C.; Shen, J.; et al. Transcriptomic analysis of seed coats in yellow-seeded brassica napus reveals novel genes that influence proanthocyanidin biosynthesis. Front. Plant Sci. 2017, 8, 1674. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Gu, L.; Ye, S.; Zhang, H.; Cai, C.; Xiang, M.; Gao, Y.; Wang, Q.; Lin, C.; Zhu, Q. Genome-wide analysis and transcriptomic profiling of the auxin biosynthesis, transport and signaling family genes in moso bamboo (Phyllostachys heterocycla). BMC Genom. 2017, 18, 870. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Jiang, S.; Zhang, T.; Xu, H.; Fang, H.; Zhang, J.; Su, M.; Wang, Y.; Zhang, Z.; Wang, N.; et al. Apple NAC transcription factor MdNAC52 regulates biosynthesis of anthocyanin and proanthocyanidin through MdMYB9 and MdMYB11. Plant Sci. 2019, 289, 110286. [Google Scholar] [CrossRef]
- Sudha, M.L.; Indumathi, K.; Sumanth, M.S.; Rajarathnam, S.; Shashirekha, M.N. Mango pulp fibre waste: Characterization and utilization as a bakery product ingredient. J. Food Meas. Charact. 2015, 9, 382–388. [Google Scholar] [CrossRef]
- Ma, X.; Wu, H.; Liu, L.; Yao, Q.; Wang, S.; Zhan, R.; Xing, S.; Zhou, Y. Polyphenolic compounds and antioxidant properties in mango fruits. Sci. Hortic. 2011, 29, 102–107. [Google Scholar] [CrossRef]
- Abrahams, S.; Tanner, G.J.; Larkin, P.J.; Ashton, A.R. Identification and biochemical characterization of mutants in the proanthocyanidin pathway in Arabidopsis. Plant Physiol. 2002, 130, 561–576. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Vimolmangkang, S.; Soria-Guerra, R.E.; Korban, S.S. Introduction of apple ANR genes into tobacco inhibits expression of both CHI and DFR genes in flowers, leading to loss of anthocyanin. J. Exp. Bot. 2012, 63, 2437–2447. [Google Scholar] [CrossRef] [PubMed]
- Grillo, S.; Blanco, A.; Cattivelli, L.; Coraggio, I.; Leone, A.; Salvi, S. Plant genetic and molecular responses to water deficit. Ital. J. Agron. 2011, 1, 617–637. [Google Scholar] [CrossRef]
- Ma, J.; Wang, L.Y.; Dai, J.X.; Wang, Y.; Lin, D. The NAC-type transcription factor CaNAC46 regulates the salt and drought tolerance of transgenic Arabidopsis thaliana. BMC Plant Biol. 2021, 21, 11. [Google Scholar] [CrossRef] [PubMed]
- Fornalé, S.; Lopez, E.; Salazar-Henao, J.E.; Fernández-Nohales, P.; Rigau, J.; Caparros-Ruiz, D. AtMYB7, a new player in the regulation of UV-sunscreens in Arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 507–516. [Google Scholar] [CrossRef]
- Zhang, D.H. Characterization of Transcription Factors MdMYB88, MdMYB124, and MdMYB3 in Response to Drought and Low Nitrate Stresses in Apple. Ph.D. Thesis, Northwest A&F University, Xianyang, China, 2021. [Google Scholar]
- Ramakrishna, A.; Ravishankar, G.A. Influence of abiotic stress signals on secondary metabolites in plants. Plant Signal. Behav. 2011, 6, 1720–1731. [Google Scholar]
- Osawa, H.; Endo, I.; Hara, Y.; Matsushima, Y.; Tange, T. Transient proliferation of proanthocyanidin-accumulating cells on the epidermal apex contributes to highly aluminum-resistant root elongation in camphor tree. Plant Physiol. 2011, 155, 433–446. [Google Scholar] [CrossRef]
- Tan, L.; Wang, M.; Kang, Y.; Azeem, F.; Zhou, Z.; Tuo, D.; María Preciado Rojo, L.; Khan, I.A.; Pan, Z. Biochemical and Functional Characterization of Anthocyanidin Reductase (ANR) from Mangifera indica L. Molecules 2018, 23, 2876. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, X.; Zhu, C.; Yang, K.; Liu, Y.; Gao, Z. Identification of Late Flavonoid Biosynthesis Genes of Moso Bamboo Reveals the Potential Function of PeANR4 Involved in Osmotic and Salt Stress. Forests 2023, 14, 1399. https://doi.org/10.3390/f14071399
Xiao X, Zhu C, Yang K, Liu Y, Gao Z. Identification of Late Flavonoid Biosynthesis Genes of Moso Bamboo Reveals the Potential Function of PeANR4 Involved in Osmotic and Salt Stress. Forests. 2023; 14(7):1399. https://doi.org/10.3390/f14071399
Chicago/Turabian StyleXiao, Xiaoyan, Chenglei Zhu, Kebin Yang, Yan Liu, and Zhimin Gao. 2023. "Identification of Late Flavonoid Biosynthesis Genes of Moso Bamboo Reveals the Potential Function of PeANR4 Involved in Osmotic and Salt Stress" Forests 14, no. 7: 1399. https://doi.org/10.3390/f14071399
APA StyleXiao, X., Zhu, C., Yang, K., Liu, Y., & Gao, Z. (2023). Identification of Late Flavonoid Biosynthesis Genes of Moso Bamboo Reveals the Potential Function of PeANR4 Involved in Osmotic and Salt Stress. Forests, 14(7), 1399. https://doi.org/10.3390/f14071399





